Hepatitis E ORF2 Blocks Trophoblast Autophagy to Induce Miscarriage via LC3B Binding Rather than PI3K/Akt/mTOR Pathway Suppression
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. Materials
2.3. Plasmid Transfection
2.4. Cell Selection
2.5. RT-PCR
2.6. Western Blot
2.7. Immunofluorescence Assay (IFA)
2.8. Co-Immunoprecipitation
2.9. Statistical Analysis
3. Result
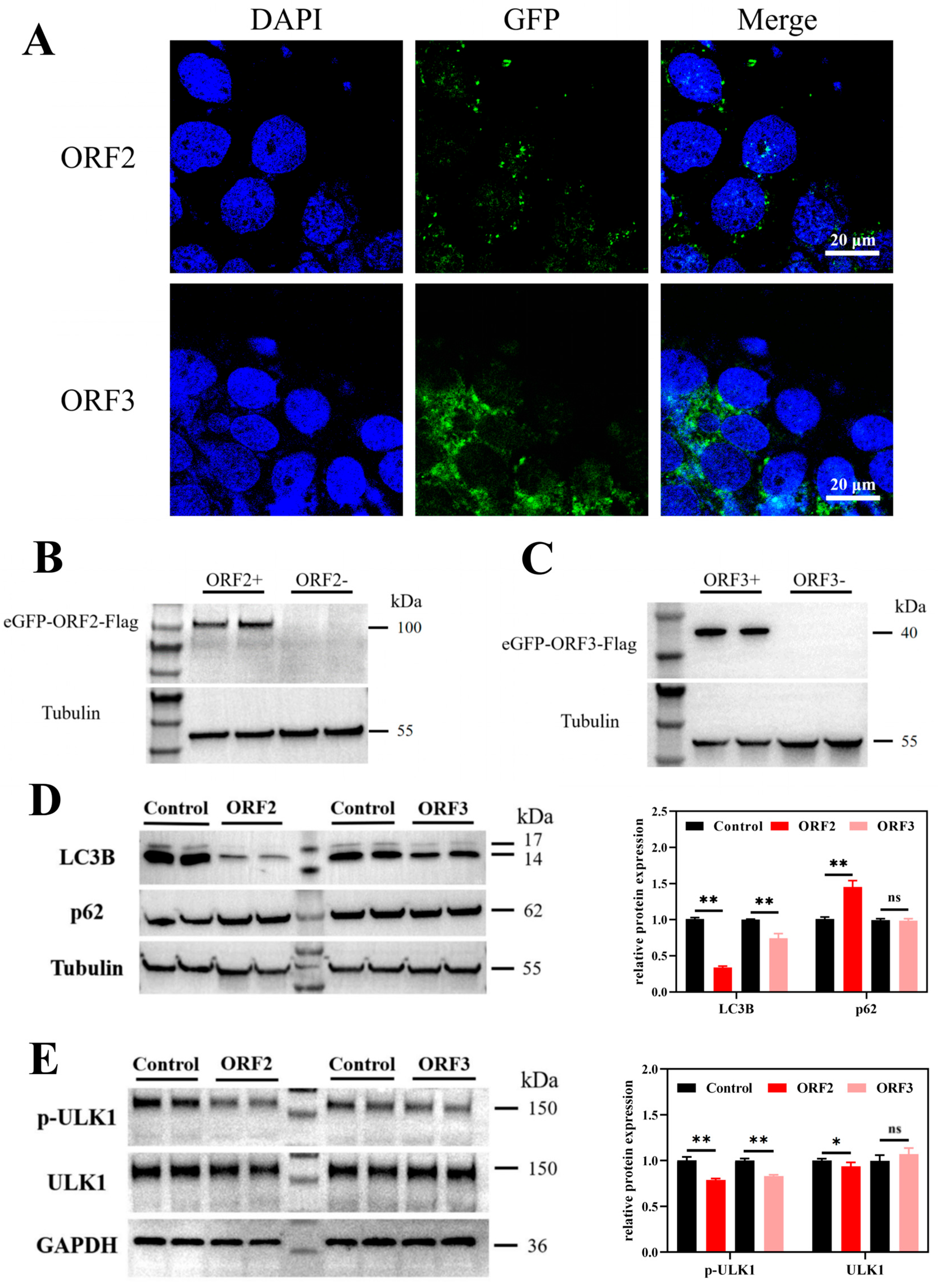
3.1. Overexpression of HEV ORF2 and ORF3 in JEG-3 Cells
3.2. HEV ORF2 Plays a Key Role in Autophagy Inhibition in JEG-3 Cells by Suppressing ULK1 Phosphorylation
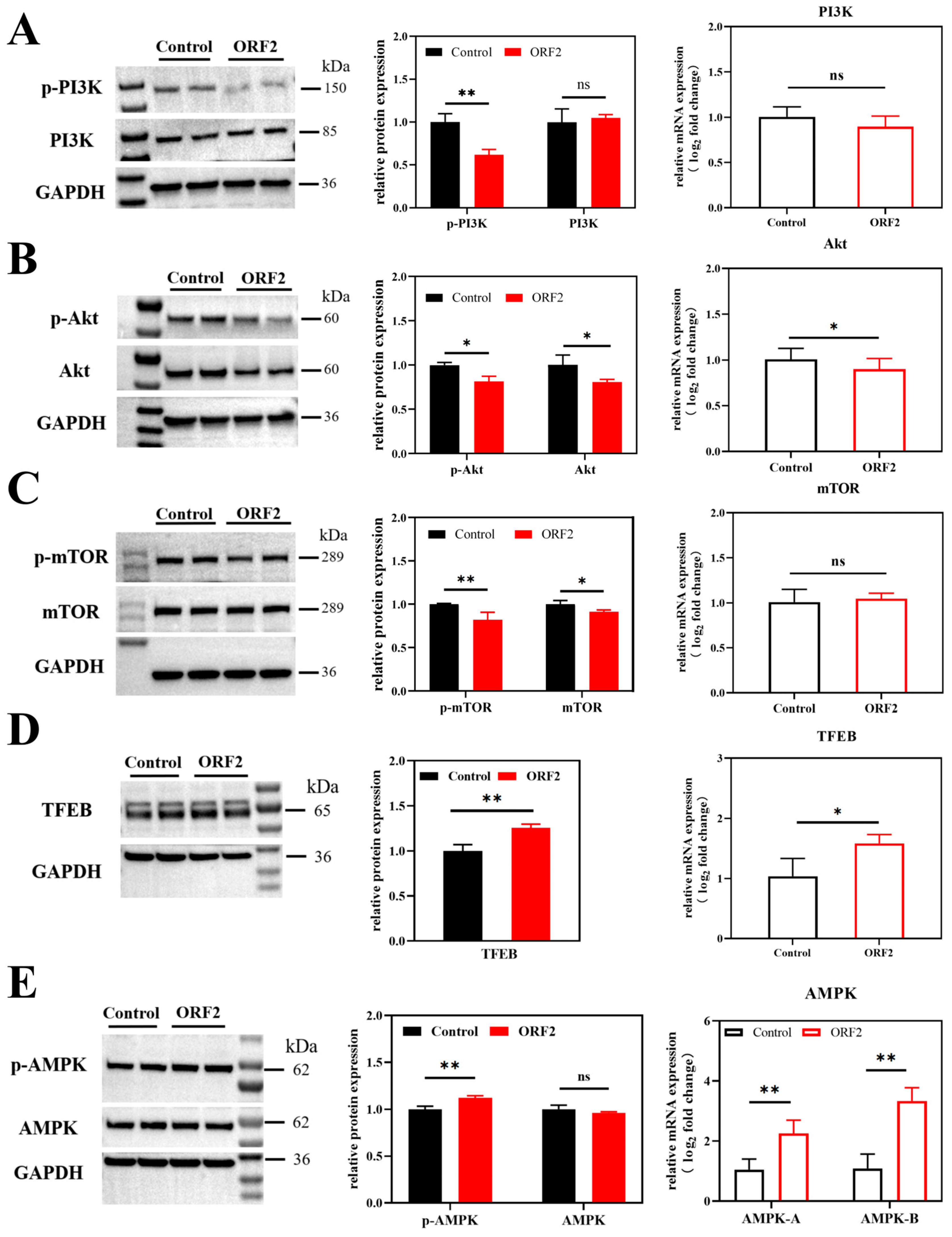
3.3. HEV ORF2 Suppresses the Upstream PI3K/Akt/mTOR Pathway in JEG-3 Cells
3.4. HEV ORF2 Activates AMPK to Stimulate Alternative Autophagy Pathways
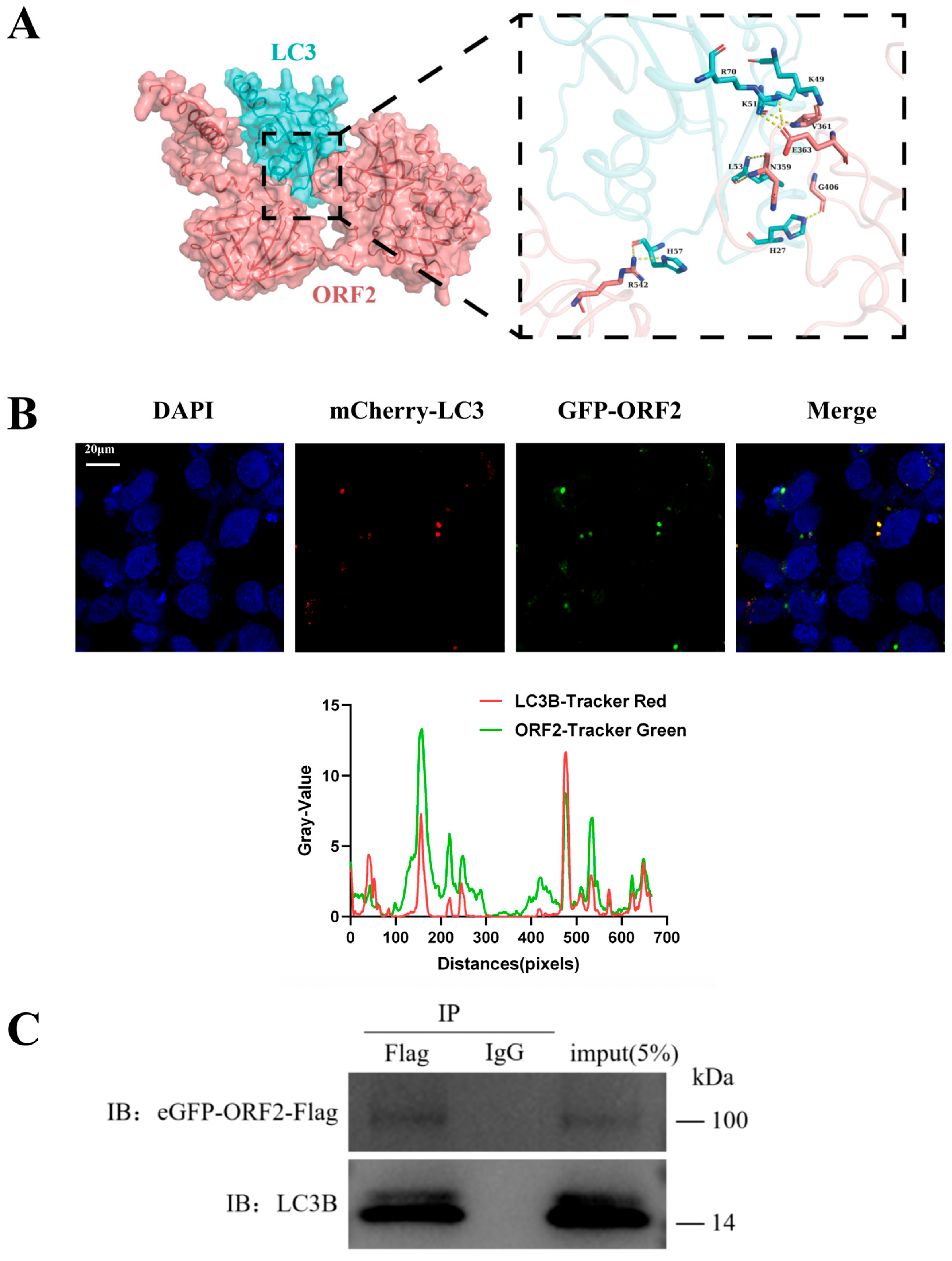
3.5. HEV ORF2 Directly Binds LC3 to Suppress Autophagy in JEG-3 Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Feng, Y.; Chen, Y.; Wu, X.; Chen, J.; Zhou, Q.; Liu, B.; Zhang, L.; Yi, C. Interplay of energy metabolism and autophagy. Autophagy 2024, 20, 4–14. [Google Scholar] [CrossRef]
- Tan, H.X.; Yang, S.L.; Li, M.Q.; Wang, H.Y. Autophagy suppression of trophoblast cells induces pregnancy loss by activating decidual NK cytotoxicity and inhibiting trophoblast invasion. Cell Commun. Signal. 2020, 18, 73. [Google Scholar] [CrossRef]
- Xiang, H.; Zhang, J.; Lin, C.; Zhang, L.; Liu, B.; Ouyang, L. Targeting autophagy-related protein kinases for potential therapeutic purpose. Acta Pharm. Sin. B 2020, 10, 569–581. [Google Scholar] [CrossRef]
- Li, M.Y.; Shen, H.H.; Cao, X.Y.; Gao, X.X.; Xu, F.Y.; Ha, S.Y.; Sun, J.S.; Liu, S.P.; Xie, F.; Li, M.Q. Targeting a mTOR/autophagy axis: A double-edged sword of rapamycin in spontaneous miscarriage. Biomed. Pharmacother. 2024, 177, 116976. [Google Scholar] [CrossRef]
- Lee, S.; Shin, J.; Kim, J.S.; Shin, J.; Lee, S.K.; Park, H.W. Targeting TBK1 Attenuates LPS-Induced NLRP3 Inflammasome Activation by Regulating of mTORC1 Pathways in Trophoblasts. Front. Immunol. 2021, 12, 743700. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zheng, F.; Wang, D.; Yang, Q. Regulation of ULK1 by WTAP/IGF2BP3 axis enhances mitophagy and progression in epithelial ovarian cancer. Cell Death Dis. 2024, 15, 97. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Napolitano, G.; de Araujo, M.E.G.; Esposito, A.; Monfregola, J.; Huber, L.A.; Ballabio, A.; Hurley, J.H. Structure of the lysosomal mTORC1-TFEB-Rag-Ragulator megacomplex. Nature 2023, 614, 572–579. [Google Scholar] [CrossRef]
- Zheng, W.; Zhang, Y.; Xu, P.; Wang, Z.; Shao, X.; Chen, C.; Cai, H.; Wang, Y.; Sun, M.A.; Deng, W.; et al. TFEB safeguards trophoblast syncytialization in humans and mice. Proc. Natl. Acad. Sci. USA 2024, 121, e2404062121. [Google Scholar] [CrossRef]
- Nwadike, C.; Williamson, L.E.; Gallagher, L.E.; Guan, J.L.; Chan, E.Y.W. AMPK Inhibits ULK1-Dependent Autophagosome Formation and Lysosomal Acidification via Distinct Mechanisms. Mol. Cell Biol. 2018, 38, e00023-18. [Google Scholar] [CrossRef]
- LeDesma, R.; Nimgaonkar, I.; Ploss, A. Hepatitis E Virus Replication. Viruses 2019, 11, 719. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Chimeno, M.; Cenalmor, A.; Garcia-Lugo, M.A.; Hernandez, M.; Rodriguez-Lazaro, D.; Avellon, A. Proline-Rich Hypervariable Region of Hepatitis E Virus: Arranging the Disorder. Microorganisms 2020, 8, 1417. [Google Scholar] [CrossRef]
- Zhou, Z.; Xie, Y.; Wu, C.; Nan, Y. The Hepatitis E Virus Open Reading Frame 2 Protein: Beyond Viral Capsid. Front. Microbiol. 2021, 12, 739124. [Google Scholar] [CrossRef]
- Peron, J.M.; Larrue, H.; Izopet, J.; Buti, M. The pressing need for a global HEV vaccine. J. Hepatol. 2023, 79, 876–880. [Google Scholar] [CrossRef]
- Gouttenoire, J.; Pollán, A.; Abrami, L.; Oechslin, N.; Mauron, J.; Matter, M.; Oppliger, J.; Szkolnicka, D.; Dao Thi, V.L.; van der Goot, F.G.; et al. Palmitoylation mediates membrane association of hepatitis E virus ORF3 protein and is required for infectious particle secretion. PLoS Pathog. 2018, 14, e1007471. [Google Scholar] [CrossRef]
- Ding, Q.; Heller, B.; Capuccino, J.M.V.; Song, B.; Nimgaonkar, I.; Hrebikova, G.; Contreras, J.E.; Ploss, A. Hepatitis E virus ORF3 is a functional ion channel required for release of infectious particles. Proc. Natl. Acad. Sci. USA 2017, 114, 1147–1152. [Google Scholar] [CrossRef]
- Van Tong, H.; Hoan, N.X.; Wang, B.; Wedemeyer, H.; Bock, C.T.; Velavan, T.P. Hepatitis E Virus Mutations: Functional and Clinical Relevance. EBioMedicine 2016, 11, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Maltepe, E.; Fisher, S.J. Placenta: The forgotten organ. Annu. Rev. Cell Dev. Biol. 2015, 31, 523–552. [Google Scholar] [CrossRef]
- Turco, M.Y.; Gardner, L.; Kay, R.G.; Hamilton, R.S.; Prater, M.; Hollinshead, M.S.; McWhinnie, A.; Esposito, L.; Fernando, R.; Skelton, H.; et al. Trophoblast organoids as a model for maternal-fetal interactions during human placentation. Nature 2018, 564, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Abbas, Y.; Turco, M.Y.; Burton, G.J.; Moffett, A. Investigation of human trophoblast invasion in vitro. Hum. Reprod. Update 2020, 26, 501–513. [Google Scholar] [CrossRef]
- Yang, H.L.; Lai, Z.Z.; Shi, J.W.; Zhou, W.J.; Mei, J.; Ye, J.F.; Zhang, T.; Wang, J.; Zhao, J.Y.; Li, D.J.; et al. A defective lysophosphatidic acid-autophagy axis increases miscarriage risk by restricting decidual macrophage residence. Autophagy 2022, 18, 2459–2480. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Shen, H.; Wang, Z.; Qin, X.; Li, M.; Zhang, X. Autophagy Inhibition in Trophoblasts Induces Aberrant Shift in CXCR4+ Decidual NK Cell Phenotype Leading to Pregnancy Loss. J. Clin. Med. 2023, 12, 7491. [Google Scholar] [CrossRef]
- Mlyczyńska, E.; Kurowska, P.; Drwal, E.; Opydo-Chanek, M.; Tworzydło, W.; Kotula-Balak, M.; Rak, A. Apelin and apelin receptor in human placenta: Expression, signalling pathway and regulation of trophoblast JEG-3 and BeWo cells proliferation and cell cycle. Int. J. Mol. Med. 2020, 45, 691–702. [Google Scholar] [CrossRef]
- Tu, W.; Qin, M.; Li, Y.; Wu, W.; Tong, X. Metformin regulates autophagy via LGMN to inhibit choriocarcinoma. Gene 2023, 853, 147090. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S. Autophagy-Based Diagnosis of Pregnancy Hypertension and Pre-Eclampsia. Am. J. Pathol. 2018, 188, 2457–2460. [Google Scholar] [CrossRef]
- Cao, B.; Macones, C.; Mysorekar, I.U. ATG16L1 governs placental infection risk and preterm birth in mice and women. JCI Insight 2016, 1, e86654. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, B.; Tian, J.; Teng, X.; Liu, T. Vital role of autophagy flux inhibition of placental trophoblast cells in pregnancy disorders induced by HEV infection. Emerg. Microbes Infect. 2023, 12, 2276336. [Google Scholar] [CrossRef]
- Lee, D.H.; Park, J.S.; Lee, Y.S.; Han, J.; Lee, D.K.; Kwon, S.W.; Han, D.H.; Lee, Y.H.; Bae, S.H. SQSTM1/p62 activates NFE2L2/NRF2 via ULK1-mediated autophagic KEAP1 degradation and protects mouse liver from lipotoxicity. Autophagy 2020, 16, 1949–1973. [Google Scholar] [CrossRef] [PubMed]
- Jiao, H.; Su, G.Q.; Dong, W.; Zhang, L.; Xie, W.; Yao, L.M.; Chen, P.; Wang, Z.X.; Liou, Y.C.; You, H. Chaperone-like protein p32 regulates ULK1 stability and autophagy. Cell Death Differ. 2015, 22, 1812–1823. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, C.; Mehra, D.; Song, D.; Mancebo, A.; Park, J.M.; Kim, D.H.; Puchner, E.M. ULK1 forms distinct oligomeric states and nanoscopic structures during autophagy initiation. Sci. Adv. 2023, 9, eadh4094. [Google Scholar] [CrossRef]
- Aslan, A.T.; Balaban, H.Y. Hepatitis E virus: Epidemiology, diagnosis, clinical manifestations, and treatment. World J. Gastroenterol. 2020, 26, 5543–5560. [Google Scholar] [CrossRef]
- Sayed, I.M.; Karam-Allah Ramadan, H.; Hafez, M.H.R.; Elkhawaga, A.A.; El-Mokhtar, M.A. Hepatitis E virus (HEV) open reading frame 2: Role in pathogenesis and diagnosis in HEV infections. Rev. Med. Virol. 2022, 32, e2401. [Google Scholar] [CrossRef]
- Khan, N.; Kakakhel, S.; Malik, A.; Nigar, K.; Akhtar, S.; Khan, A.A.; Khan, A. Genetic substructure and host-specific natural selection trend across vaccine-candidate ORF-2 capsid protein of hepatitis-E virus. J. Viral Hepat. 2024, 31, 524–534. [Google Scholar] [CrossRef]
- Maas, M.; Neumann-Haefelin, C. Deciphering the role of soluble ORF2 protein in virus-host interaction in HEV infection. Hepatology 2023, 78, 1692–1694. [Google Scholar] [CrossRef]
- Yao, Y.; Li, S.; Zhu, Y.; Xu, Y.; Hao, S.; Guo, S.; Feng, W.H. miR-204 suppresses porcine reproductive and respiratory syndrome virus (PRRSV) replication via inhibiting LC3B-mediated autophagy. Virol. Sin. 2023, 38, 690–698. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Ma, H.; Liu, H.; Ye, W.; Li, Z.; Cheng, L.; Zhang, L.; Lei, Y.; Shen, L.; Zhang, F. The Glycoprotein and Nucleocapsid Protein of Hantaviruses Manipulate Autophagy Flux to Restrain Host Innate Immune Responses. Cell Rep. 2019, 27, 2075–2091.e5. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhu, Y.; Ren, C.; Yang, S.; Tian, S.; Chen, H.; Jin, M.; Zhou, H. Influenza A virus protein PB1-F2 impairs innate immunity by inducing mitophagy. Autophagy 2021, 17, 496–511. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Wang, Y.; Yin, L.; Liang, W.; Zhang, J.; Ma, C.; Zhang, Y.; Liu, B.; Wang, J.; Zhao, W.; et al. The nonstructural protein 1 of respiratory syncytial virus hijacks host mitophagy as a novel mitophagy receptor to evade the type I IFN response in HEp-2 cells. mBio 2023, 14, e01480-23. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Chen, Y.; Yang, Y.; Bai, Q.; Tian, X.; Zhou, C.; Lu, X.; Liu, T. Hepatitis E ORF2 Blocks Trophoblast Autophagy to Induce Miscarriage via LC3B Binding Rather than PI3K/Akt/mTOR Pathway Suppression. Microorganisms 2026, 14, 181. https://doi.org/10.3390/microorganisms14010181
Chen Y, Yang Y, Bai Q, Tian X, Zhou C, Lu X, Liu T. Hepatitis E ORF2 Blocks Trophoblast Autophagy to Induce Miscarriage via LC3B Binding Rather than PI3K/Akt/mTOR Pathway Suppression. Microorganisms. 2026; 14(1):181. https://doi.org/10.3390/microorganisms14010181
Chicago/Turabian StyleChen, Yinzhu, Yifei Yang, Qianyu Bai, Xinyuan Tian, Chaoyu Zhou, Xuancheng Lu, and Tianlong Liu. 2026. "Hepatitis E ORF2 Blocks Trophoblast Autophagy to Induce Miscarriage via LC3B Binding Rather than PI3K/Akt/mTOR Pathway Suppression" Microorganisms 14, no. 1: 181. https://doi.org/10.3390/microorganisms14010181
APA StyleChen, Y., Yang, Y., Bai, Q., Tian, X., Zhou, C., Lu, X., & Liu, T. (2026). Hepatitis E ORF2 Blocks Trophoblast Autophagy to Induce Miscarriage via LC3B Binding Rather than PI3K/Akt/mTOR Pathway Suppression. Microorganisms, 14(1), 181. https://doi.org/10.3390/microorganisms14010181






