Metabolic Interactions in the Gastrointestinal Tract (GIT): Host, Commensal, Probiotics, and Bacteriophage Influences
Abstract
:1. Introduction
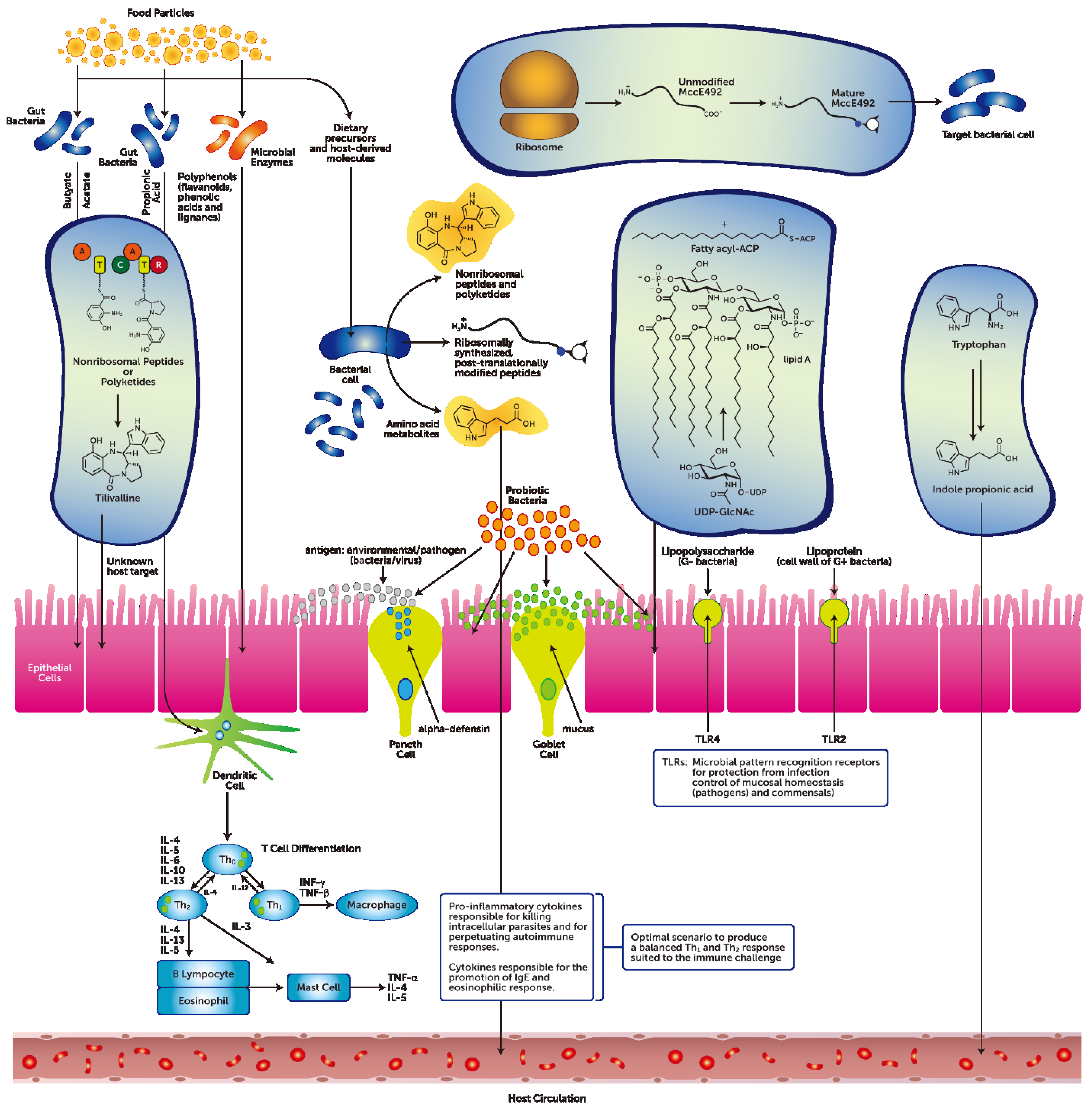
2. What this Review Proposes to Add
3. The Gastrointestinal Tract Barrier and Chemical Transmitters
3.1. Diet-Independent Metabolites from the GIT Microbiota
3.2. Diet-Dependent Metabolites from the GIT Microbiota
3.3. The Metabolic Effects of Probiotics
3.4. Metabolites that Risk Disrupting GIT Host-Microbial Homeostasis
3.5. GIT Bacteriophages Chemical Modulators of the GIT Microbiome
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Morgan, X.C.; Segata, N.; Huttenhower, C. Biodiversity and functional genomics in the human microbiome. Trends Genet. 2013, 29, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Sommer, F.; Backhed, F. The gut microbiota—Masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.E.; Sjovall, H.; Hansson, G.C. The gastrointestinal mucus system in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, A.; Loening-Baucke, V.; Lochs, H.; Hale, L.P. Spatial organization of bacterial flora in normal and inflamed intestine: A fluorescence in situ hybridization study in mice. World J. Gastroenterol. 2005, 11, 1131–1140. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Wang, Y.; Zhu, L.; Liu, W.; Liao, N.; Jiang, M.; Zhu, B.; Yu, H.D.; Xiang, C.; Wang, X. Comparative analysis of the distribution of segmented filamentous bacteria in humans, mice and chickens. ISME J. 2013, 7, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Klaasen, H.L.; Koopman, J.P.; Poelma, F.G.; Beynen, A.C. Intestinal, segmented, filamentous bacteria. FEMS Microbiol. Rev. 1992, 8, 165–180. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, I.I.; Frutos, R.L.; Manel, N.; Yoshinaga, K.; Rifkin, D.B.; Sartor, R.B.; Finlay, B.B. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 2008, 4, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar]
- Relman, D.A. Microbiology: Learning about who we are. Nature 2012, 486, 194–195. [Google Scholar] [CrossRef] [PubMed]
- Donia, M.S.; Fischbach, M.A. Small molecules from the human microbiota. Science 2015. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Akira, S. Toll-like receptors. Curr. Protoc. Immunol. 2015. [Google Scholar] [CrossRef]
- Jimenez-Dalmaroni, M.J.; Gerswhin, M.E.; Adamopoulos, I.E. The critical role of toll-like receptors—From microbial recognition to autoimmunity: A comprehensive review. Autoimmun. Rev. 2015, 15, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Arpaia, N.; Rudensky, A.Y. Microbial metabolites control gut inflammatory responses. Proc. Natl. Acad. Sci. 2014, 111, 2058–2059. [Google Scholar] [CrossRef] [PubMed]
- Russell, W.R.; Hoyles, L.; Flint, H.J.; Dumas, M.E. Colonic bacterial metabolites and human health. Curr. Opin. Microbiol. 2013, 16, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Shimada, Y.; Kinoshita, M.; Harada, K.; Mizutani, M.; Masahata, K.; Kayama, H.; Takeda, K. Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS ONE 2013, 8, e80604. [Google Scholar] [CrossRef] [PubMed]
- Coskun, M. Intestinal epithelium in inflammatory bowel disease. Front. Med. 2014. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.M.; Duckworth, C.A.; Burkitt, M.D.; Watson, A.J.; Campbell, B.J.; Pritchard, D.M. Epithelial cell shedding and barrier function: A matter of life and death at the small intestinal villus tip. Vet. Pathol. 2015, 52, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Holzer, P.; Michl, T.; Danzer, M.; Jocic, M.; Schicho, R.; Lippe, I.T. Surveillance of the gastrointestinal mucosa by sensory neurons. J. Physiol. Pharmacol. 2001, 52, 505–521. [Google Scholar] [PubMed]
- Furness, J.B.; Clerc, N. Responses of afferent neurons to the contents of the digestive tract, and their relation to endocrine and immune responses. Prog. Brain Res. 2000, 122, 159–172. [Google Scholar] [PubMed]
- DelValle, J.; Yamada, T. The gut as an endocrine organ. Annu. Rev. Med. 1990, 41, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Ahlman, H.; Nilsson, O. The gut as the largest endocrine organ in the body. Ann. Oncol. 2001, 12, S63–S68. [Google Scholar] [CrossRef] [PubMed]
- Liddel, R. Gastrointestinal hormones and neurotransmitters. In Sleisenger and Fordtran’s Gastrointestinal and Liver Disease; Elsevier: Amsterdam, The Netherlands, 2010. [Google Scholar]
- Vitetta, L.; Manuel, R.; Zhou, J.Y.; Linnane, A.W.; Hall, S.; Coulson, S. The overarching influence of the gut microbiome on end-organ function: The role of live probiotic cultures. Pharmaceuticals 2014, 7, 954–989. [Google Scholar] [CrossRef] [PubMed]
- Mandicourt, G.; Iden, S.; Ebnet, K.; Aurrand-Lions, M.; Imhof, B.A. JAM-C regulates tight junctions and integrin-mediated cell adhesion and migration. J. Biol. Chem. 2007, 282, 1830–1837. [Google Scholar] [CrossRef] [PubMed]
- Vitetta, L.; Hall, S.; Linnane, A.W. Live probiotic cultures and the gastrointestinal tract: Symbiotic preservation of tolerance whilst attenuating pathogenicity. Front. Cell Infect. Microbiol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Packey, C.D.; Sartor, R.B. Commensal bacteria, traditional and opportunistic pathogens, dysbiosis and bacterial killing in inflammatory bowel diseases. Curr. Opin. Infect. Dis. 2009, 22, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Ubelmann, F.; Chamaillard, M.; El-Marjou, F.; Simon, A.; Netter, J.; Vignjevic, D.; Nichols, B.L.; Quezada-Calvillo, R.; Grandjean, T.; Louvard, D.; et al. Enterocyte loss of polarity and gut wound healing rely upon the F-actin-severing function of villin. Proc. Natl. Acad. Sci. 2013, 110, E1380–E1389. [Google Scholar] [CrossRef] [PubMed]
- Young, R.P.; Hopkins, R.J.; Marsland, B. The gut-liver-lung axis: Modulation of the innate immune response and its possible role in COPD. Am. J. Respir. Cell Mol. Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Schuijt, T.J.; Lankelma, J.M.; Scicluna, B.P.; de Sousa, E.M.F.; Roelofs, J.J.; de Boer, J.D.; Hoogendijk, A.J.; de Beer, R.; de Vos, A.; Belzer, C.; et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut 2015. [Google Scholar] [CrossRef] [PubMed]
- Trompette, A.; Gollwitzer, E.S.; Yadava, K.; Sichelstiel, A.K.; Sprenger, N.; Ngom-Bru, C.; Blanchard, C.; Junt, T.; Nicod, L.P.; Harris, N.L.; et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014, 20, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Kasubuchi, M.; Hasegawa, S.; Hiramatsu, T.; Ichimura, A.; Kimura, I. Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients 2015, 7, 2839–2849. [Google Scholar] [CrossRef] [PubMed]
- Blaser, M.J. The microbiome revolution. J. Clin. Invest. 2014, 124, 4162–4165. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, T.E.; Morton, J.M. The human gut microbiome: A review of the effect of obesity and surgically induced weight loss. JAMA Surg. 2013, 148, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.J.; Hase, K. Gut microbiota-generated metabolites in animal health and disease. Nat. Chem Biol. 2014, 10, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Brestoff, J.R.; Artis, D. Commensal bacteria at the interface of host metabolism and the immune system. Nat. Immunol. 2013, 14, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Bouskra, D.; Brezillon, C.; Berard, M.; Werts, C.; Varona, R.; Boneca, I.G.; Eberl, G. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 2008, 456, 507–510. [Google Scholar] [CrossRef] [PubMed]
- Moreau, M.C.; Ducluzeau, R.; Guy-Grand, D.; Muller, M.C. Increase in the population of duodenal immunoglobulin A plasmocytes in axenic mice associated with different living or dead bacterial strains of intestinal origin. Infect. Immun. 1978, 21, 532–539. [Google Scholar] [PubMed]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Kamada, N.; Chen, G.Y.; Inohara, N.; Nunez, G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 2013, 14, 685–690. [Google Scholar] [CrossRef] [PubMed]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Round, J.L.; Lee, S.M.; Li, J.; Tran, G.; Jabri, B.; Chatila, T.A.; Mazmanian, S.K. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 2011, 332, 974–947. [Google Scholar] [CrossRef] [PubMed]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Young, K.D. Indole production by the tryptophanase TnaA in Escherichia coli is determined by the amount of exogenous tryptophan. Microbiology 2013, 159, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Bansal, T.; Alaniz, R.C.; Wood, T.K.; Jayaraman, A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc. Natl. Acad. Sci. 2010, 107, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Menard, S.; Cerf-Bensussan, N.; Heyman, M. Multiple facets of intestinal permeability and epithelial handling of dietary antigens. Mucosal Immunol. 2010, 3, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Baumgart, D.C.; Dignass, A.U. Intestinal barrier function. Curr. Opin. Clin. Nutr. Metab. Care 2002, 5, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Greer, R.L.; Morgun, A.; Shulzhenko, N. Bridging immunity and lipid metabolism by gut microbiota. J. Allergy Clin. Immunol. 2013, 132, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Maslowski, K.M.; Vieira, A.T.; Ng, A.; Kranich, J.; Sierro, F.; Yu, D.; Schilter, H.C.; Rolph, M.S.; Mackay, F.; Artis, D.; et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009, 461, 1282–1286. [Google Scholar] [CrossRef] [PubMed]
- Kimura, I.; Inoue, D.; Hirano, K.; Tsujimoto, G. The SCFA receptor GPR43 and energy metabolism. Front. Endocrinol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.H.; Pomare, E.W.; Branch, W.J.; Naylor, C.P.; Macfarlane, G.T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 1987, 28, 1221–1227. [Google Scholar] [CrossRef] [PubMed]
- Ai, T.L.; Solomon, B.D.; Hsieh, C.S. T-cell selection and intestinal homeostasis. Immunol. Rev. 2014, 259, 60–74. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, S.; Yamaguchi, T.; Nomura, T.; Ono, M. Regulatory T cells and immune tolerance. Cell 2008, 133, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, K.; Tunaru, S.; Offermanns, S. GPR109A, GPR109B and GPR81, a family of hydroxy-carboxylic acid receptors. Trends Pharmacol. Sci. 2009, 30, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Le Poul, E.; Loison, C.; Struyf, S.; Springael, J.Y.; Lannoy, V.; Decobecq, M.E.; Brezillon, S.; Dupriez, V.; Vassart, G.; Damme, J.V.; et al. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J. Biol. Chem. 2003, 278, 25481–25489. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, D.R.; Garge, N.; Zhang, X.; Sun, W.; O’Connell, T.M.; Bunger, M.K.; Bultman, S.J. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011, 13, 517–526. [Google Scholar] [CrossRef] [PubMed]
- De Vadder, F.; Kovatcheva-Datchary, P.; Goncalves, D.; Vinera, J.; Zitoun, C.; Duchampt, A.; Bäckhed, F.; Mithieux, G. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 2014, 156, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Tolhurst, G.; Heffron, H.; Lam, Y.S.; Parker, H.E.; Habib, A.M.; Diakogiannaki, E.; Cameron, J.; Grosse, J.; Reimann, F.; Gribble, F.M. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes 2012, 61, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Kimura, I.; Inoue, D.; Maeda, T.; Hara, T.; Ichimura, A.; Miyauchi, S.; Kobayashi, M.; Hirasawa, A.; Tsujimoto, G. Short-chain fatty acids and ketones directly regulate sympathetic nervous system via G protein-coupled receptor 41 (GPR41). Proc. Natl. Acad. Sci. 2011, 108, 8030–8035. [Google Scholar] [CrossRef] [PubMed]
- Inoue, D.; Kimura, I.; Wakabayashi, M.; Tsumoto, H.; Ozawa, K.; Hara, T.; Takei, Y.; Hirasawa, A.; Ishihama, Y.; Tsujimoto, G. Short-chain fatty acid receptor GPR41-mediated activation of sympathetic neurons involves synapsin 2b phosphorylation. FEBS Lett. 2012, 586, 1547–1554. [Google Scholar] [CrossRef] [PubMed]
- Cresci, G.A.; Thangaraju, M.; Mellinger, J.D.; Liu, K.; Ganapathy, V. Colonic gene expression in conventional and germ-free mice with a focus on the butyrate receptor GPR109A and the butyrate transporter SLC5A8. J. Gastrointest. Surg. 2010, 14, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Lopetuso, L.R.; Scaldaferri, F.; Petito, V.; Gasbarrini, A. Commensal Clostridia: Leading players in the maintenance of gut homeostasis. Gut Pathogs. 2013, 14, 31–33. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Goswami, K.; Chen, C.; Xiaoli, L.; Eaton, K.A.; Dudley, E.G. Co-culturing Escherichia coli O157:H7 with a non-pathogenic E. coli strain increases toxin production and virulence in a germ-free mouse model. Infect. Immun. 2015. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Lee, K.S.; Lee, D.K.; Lee, S.Y.; Chang, H.Y.; Choi, J.; Lee, J.I. Omega-3 polyunsaturated fatty acid and ursodeoxycholic acid have an additive effect in attenuating diet-induced nonalcoholic steatohepatitis in mice. Exp. Mol. Med. 2014. [Google Scholar] [CrossRef] [PubMed]
- El Aidy, S.; van den Abbeele, P.; van de Wiele, T.; Louis, P.; Kleerebezem, M. Intestinal colonization: How key microbial players become established in this dynamic process: Microbial metabolic activities and the interplay between the host and microbes. Bioessays 2013, 35, 913–923. [Google Scholar] [CrossRef] [PubMed]
- Wopereis, H.; Oozeer, R.; Knipping, K.; Belzer, C.; Knol, J. The first thousand days—Intestinal microbiology of early life: Establishing a symbiosis. Pediatr. Allergy Immunol. 2014, 25, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Mizock, B.A. Probiotics. Dis. Month 2015, 61, 259–290. [Google Scholar] [CrossRef] [PubMed]
- Mondel, M.; Schroeder, B.O.; Zimmermann, K.; Huber, H.; Nuding, S.; Beisner, J.; Fellermann, K.; Stang, E.F.; Wehkamp, J. Probiotic E. coli treatment mediates antimicrobial human beta-defensin synthesis and fecal excretion in humans. Mucosal Immunol. 2009, 2, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Schlee, M.; Harder, J.; Koten, B.; Stange, E.F.; Wehkamp, J.; Fellermann, K. Probiotic lactobacilli and VSL#3 induce enterocyte beta-defensin 2. Clin. Exp. Immunol. 2008, 151, 528–535. [Google Scholar] [PubMed]
- Segawa, S.; Fujiya, M.; Konishi, H.; Ueno, N.; Kobayashi, N.; Shigyo, T.; Kohgo, Y. Probiotic-derived polyphosphate enhances the epithelial barrier function and maintains intestinal homeostasis through integrin-p38 MAPK pathway. PLoS ONE 2011, 6, e23278. [Google Scholar] [CrossRef] [PubMed]
- Etzold, S.; MacKenzie, D.A.; Jeffers, F.; Walshaw, J.; Roos, S.; Hemmings, A.M.; Juge, N. Structural and molecular insights into novel surface-exposed mucus adhesins from Lactobacillus reuteri human strains. Mol. Microbiol. 2014, 92, 543–556. [Google Scholar] [CrossRef] [PubMed]
- Makras, L.; Triantafyllou, V.; Fayol-Messaoudi, D.; Adriany, T.; Zoumpopoulou, G.; Tsakalidou, E.; Servin, A.; Vuyst, L.D. Kinetic analysis of the antibacterial activity of probiotic lactobacilli towards Salmonella enterica serovar Typhimurium reveals a role for lactic acid and other inhibitory compounds. Res. Microbiol. 2006, 157, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Bermudez-Brito, M.; Plaza-Diaz, J.; Munoz-Quezada, S.; Gomez-Llorente, C.; Gil, A. Probiotic mechanisms of action. Ann. Nutr. Metab. 2012, 61, 160–174. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, D.S.; Cho, G.S.; Hanak, A.; Huch, M.; Franz, C.M.; Arneborg, N. The effect of bacteriocin-producing Lactobacillus plantarum strains on the intracellular pH of sessile and planktonic Listeria monocytogenes single cells. Int. J. Food Microbiol. 2010, 141, S53–S59. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, N.; Seto, H.; Koga, S.; Zendo, T.; Sonomoto, K. Identification of lactococcus-specific bacteriocins produced by lactococcal isolates, and the discovery of a novel bacteriocin, lactococcin Z. Probiotics Antimicrob. Proteins 2015, 7, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Makras, L.; de Vuyst, L. The in vitro inhibition of Gram-negative pathogenic bacteria by bifidobacteria is caused by the production of organic acids. Int. Dairy J. 2006, 16, 1049–1057. [Google Scholar] [CrossRef]
- Fujiwara, S.; Hashiba, H.; Hirota, T.; Forstner, J.F. Purification and characterization of a novel protein produced by Bifidobacterium longum SBT2928 that inhibits the binding of enterotoxigenic Escherichia coli Pb176 (CFA/II) to gangliotetraosylceramide. J. App. Microbiol. 1999, 86, 615–621. [Google Scholar] [CrossRef]
- Oelschlaeger, T.A. Mechanisms of probiotic actions—A review. Int. J. Med. Microbiol. 2010, 300, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Coloretti, F.; Carri, S.; Armaforte, E.; Chiavari, C.; Grazia, L.; Zambonelli, C. Antifungal activity of lactobacilli isolated from salami. FEMS Microbiol. Lett. 2007, 271, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Zavaleta, O.; Lopez-Malo, A.; Hernandez-Mendoza, A.; Garcia, H.S. Antifungal activity of lactobacilli and its relationship with 3-phenyllactic acid production. Int. J. Food Microbiol. 2014, 173, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Briskey, D.; Peake, J.; Coulson, S.; Vitetta, L. High fat diet, probiotics and non-alcoholic fatty liver disease. In Proceedings of the 4th Beneficial Microbes Conference, The Hague, The Netherlands, 16–18 March 2015.
- Ma, Y.Y.; Li, L.; Yu, C.H.; Shen, Z.; Chen, L.H.; Li, Y.M. Effects of probiotics on nonalcoholic fatty liver disease: A meta-analysis. World J. Gastroenterol. 2013, 19, 6911–6918. [Google Scholar] [CrossRef] [PubMed]
- Vitetta, L.; Bambling, M.; Alford, H. The gastrointestinal tract microbiome, probiotics, and mood. Inflammopharmacology 2014, 22, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Tillisch, K.; Labus, J.; Kilpatrick, L.; Jiang, Z.; Stains, J.; Ebrat, B.; Guyonnet, D.; Legrain-Raspaud, S.; Trotin, B.; Naliboff, B.; et al. Consumption of fermented milk product with probiotic modulates brain activity. Gastroenterology 2013, 144, 1394–1401. [Google Scholar] [CrossRef] [PubMed]
- Neish, A.S. Redox signaling mediated by the gut microbiota. Free Rad. Res. 2013, 47, 950–957. [Google Scholar] [CrossRef] [PubMed]
- Vitetta, L.; Linnane, A.W.; Gobe, G.C. From the gastrointestinal tract (GIT) to the kidneys: Live bacterial cultures (probiotics) mediating reductions of uremic toxin levels via free radical signaling. Toxins 2013, 5, 2042–2057. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, M.; Floch, M.H. Prebiotics, probiotics and digestive health. Curr. Opin. Clin. Nutr. Metab. Care 2012, 15, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Brisbin, J.T.; Gong, J.; Sharif, S. Interactions between commensal bacteria and the gut-associated immune system of the chicken. Anim. Health Res. 2008, 9, 101–110. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, A.M.; Shanahan, F. Gut microbiota: Mining for therapeutic potential. Clin. Gastroenterol. Hepatol. 2007, 5, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Clavel, T.; Haller, D. Molecular interactions between bacteria, the epithelium, and the mucosal immune system in the intestinal tract: Implications for chronic inflammation. Curr. Issues Intest. Microbiol. 2007, 8, 25–43. [Google Scholar] [PubMed]
- Blumberg, R.; Powrie, F. Microbiota, disease, and back to health: A metastable journey. Sci. Transl. Med. 2012. [Google Scholar] [CrossRef] [PubMed]
- Chassaing, B.; Koren, O.; Goodrich, J.K.; Poole, A.C.; Srinivasan, S.; Ley, R.E.; Gewirtz, A.T. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 2015, 519, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Ierardi, E.; Sorrentino, C.; Principi, M.; Giorgio, F.; Losurdo, G.; di Leo, A. Intestinal microbial metabolism of phosphatidylcholine: A novel insight in the cardiovascular risk scenario. Hepatobiliary Surg. Nutr. 2015, 4, 289–292. [Google Scholar] [PubMed]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.M.; Wu, Y.P.; Li, L.; et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, S.; Loo, T.M.; Atarashi, K.; Kanda, H.; Sato, S.; Oyadomari, S.; Iwakura, Y.; Oshima, K.; Morita, H.; Hattori, M.; et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013, 499, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Angley, M.T.; Gerber, J.P.; Sorich, M.J. A review of candidate urinary biomarkers for autism spectrum disorder. Biomarkers 2011, 16, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Vitetta, L.; Gobe, G. Uremia and chronic kidney disease: The role of the gut microflora and therapies with pro- and prebiotics. Mol. Nutr. Food Res. 2013, 57, 824–832. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.J.; Small, D.M.; Vesey, D.A.; Johnson, D.W.; Francis, R.; Vitetta, L.; Gobe, G.C.; Morais, C. Indoxyl sulphate and kidney disease: Causes, consequences and interventions. Nephrology 2015. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D. Metabolism: Dietary emulsifiers—Sweepers of the gut lining? Nat. Rev. Endocrinol. 2015, 11, 319–320. [Google Scholar] [CrossRef] [PubMed]
- Bradlow, H.L. Obesity and the gut microbiome: Pathophysiological aspects. Horm. Mol. Biol. Clin. Invest. 2014, 17, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Greiner, A.K.; Papineni, R.V.; Umar, S. Chemoprevention in gastrointestinal physiology and disease. Natural products and microbiome. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 307, G1–G15. [Google Scholar] [CrossRef] [PubMed]
- Weinbauer, M.G. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 2004, 28, 127–181. [Google Scholar] [CrossRef] [PubMed]
- Chibani-Chennoufi, S.; Bruttin, A.; Dillmann, M.L.; Brussow, H. Phage-host interaction: An ecological perspective. J. Bacteriol. 2004, 186, 3677–3686. [Google Scholar] [CrossRef] [PubMed]
- Wommack, K.E.; Colwell, R.R. Virioplankton: Viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 2000, 64, 69–114. [Google Scholar] [CrossRef] [PubMed]
- Bergh, O.; Borsheim, K.Y.; Bratbak, G.; Heldal, M. High abundance of viruses found in aquatic environments. Nature 1989, 340, 467–468. [Google Scholar] [CrossRef] [PubMed]
- Mills, S.; Shanahan, F.; Stanton, C.; Hill, C.; Coffey, A.; Ross, R.P. Movers and shakers: Influence of bacteriophages in shaping the mammalian gut microbiota. Gut microb. 2013, 4, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Koskella, B.; Meaden, S. Understanding bacteriophage specificity in natural microbial communities. Viruses 2013, 5, 806–823. [Google Scholar] [CrossRef] [PubMed]
- Dalmasso, M.; Hill, C.; Ross, R.P. Exploiting gut bacteriophages for human health. Trends Microbiol. 2014, 22, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Reyes, A.; Haynes, M.; Hanson, N.; Angly, F.E.; Heath, A.C.; Rohwer, F.; Gordon, J.I. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 2010, 466, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, M.; Haynes, M.; Kelley, S.; Angly, F.; Edwards, R.A.; Felts, B.; Mahaffy, J.M.; Mueller, J.; Nulton, J.; Rayhawk, S.; et al. Viral diversity and dynamics in an infant gut. Res. Microbiol. 2008, 159, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Pride, D.T.; Salzman, J.; Haynes, M.; Rohwer, F.; Davis-Long, C.; White, R.A., III; Loomer, P.; Armitage, G.C.; Relman, D.A. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 2012, 6, 915–926. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vitetta, L.; Hall, S.; Coulson, S. Metabolic Interactions in the Gastrointestinal Tract (GIT): Host, Commensal, Probiotics, and Bacteriophage Influences. Microorganisms 2015, 3, 913-932. https://doi.org/10.3390/microorganisms3040913
Vitetta L, Hall S, Coulson S. Metabolic Interactions in the Gastrointestinal Tract (GIT): Host, Commensal, Probiotics, and Bacteriophage Influences. Microorganisms. 2015; 3(4):913-932. https://doi.org/10.3390/microorganisms3040913
Chicago/Turabian StyleVitetta, Luis, Sean Hall, and Samantha Coulson. 2015. "Metabolic Interactions in the Gastrointestinal Tract (GIT): Host, Commensal, Probiotics, and Bacteriophage Influences" Microorganisms 3, no. 4: 913-932. https://doi.org/10.3390/microorganisms3040913
APA StyleVitetta, L., Hall, S., & Coulson, S. (2015). Metabolic Interactions in the Gastrointestinal Tract (GIT): Host, Commensal, Probiotics, and Bacteriophage Influences. Microorganisms, 3(4), 913-932. https://doi.org/10.3390/microorganisms3040913





