1. Introduction
The family
Paenibacillaceae of the order
Bacillales was created by De Vos in 2009 [
1]. Most of the members of the family
Paenibacillaceae have rod-shaped cells with different physiological characteristics, such as being Gram-positive or Gram-negative, having ellipsoidal endospores in swollen sporangia, and being aerobic or facultatively anaerobic [
1]. At the time of writing, the family
Paenibacillaceae currently comprises 19 recognized genera:
Ammoniibacillus [
2],
Ammoniphilus [
3],
Aneurinibacillus [
4],
Brevibacillus [
4],
Chengkuizengella [
5],
Cohnella [
6],
Ferviditalea [
7],
Fontibacillus [
8],
Gordoniibacillus [
9],
Gorillibacterium [
10],
Insulibacter [
11],
Longirhabdus,
Marinicrinis [
12],
Oxalophagus [
13],
Paenibacillus [
14],
Paludirhabdus [
15],
Saccharibacillus [
16],
Thermobacillus [
17], and
Xylanibacillus [
18] (
https://lpsn.dsmz.de/family/paenibacillaceae, accessed on 12 November 2025).
Within the family
Paenibacillaceae, the genus
Fontibacillus is very closely related to the genus
Paenibacillus. The genus
Paenibacillus with
Paenibacillus polymyxa as the type species was created in 1993, and at that time it encompassed eleven species, three of which,
Paenibacillus polymyxa,
Paenibacillus azotofixans, and
Paenibacillus macerans, were N
2-fixing species [
14]. The genus
Paenibacillus has developed as a large genus comprising 400 species (
https://lpsn.dsmz.de/genus/paenibacillus, accessed on 12 November 2025). Many novel species and strains with N
2-fixing ability have been described [
19,
20,
21]. A
nif (
nitrogen
fixation) gene cluster composed of 9–10 genes [
nifB nifH nifD nifK nifE nifN nifX hesA (
orf1)
nifV] encoding Mo-nitrogenase is conserved in N
2-fixing
Paenibacillus strains [
19,
20,
21,
22]. In addition, some N
2-fixing
Paenibacillus species have V-nitrogenase or Fe-nitrogenase [
19]. Some members of
Paenibacillus can promote plant growth via nitrogen fixation [
23]. Inoculation of N
2-fixng
Paenibacillus triticisoli BJ-18 as a bio-fertilizer resulted in changes to the microbial community structure in the rhizosphere of wheat in fields [
24].
Genus
Fontibacillus with
F. aquaticusas as the type species was created in 2010, based on chemotaxonomic characteristics and the 16S rRNA gene [
8]. Compared to
Paenibacillus, the genus
Fontibacillus is a small genus which includes five recognized species.
F. aquaticus gen. nov., sp. nov., was isolated from a warm spring [
8].
Fontibacillus panacisegetis sp. nov. was isolated from soil of a ginseng field [
25].
Fontibacillus phaseoli sp. nov. was isolated from
Phaseolus vulgaris nodules [
26].
Fontibacillus solani sp. nov. was isolated from potato root [
27].
Fontibacillus pullulanilyticus sp. nov. was isolated from soil [
28]. Among the five
Fontibacillus species, only
F. phaseoli is a N
2-fixing bacterium which has Mo-nitrogenase encoded by 10 genes (
nifB nifH nifD nifK nifE nifN nifX hesA orf1 nifV).
Nitrogen (N) is the most important nutrient for plant growth. Growth of non-legume plants, such as rice wheat and maize, depends highly on chemical N fertilizers. However, production and over-application of chemical N fertilizers results in economic costs and environmental pollution. One approach to reduce use of N fertilizers is inoculation of non-legume plants with N2-fixing bacteria (biofertilizer), and another approach is the direct transfer of nif genes into cereal crops so that they can fix their nitrogen by using synthetic biology. Thus, it is necessary to isolate the novel N2-fixing microorganisms for application as biofertilizer and in engineering nif genes into non-legume plants.
In this study, 118 soil samples were taken from the rhizospheres of 17 plants, including rice, maize, wheat, oat, cowpea, onion, cabbage, spinach, rapeseed, eggplant, coriander, cluster mallow, fragrant plantain lily, peppermint, tall fescue, poplar, and ash tree (Fraxinus chinensis) in different regions of China. These soil samples were individually suspended in sterile water, and then these suspensions were individually spread on nitrogen-free medium agar plates for the growth of bacterial colonies. Twenty-four strains with the nifH gene encoding Fe protein of nitrogenase were obtained by screening 3200 bacterial colonies by PCR amplification using the nifH gene as a probe. Analysis of 16S rRNA gene revealed that among the 24 strains, only strain BL-9T, isolated from the rhizosphere of Fraxinus chinensis, belongs to Fontibacillus genus and the other 23 strains are members of the Klebsiella and Paenibacillus genera. Based on genomic, phylogenetic, chemotaxonomic, and phenotypic features, strain BL-9T is demonstrated to be a novel species of the genus Fontibacillus and the name proposed for this species is Fontibacillus forbon sp. nov. As we know, Fontibacillus forbon sp. nov is the second N2-fixing species within the Fontibacillus genus. The N2-fixing Fontibacillus species might have potential application as biofertilizer just as some N2-fixing Paenibacillus species did.
2. Materials and Methods
2.1. Isolation of Strains
N
2-fixing microorganisms were isolated by using nitrogen-free medium which was composed of 0.1 g NaCl, 0.01 g FeCl
3, 0.2 g MgSO
4·7H
2O, 0.002 g Na
2MoO
4, 0.1 g K
2HPO
4, 0.4 g KH
2PO
4, and 20 g sucrose per liter [
21]. A total of 118 soil samples were taken from the rhizospheres of 17 plants, including rice, maize, wheat, oat, cowpea, onion, cabbage, spinach, rapeseed, eggplant, coriander, cluster mallow, fragrant plantain lily, peppermint, tall fescue, poplar, and ash tree (
Fraxinus chinensis) in Hebei province, Henan province, Jiangsu province, Sichuan province and Beijing suburbs, China. Each soil sample was suspended in sterile water, and 100 μL suspension was spread on nitrogen-free medium agar plates. After incubation at 30 °C for 3 days, single colonies were identified by PCR amplification with the
nif gene probe.
Strain BL-9T was isolated from the rhizosphere of Fraxinus chinensis in Haidian District of Beijing, China (39°57′52.84′′ N 116°17′52.84′′ E). The soil type is sandy loam, and the organic matter content is about 18 g/kg with pH 6.0–8.0.
2.2. Sequence Analysis of nifH and 16S rRNA Genes and Construction of Phylogenetic Trees
NifH protein encoded by the
nifH gene is a structural unit of nitrogenase and
nifH is a key gene used for identification of N
2-fixing microorganisms. The
nifH gene of strain BL-9
T was PCR amplified with primers
nifH-P1 (5′-GGCTGCGATCCVAAGGCCGAYTCVACCCG-3′) and
nifH-P2 (5′-CTGVGCCTTGTTYT CGCGGATSGGCATGGC-3′) [
21]. The 16S rRNA gene of strain BL-9
T was PCR-amplified with primers 16S P1 (5′-AGAGTTTGATCCTGGCTCAGAACGAACGCT-3′) and 16S P2 (5′-TACGGCTACCTTGTTACGACTTCACCCC-3′) [
29]. The PCR-amplified
nifH and 16S rRNA gene products were sequenced. The sequences of the
nifH gene and 16S rRNA gene were analyzed on NCBI (
https://blast.ncbi.nlm.nih.gov, accessed on 12 November 2025). The phylogenetic tree was constructed with the maximum likelihood (ML) in the software MEGA7 [
30]. Bootstrap analysis with 500 replicates was performed, and bootstrap values were calculated to evaluate the confidence levels of tree branches.
2.3. Genome Sequencing
The genomic DNA of strain BL-9
T was extracted using the TIANamp Bacteria DNA Kit (DP302-02) made by TIANGEN BIOTECH Co., Ltd., Beijing, China. Genome sequencing was performed in Biomarker Technologies, Beijing, China by using the Illumina PE150 platform. Fragments below 500 bp were filtered out, and contaminated samples were further decontaminated. For genome assembly, the filtered reads were assembled by Spades v3.6.2 software. The assembled genome was then evaluated, statistically analyzed, and subject to subsequent gene prediction. The GeneMarkS software (Version 4.17) was employed to predict protein-coding genes in the sequenced genome [
31]. The protein sequences of predicted genes were aligned with various functional databases using Diamond (e value ≤ 1 × 10
−5). Genomic assembly metrics of strain BL-9
T are shown in
Table 1.
2.4. Genomic Feature Analysis
For construction of a genome based phylogenetic tree, the 92 single-copy core genes were extracted from the genome sequences using the UBCG program [
34]. Then, the phylogenomic tree was reconstructed using IQ-TREE v2.0.7 software [
35] based on the concatenated sequence dataset with 1000 bootstrap replicates.
2.5. Nitrogenase Activity Assay
The acetylene reduction assay was used to measure the nitrogenase activity of strain BL-9
T as described previously [
20,
36], and
Paenibacillus polymyxa WLY78 was used as a positive control.
Each strain of BL-9T and P. polymyxa WLY78 was anaerobically grown in nitrogen-deficient medium supplemented with 2 mM glutamate to a final OD600 of 0.2–0.4. Then, 1 mL of culture was transferred to a 25 mL test tube, and then the test tube was sealed with a robber stopper. The headspace in the tube was evacuated and replaced with argon gas. After incubating the cultures for 6–8 h at 30 °C with shaking, C2H2 (10% of the headspace volume) was injected into the test tubes. Incubating for another 3 h, 100 μL gas was withdrawn from the test tube through the rubber stopper with a gas tight syringe and then injected into gas chromatograph to quantify C2H4 production. The nitrogen activity was expressed in nmol C2H4/mg protein/h. All treatments were in three replicates.
2.6. Chemotaxonomic Characterization
Whole-cell fatty acids, polar lipids, and respiratory quinones were analyzed by Preservation Center of China Agricultural Microbial Strains in Chinese Academy of Agricultural Sciences, Beijing, China. Polar lipid was extracted according to the method described by Minnikin et al. [
37] and was identified by two-dimensional TLC (Thin Layer Chromatography). Analysis of compositions of cellular fatty acids was performed by the method described by Komagata and Suzuki [
38] using the Sherlock Identification System (MIDI) [
39]. Cellular menaquinones and isoprenoid quinones were extracted and analyzed using HPLC (High Performance Liquid Chromatography) [
40].
2.7. Morphological, Physiological, and Biochemical Analysis
Strain BL-9T and reference strains were routinely grown on LB agar at 30 °C for 2–3 days.
Cell morphology was observed by scanning electrical microscopy (SEM). Physiological and biochemical tests, such as starch hydrolysis, nitrate reduction, and NaCl tolerance, were performed as described by Zhang et al. [
21].
Gram staining of strain BL-9T grown on LB medium for 14 h was performed with E. coli and Bacillus subtilis as controls. Bacterial cells were spread on a glass slide and dried in air. Then, crystal purple solution was added to smear on the glass slide. After 1 min, the smear was rinsed with distilled water. Iodine solution was added to the smear for 1 min and then the smear on the glass slide was rinsed with distilled water. 95% ethanol was added to the smear on the glass slide for 20 s. Sarranine solution was added to the smear on the glass slide for 1 min and the smear on the glass slide was rinsed with distilled water. Finally, bacterial cells were observed under a microscope. All treatments were in three replicates.
3. Results and Discussion
3.1. Isolation of N2-Fixing Microorganisms
A total of 118 soil samples were taken from the rhizospheres of 17 plants, including rice, maize, wheat, oat, cowpea, onion, cabbage, spinach, rapeseed, eggplant, coriander, cluster mallow, fragrant plantain lily, peppermint, tall fescue, poplar, and ash tree (Fraxinus chinensis) in different regions of China. These soil samples were individually resuspended in sterile water, and these resuspensions were individually spread on nitrogen-free medium for the growth of bacterial colonies. Twenty-four strains with nifH gene encoding Fe protein of nitrogenase were obtained by screening 3200 bacterial colonies using PCR amplification with the nifH gene as a probe. Analysis of the 16S rRNA gene revealed that among the 24 strains, only strain BL-9T, isolated from the rhizosphere of Fraxinus chinensis, belongs to Fontibacillus genus, and the other 23 strains belong to Klebsiella and Paenibacillus genera. Strain BL-9T was then selected to be further investigated.
3.2. Phylogenetic Analysis of 16S rRNA Gene
Homology comparison of the 16S rRNA gene sequence of strain BL-9
T with those held in the GenBank database revealed that strain BL-9
T had high similarity to members of both
Fontibacillus and
Paenibacillus genera. Strain BL-9
T showed the highest sequence similarity with
Fontibacillus phaseoli BAPVE7B (98.03%), followed by
Fontibacillus solani A4STR04 (96.72%),
Fontibacillus panacisegetis (96.6%),
Paenibacillus vini (96.6%),
Paenibacillus anaericanus (96.6%), and
Paenibacillus segetis DB13260 (96.57%). A 98.65% similarity of 16S rRNA gene sequences is the threshold to differentiate bacterial species [
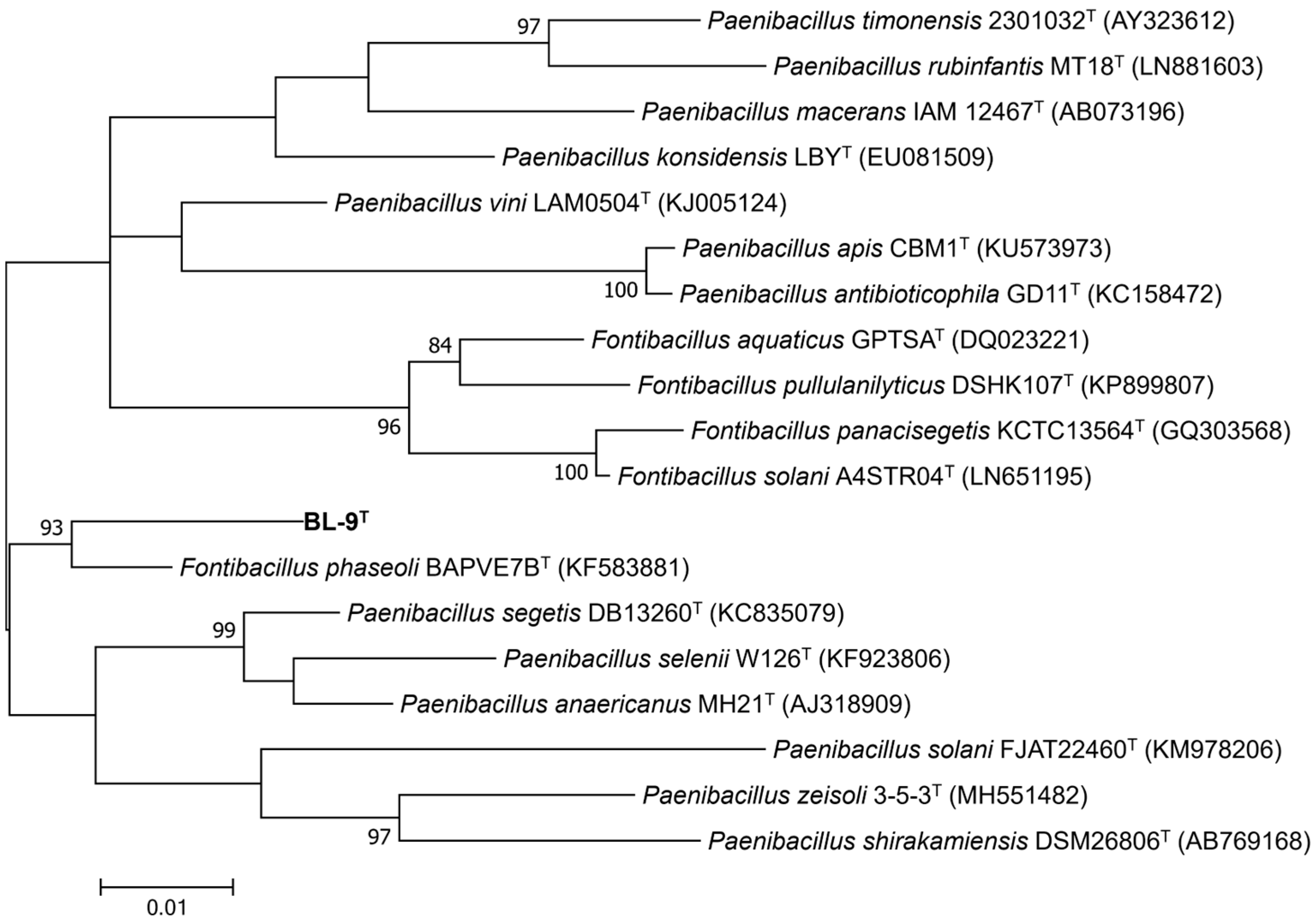
41]. The phylogenetic tree based on the 16S rRNA gene sequences exhibited that strain BL-9
T formed a distinct monophyletic group with
F. phaseoli BAPVE7B
T, supported by a high bootstrap value of 93% (
Figure 1). These results indicate that strain BL-9
T is a novel species within the genus
Fontibacillus.
3.3. Genomic Features and Phylogenomic Tree
Strain BL-9
T was genome-sequenced to evaluate its genome features. The genome size of strain BL-9
T is 5.5 Mb with 5081 genes. There are 91 tRNA genes and a 16S rRNA gene. The DNA G+C content of strain BL-9
T is 49.7% (
Table 2). Whereas its closest relative strain
F. phaseoli has a genome size of 5.4 Mb with 5071 genes. The DNA G+C content of
F. phaseoli is 49.5%. The genome sequence of strains BL-9
T was deposited in the GenBank under accession number GCA_046559655.1 (
Table 1).
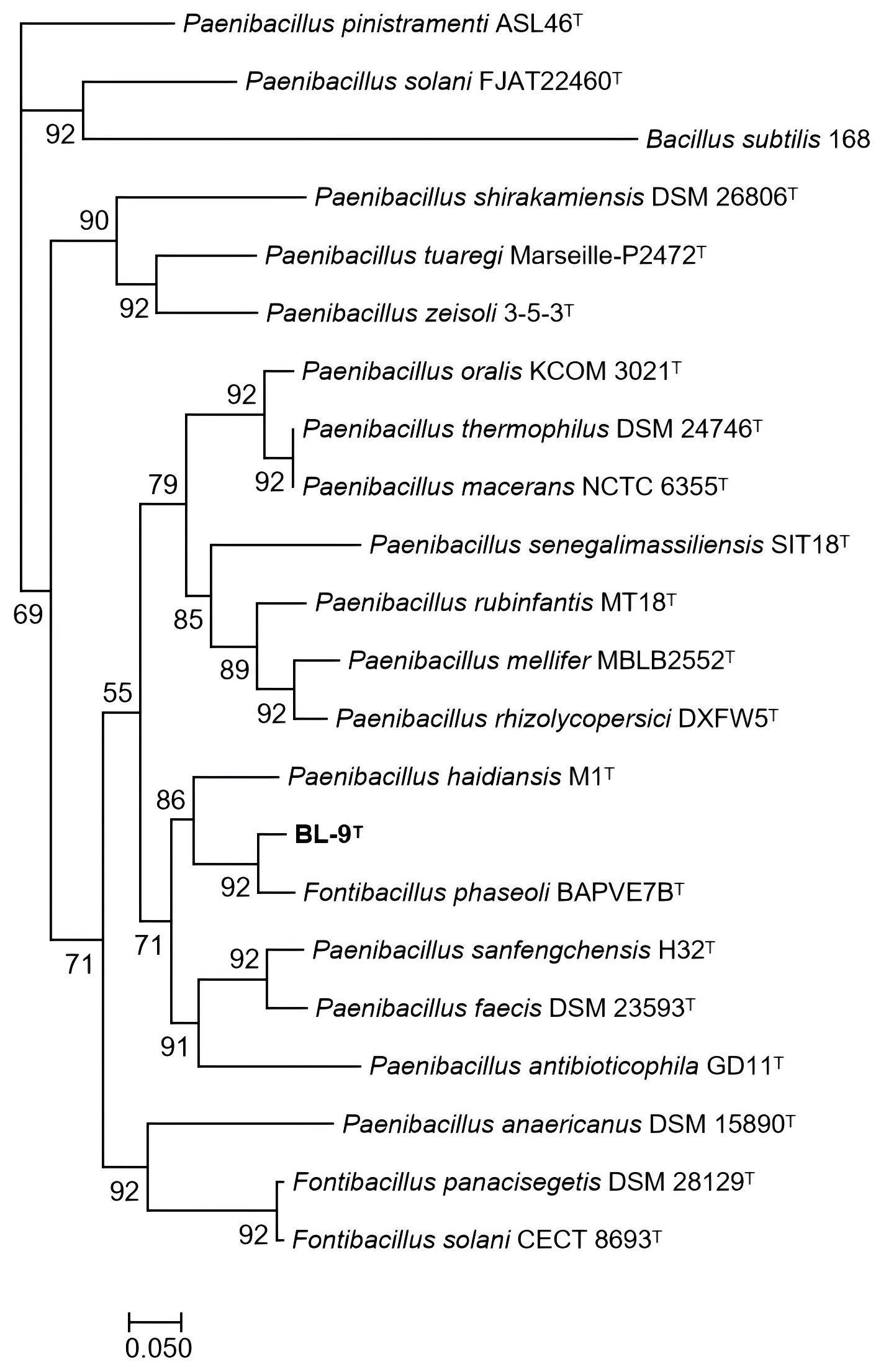
The phylogenomic tree (
Figure 2), based on 92 single-cope core genes, demonstrated that strain BL-9
T was clustered together with
F. phaseoli BAPVE7B
T, consistent with the phylogenetic analysis of the 16S rRNA gene described above.
It is recognized that ANI and dDDH for the species threshold are 95% and 70%, respectively [
33,
42,
43]. ANI and dDDH values were obtained by comparing genome sequence of strain BL-9
T with those of the closely related species of
Fontibacillus and
Paenibacillus genera (
Table 3). The highest ANI (42.5%) and the highest dDDH (90.94%) were between strain BL-9
T and reference strain
F. phaseoli BAPVE7B. The ANI and dDDH values between strain BL-9
T and its closely related species of
Fontibacillus and
Paenibacillus genera were 69.72–82.32% and 18.0–25.7%, respectively (
Table 3). These genomic relatedness data of strain BL-9
T are below the thresholds of ANI (95.0%) and dDDH (70.0%), indicating that strain BL-9
T is a novel species of
Fontibacillus genus.
3.4. Nitrogen Fixation (nif) Genes and Nitrogenase Activity
Nitrogenase activity was measured by the acetylene reduction assay as described in the methods section. Strain BL-9
T exhibited nitrogenase activity with 4802 (nmol C
2H
4/mg protein/h), while
P. polymyxa WLY78 (positive control) had nitrogenase activity with 3479 (nmol C
2H
4/mg protein/h). The results are consistent with the reports that nitrogenase activities exhibited variation among different N
2-fxing strains [
20,
21].
The
nifH gene encoding subunit of nitrogenase is highly conserved among N
2-fixing organisms and it is used as an indicator for identifying nitrogen-fixing bacteria. The
nifH gene of strain BL-9
T exhibited the highest similarity with
Paenibacillus abekawaensis MG1 (74.6%), followed by
Paenibacillus stellifer DSM14472 (73.4%), and
Paenibacillus durus DSM1735 (73.1%). The
nifH gene of strain BL-9
T also had high similarity with those of other N
2-fixing bacteria (e.g.,
Methylococcus capsulatus,
Rhodobacter sphaeroides,
Bradyrhizobium amphicarpzeae). The phylogenetic tree based on
nifH gene sequences showed that strain BL-9
T is clustered together with the N
2-fixing
Paenibacillus species (
Figure S1).
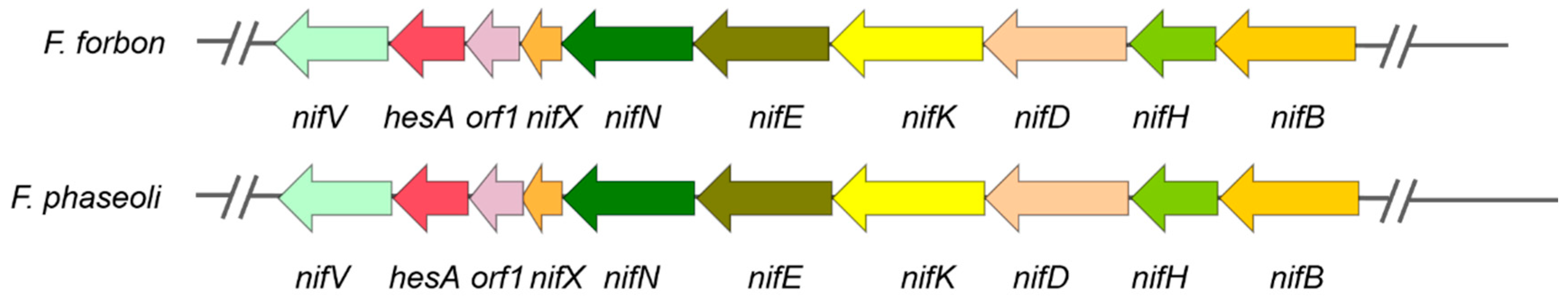
Genome sequence analysis showed that strain Bl-9
T has a Mo-nitrogenase encoded by a
nif gene cluster containing 10 genes (
nifB, nifH, nifD, nifK, nifE, nifN, nifX, orf1, hesA, and
nifV), just as observed in
F. phaseoli BAPVE7B (
Figure 3). The
nif gene cluster of strain Bl-9
T has 91.5% identity with that of
F. phaseoli BAPVE7B (
Table S1). These data supported that strain BL-9
T is a N
2-fixing bacterium which was isolated from the rhizosphere of
Fraxinus chinensis, whereas
F. phaseoli BAPVE7B was isolated from
Phaseolus vulgaris nodules [
26]. In N
2-fixing
Paenibacillus spp., some species (e.g.,
P. ploymyxa,
P. beijingensis,
P. massiliensis) have nine genes (
nifB, nifH, nifD, nifK, nifE, nifN, nifX, hesA and
nifV), while some species (e.g.,
Paenibacillus sabinae,
Paenibacillus forsythiae,
Paenibacillus graminis) have ten genes, which have an additional
orf1 [
22,
26].
3.5. Morphological and Physiological Characteristics
Strain BL-9
T was Gram-negative, facultatively anaerobic, motile, and rod-shaped. Our results are in agreement with the reports that some species of the order
Bacillales are Gram-variable, although most of the species from the order
Bacillales are Gram-positive. For example,
Paenibacillus favisporus was Gram-variable [
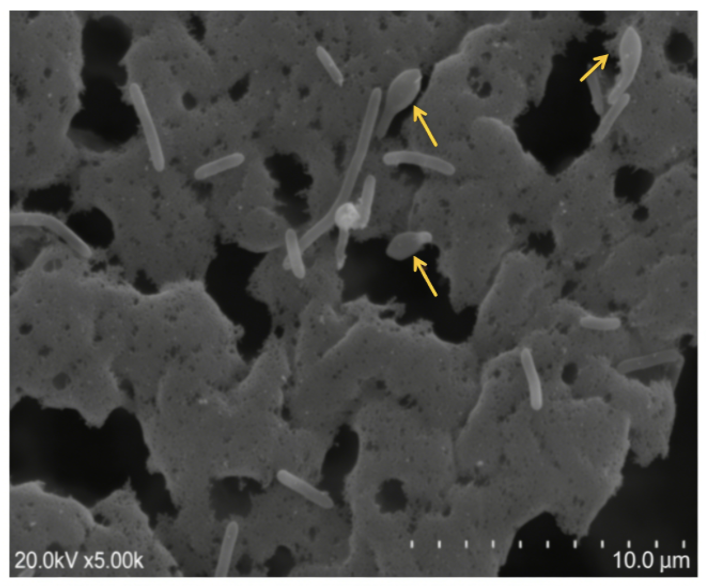
44]. Scanning electron microscopy of strain BL-9
T grown on LB agar for three days revealed ellipsoidal spores in swollen sporangia (
Figure 4).
Strain BL-9
T and the reference strains
F. phaseoli BAPVE7B
T,
F. solani A4STR04
T, and
P. segetis DB13260
T were tested for a range of physiological and biochemical characteristics. Strain BL-9
T,
F. phaseoli BAPVE7B
T, and
F. solani A4STR04
T exhibited positive nitrate reductase activity and starch hydrolysis, while
P. segetis DB13260
T had negative nitrate reductase activity and did not hydrolyze starch. Strain BL-9
T utilized fructose, glucose, galactose, maltose, mannitol, sorbitol, sucrose, and lactose to produce acid, while
P. segetis DB13260
T utilized all of these compounds except for inositol. However, reference strain
F. phaseoli BAPVE7B
T only utilized glucose, galactose, maltose, sucrose, and lactose to produce acid. Reference strain
F. solani A4STR04
T utilized fructose, maltose, and lactose, and utilized weakly galactose and sucrose. Differentiatial phenotypic characteristics among strain BL-9
T and reference strains are summarized in
Table 4.
3.6. Chemotaxonomic Characteristics
Chemotaxonomic features of strain BL-9
T were determined according to the previously described methods [
22,
41,
42,
43]. Analysis of the cellular fatty acid revealed that anteiso-C15:0 is the major fatty acid component of strain BL-9
T (55.14%) (
Table 3). Anteiso-C15:0 is also the predominant fatty acid for its reference strains
F. phaseoli BAPVE7B
T (53.1%),
F. solani A4STR04
T (61.5%), and
P. segetis DB13260
T (43.9%). The fatty acid contents showed significant variation among strain BL-9
T and reference strains, although anteiso-C15:0 is the major fatty acid for strain BL-9 and its reference strains (
Table 5). These data indicate that strain BL-9
T is distinguished from other members of both genera
Fontibacillus and
Paenibacillus.Strain BL-9
T contains the major polar lipids, including DPG (diphosphatidylglycerol), APL (aminophospholipids), PG (phosphatidylglycerol), APGL (unidentified aminophosphoglycolipid), and unidentified phospholipids (PL2, PL3 and PL4) (
Figure S2). The results showed that the polar lipid profiles of strain BL-9
T are similar to those of
Fontibacillus spp. (e.g.,
F. aquaticus and
F. phaseoli) [
8,
19], supporting that strain BL-9
T is a member of the
Fontibacillus genus.
The major respiratory quinone component of strain BL-9
T is menaquinone-7 (MK-7) (
Figure S3). MK-7 is also the only menaquinone in the
Fontibacillus species, such as
F. aquaticus and
F. panacisegetis [
8,
23]. The data support that strain BL-9
T is a member of the
Fontibacillus genus.
4. Conclusions
A novel species Fontibacillus forbon sp. nov., with strain BL-9T as the type strain was isolated from the rhizosphere of Fraxinus chinensis. Strain BL-9T is a facultatively anaerobic, rod-shaped, endospore-forming, and motile bacterium. Strain BL-9T was able to fix nitrogen and grew on nitrogen-free medium. Genome of strain BL-9T has a nif (nitrogen fixation) gene cluster containing 10 genes (nifB nifH nifD nifK nifE nifN nifX orf1 hesA nifV). DNA G+C content of strain BL-9T is 49.7%. The predominant fatty acid is anteiso-C15:0, the major menaquinone is MK-7, and the major polar lipid is diphosphatidylglycerol. Chemotaxonomic analyses demonstrated that strain BL-9T and species of the genus Fontibacillus have common features, with anteiso-C15:0 as the predominant fatty acid, MK-7 as the major menaquinone, and diphosphatidylglycerol as the major polar lipid. Strain BL-9T and its closely related species of Fontibacillus have some common and distinguished physiological characteristics. Phylogenies, based on the 16S rRNA gene and core genome, revealed that strain BL-9T was most closely related to Fontibacillus phaseoli BAPVE7B. However, the digital DNA-DNA hybridization (dDDH) and average nucleotide identity (ANI) between strain BL-9T and its closely related type strain F. phaseoli BAPVE7B were 42.5% and 90.94%, respectively, indicating that strain BL-9T represents a novel species of the genus Fontibacillus. The name proposed for this species is Fontibacillus forbon sp. nov., with the type strain BL-9T (=GDMCC 1.5526T = JCM 37804T).
5. Description of Fontibacillus forbon sp. nov
Fontibacillus forbon (forbon, named after Forbon Technology Co., Ltd., Wuhan, China).
Cells are Gram-negative, facultatively anaerobic, rod-shaped, and motile. An ellipsoidal spore is formed in swollen sporangia. The colonies on the LB medium are cream white, convex, and circular, with a diameter of 1.0–2.0 mm. Cells grow at 25–35 °C, with optimum growth at 26 °C. The pH range for growth is a pH of 6.0–8.0 (optimum pH 7.0). The optimum concentration of NaCl for growth is 1.0%. The various substrates: D-fructose, D-galactose, D-glucose, D-xylose, lactose, maltose, sucrose, inositol, D-mannitol, and D-sorbitol, are utilized. Nitrate is reduced to nitrite. Starch is hydrolyzed. The predominant fatty acid is anteiso-C15:0. The major polar lipids are DPG (diphosphatidylglycerol), APL (aminophospholipids), PG (phosphatidylglycerol), and APGL (unidentified aminophosphoglycolipid). The major menaquinone is MK-7. Strain BL-9T exhibits nitrogenase activity and has a nif gene cluster composed of 10 genes (nifB, nifH, nifD, nifK, nifE, nifN, nifX, orf1, hesA, nifV). The genome size is 5.5 Mb, and the G+C content is 49.7%.
The type strain BL-9T (=CGMCC 1.5526T = JCM 37804T) was isolated from the rhizosphere soil of Fraxinus chinensis in the Haidian District of Beijing, China. The GenBank accession numbers for the 16S rRNA sequence and for the genome sequence are PQ803957.1. and GCA_046559655.1., respectively. The type strain BL-9T was deposited in the Japan Collection of Microorganisms with No. JCM 37804T and in the Chinese Guangdong Microbial Culture Collection Center with No. CGMCC 1.5526T.
Supplementary Materials
The following supporting information can be downloaded at:
https://www.mdpi.com/article/10.3390/microorganisms14010049/s1, Figure S1: The maximum-likelihood phylogenetic tree based on
nifH gene sequences (823 base pairs) of strain BL-9
T together with its closely related taxonomic groups. Bar, 0.05 nucleotide substitutions per site. Bootstrap values > 70% (based on 500 replications) are shown at branch points; Figure S2: Two-dimensional TLC plate of polar lipids extracted from strain BL-9
T. The plate was sprayed with 10% (
v/
v) molybdophosphoric aicd to show all polar lipids present. DPG, diphosphatidylglycerol; PG, phosphatidylglycerol; APL, aminophospholipids; PL, unidentified phosphoglycolipids; L, unknown polar lipids; APGL, unidentified aminophosphoglycolipid; Figure S3: HPLC analysis shows MK-7 as the major respiratory quinone component for strain BL-9
T. Mobile phase, methanol:isopropanol = 65:35; Chromatographic column: Zorbax Eclipse XDB-C18 (4.6 * 250 mm, 5 μm; Agilent); Column temperature: 40 °C; Flow rate: 1.0 mL/min; Injection volume: 10 μL; Detection wavelength: 270 nm; Table S1: Comparison of
nif (
nitrogen
fixation) genes from the novel species
Fontibacillus forbon BL-9
T and
Fontibacillus phaseoli BAPVE7B.
Author Contributions
S.C. supervised the research work and S.C. and Y.S. wrote the main manuscript. R.H. performed the main research work. R.W., R.H., C.S., and Y.S. collected soil samples. Y.S., W.Z., R.W., and S.C. analyzed data. Y.S. and C.S. prepared figures. All authors reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The work was supported by the grant “Non leguminous nitrogen fixing bacteria and comprehensive application technology” (No. 2023110002000387) provided by Forbon Technology Co., Ltd., Wuhan, China.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article/
Supplementary Materials. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
Renzong Wang was employed by the company Forbon Technology Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Besides, the authors declare that this study received funding from Forbon Technology Co., Ltd. The funder was not involved in the study design, collection, analysis.
References
- De Vos, P.; Ludwig, W.; Schleifer, K.H.; Whitman, W.B. Family IV. Paenibacillaceae fam. nov. In The Firmicutes, Bergey’s Manual of Systematic Bacteriology, 2nd ed.; De Vos, P., Garrity, G., Jones, D., Krieg, N.R., Ludwig, W., Rainey, F.A., Schleifer, K.H., Whitman, W.B., Eds.; Springer: New York, NY, USA, 2009; Volume 2, pp. 269–296. [Google Scholar]
- Sakai, M.; Deguchi, D.; Hosoda, A.; Kawauchi, T.; Ikenaga, M. Ammoniibacillus agariperforans gen. nov., sp. nov., a thermophilic, agar-degrading bacterium isolated from compost. Int. J. Syst. Evol. Microbiol. 2015, 65, 570–577. [Google Scholar] [CrossRef]
- Zaitsev, G.M.; Tsitko, I.V.; Rainey, F.A.; Trotsenko, Y.A.; Uotila, J.S.; Stackebrandt, E.; Salkinoja-Salonen, M.S. New aerobic ammonium-dependent obligately oxalotrophic bacteria: Description of Ammoniphilus oxalaticus gen. nov., sp. nov. and Ammoniphilus oxalivorans gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 1998, 48, 151–163. [Google Scholar] [CrossRef]
- Shida, O.; Takagi, H.; Kadowaki, K.; Komagata, K. Proposal for two new genera, Brevibacillus gen. nov. and Aneurinibacillus gen. nov. Int. J. Syst. Evol. Microbiol. 1996, 46, 939–946. [Google Scholar] [CrossRef]
- Cao, W.R.; Guo, L.Y.; Du, Z.J.; Das, A.; Saren, G.; Jiang, M.Y.; Dunlap, C.A.; Rooney, A.P.; Yu, X.K.; Li, T.G. Chengkuizengella sediminis gen. nov. sp. nov., isolated from sediment. Int. J. Syst. Evol. Microbiol. 2017, 67, 2672–2678. [Google Scholar] [CrossRef]
- Kämpfer, P.; Rosselló-Mora, R.; Falsen, E.; Busse, H.J.; Tindall, B.J. Cohnella thermotolerans gen. nov., sp. nov., and classification of ‘Paenibacillus hongkongensis’ as Cohnella hongkongensis sp. nov. Int. J. Syst. Evol. Microbiol. 2006, 56, 781–786. [Google Scholar] [CrossRef]
- Chen, Y.; Lv, A.P.; Li, M.M.; OuYang, Y.T.; Lian, Z.H.; Chen, L.B.; Liu, Z.T.; Liu, L.; Jiao, J.Y.; Li, W.J. Ferviditalea candida gen. nov., sp. nov., a novel member of the family Paenibacillaceae isolated from a geothermal area. Anaerobe 2024, 88, 102866. [Google Scholar] [CrossRef]
- Saha, P.; Krishnamurthi, S.; Bhattacharya, A.; Sharma, R.; Chakrabarti, T. Fontibacillus aquaticus gen. nov., sp. nov., isolated from a warm spring. Int. J. Syst. Evol. Microbiol. 2010, 60, 422–428. [Google Scholar] [CrossRef]
- Kudryashova, E.B.; Ariskina, E.V.; Karlyshev, A.V.; Potekhina, N.V.; Shashkov, A.; Suzina, N.E.; Evtushenko, L.I. Gordonibacillus kamchatkensis gen. nov., sp. nov. from the frozen volcanic ash. Microbiology 2024, 93, 269–281. [Google Scholar] [CrossRef]
- Keita, M.B.; Padhmanabhan, R.; Caputo, A.; Robert, C.; Delaporte, E.; Raoult, D. Non-contiguous finished genome sequence and description of Gorillibacterium massiliense gen. nov, sp. nov., a new member of the family Paenibacillaceae. Stand. Genom. Sci. 2024, 9, 807–820. [Google Scholar] [CrossRef]
- Chhe, C.; Uke, A.; Baramee, S.; Tachaapaikoon, C.; Pason, P.; Waeonukul, R.; Ratanakhanokchai, K.; Kosugi, A. Insulambacter thermoxylanivorax sp. nov., a thermophilic xylanolytic bacterium isolated from compost. Int. J. Syst. Evol. Microbiol. 2023, 73, 005724. [Google Scholar] [CrossRef]
- Chen, R.W.; Zhang, J.; He, Y.Q.; Wang, K.X.; Li, C.; Long, L.J. Longirhabdus pacifica gen. nov., sp. nov., isolated from a deep-sea hydrothermal sediment in the West Pacific Ocean. Int. J. Syst. Evol. Microbiol. 2023, 69, 3362–3367. [Google Scholar] [CrossRef]
- Collins, M.D.; Lawson, P.A.; Willems, A.; Cordoba, J.J.; Fernandez-Garayzabal, J.; Garcia, P.; Cai, J.; Hippe, H.; Farrow, J.A. The phylogeny of the genus Clostridium: Proposal of five new genera and eleven new species combinations. Int. J. Syst. Evol. Microbiol. 1994, 44, 812–826. [Google Scholar] [CrossRef]
- Ash, C.; Priest, F.G.; Collins, M.D. Molecular identification of rRNA group 3 bacilli (ash, farrow, wallbanks and collins) using a PCR probe test: Proposal for the creation of a new genus Paenibacillus. Antonie Van Leeuwenhoek 1993, 64, 253–260. [Google Scholar] [CrossRef]
- Hwang, W.M.; Ko, Y.; Kang, K.; Ahn, T.Y. Paludirhabdus telluriireducens gen. nov., sp. nov. and Paludirhabdus pumila sp. nov., isolated from soil of a mountain wetland and emended description of Gorillibacterium massiliense. Int. J. Syst. Evol. Microbiol. 2018, 68, 3040–3046. [Google Scholar] [CrossRef]
- Rivas, R.; García-Fraile, P.; Zurdo-Piñeiro, J.L.; Mateos, P.F.; Martínez-Molina, E.; Bedmar, E.J.; Sánchez-Raya, J.; Velázquez, E. Saccharibacillus sacchari gen. nov., sp. nov., isolated from sugar cane. Int. J. Syst. Evol. Microbiol. 2008, 58, 1850–1854. [Google Scholar] [CrossRef]
- Touzel, J.P.; O’Donohue, M.; Debeire, P.; Samain, E.; Breton, C. Thermobacillus xylanilyticus gen. nov., sp. nov. a new aerobic thermophilic xylan-degrading bacterium isolated from farm soil. Int. J. Syst. Evol. Microbiol. 2000, 50, 315–320. [Google Scholar] [CrossRef]
- Kukolya, J.; Bata-Vidács, I.; Luzics, S.; Tóth, E.; Kéki, Z.; Schumann, P.; Táncsics, A.; Nagy, I.; Olasz, F.; Tóth, Á. Xylanibacillus composti gen. nov., sp. nov., isolated from compost. Int. J. Syst. Evol. Microbiol. 2018, 68, 698–702. [Google Scholar] [CrossRef]
- Xie, J.B.; Du, Z.; Bai, L.; Tian, C.; Zhang, Y.; Xie, J.; Wang, T.; Liu, X.; Chen, X.; Cheng, Q.; et al. Comparative genomic analysis of N2-fixing and non-N2-fixing Paenibacillus spp.: Organization, evolution and expression of the nitrogen fixation genes. PLoS Genet. 2014, 10, e1004231. [Google Scholar] [CrossRef]
- Li, Q.; Li, Y.; Liu, X.; Chen, S. Paenibacillus sinensis sp. nov., a nitrogen-fixing species isolated from plant rhizospheres. Antonie Van Leeuwenhoek 2022, 115, 7–18. [Google Scholar] [CrossRef]
- Zhang, W.; Gao, M.; Hu, R.; Shang, Y.; Liu, M.; Lan, P.; Jiao, S.; Wei, G.; Chen, S. Nitrogen-fixing Paenibacillus haidiansis and Paenibacillus sanfengchensis: Two novel species from plant rhizospheres. Microorganisms 2024, 12, 2561. [Google Scholar] [CrossRef]
- Li, Q.; Liu, X.M.; Zhang, H.W.; Chen, S. Evolution and functional analysis of orf1 within nif gene cluster from Paenibacillus graminis RSA19. Int. J. Mol. Sci. 2019, 20, 1145. [Google Scholar] [CrossRef]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.C. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Factories 2016, 15, 203. [Google Scholar] [CrossRef]
- Li, Y.; Wang, M.; Chen, S. Application of N2-fixing Paenibacillus triticisoli BJ-18 changes the compositions and functions of the bacterial, diazotrophic, and fungal microbiomes in the rhizosphere and root/shoot endosphere of wheat under field conditions. Biol. Fertil. Soils 2021, 57, 347–362. [Google Scholar] [CrossRef]
- Lee, K.C.; Kim, K.K.; Eom, M.K.; Kim, M.J.; Lee, J.S. Fontibacillus panacisegetis sp. nov., isolated from soil of a ginseng field. Int. J. Syst. Evol. Microbiol. 2011, 61, 369–374. [Google Scholar] [CrossRef]
- Flores-Félix, J.D.; Mulas, R.; Ramírez-Bahena, M.H.; Cuesta, M.J.; Rivas, R.; Brañas, J.; Mulas, D.; González-Andrés, F.; Peix, A.; Velázquez, E. Fontibacillus phaseoli sp. nov. isolated from Phaseolus vulgaris nodules. Antonie Van Leeuwenhoek 2014, 105, 23–28. [Google Scholar] [CrossRef]
- Ramírez-Bahena, M.H.; Flores-Félix, J.D.; Cuesta, M.J.; Tejedor Gil, C.; Palomo, J.L.; García Benavides, P.; Igual, J.M.; Fernández Pascual, M.; Velázquez, E.; Peix, A. Fontibacillus solani sp. nov. isolated from potato (Solanum tuberosum L.) root. Antonie Van Leeuwenhoek 2015, 107, 1315–1321. [Google Scholar] [CrossRef] [PubMed]
- Bektas, K.I.; Belduz, A.O.; Guvenmez, H.K.; Sihay, D. Fontibacillus pullulanilyticus sp.nov. isolated from soil. J. Basic Microbiol. 2016, 56, 857–863. [Google Scholar] [CrossRef] [PubMed]
- Frank, J.A.; Reich, C.I.; Sharma, S.; Weisbaum, J.S.; Wilson, B.A.; Olsen, G.J. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 2008, 74, 2461–2470. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthof, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Na, S.I.; Kim, Y.O.; Yoon, S.H.; Ha, S.M.; Baek, I.; Chun, J. UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 2018, 56, 280–285. [Google Scholar] [CrossRef]
- Wang, T.; Zhao, X.; Shi, H.; Sun, L.; Li, Y.; Li, Q.; Zhang, H.; Chen, S.; Li, J. Positive and negative regulation of transferred nif genes mediated by indigenous GlnR in Gram-positive Paenibacillus polymyxa. PLoS Genet. 2018, 14, e1007629. [Google Scholar] [CrossRef]
- Minnikin, D.E.; O’donnell, A.G.; Goodfellow, M.; Alderson, G.; Athalye, M.; Schaal, A.; Parlett, J.H. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 1984, 2, 233–241. [Google Scholar] [CrossRef]
- Komagata, K.; Suzuki, K. Lipid and cell-wall analysis in bacterial systematics. In Methods in Microbiology; Academic Press: Cambridge, MA, USA, 1988; Volume 19, pp. 161–207. [Google Scholar] [CrossRef]
- Sasser, M.; Kunitsky, C.; Jackoway, G.; Ezzell, J.W.; Teska, J.D.; Harper, B.; Parker, S.; Barden, D.; Blair, H.; Breezee, J.; et al. Identification of Bacillus anthracis from culture using gas chromatographic analysis of fatty acid methyl esters. J. AOAC Int. 2005, 88, 178–181. [Google Scholar] [CrossRef]
- Collins, M.D.; Goodfellow, M.; Minnikin, D.E. Fatty acid, isoprenoid quinone and polar lipid composition in the classification of Curtobacterium and related taxa. Microbiology 1980, 118, 29–37. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Tiedje, J.M. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 2567–2572. [Google Scholar] [CrossRef]
- Velázquez, E.; de Miguel, T.; Poza, M.; Rivas, R.; Rosselló-Mora, R.; Villa, T.G. Paenibacillus favisporus sp. nov., a xylanolytic bacterium isolated from cow faeces. Int. J. Syst. Evol. Microbiol. 2004, 54, 59–64. [Google Scholar] [CrossRef] [PubMed]
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |