Role of NaCl and Glutamine on Biofilm Production from Pseudomonas aeruginosa
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteria Strain, Media, and Growth Conditions
2.2. Antimicrobial Resistance Profiles of P. aeruginosa Strains
2.3. Bacterial Growth Assessment
2.4. Pyoverdine Assay
2.5. Phenazine Assay
2.6. Biofilm Assay
2.7. RNA Isolation
2.8. Reverse Transcription PCR (RT-PCR)
2.9. Primer Design and Quantitative Real-Time PCR
2.10. Statistical Analysis
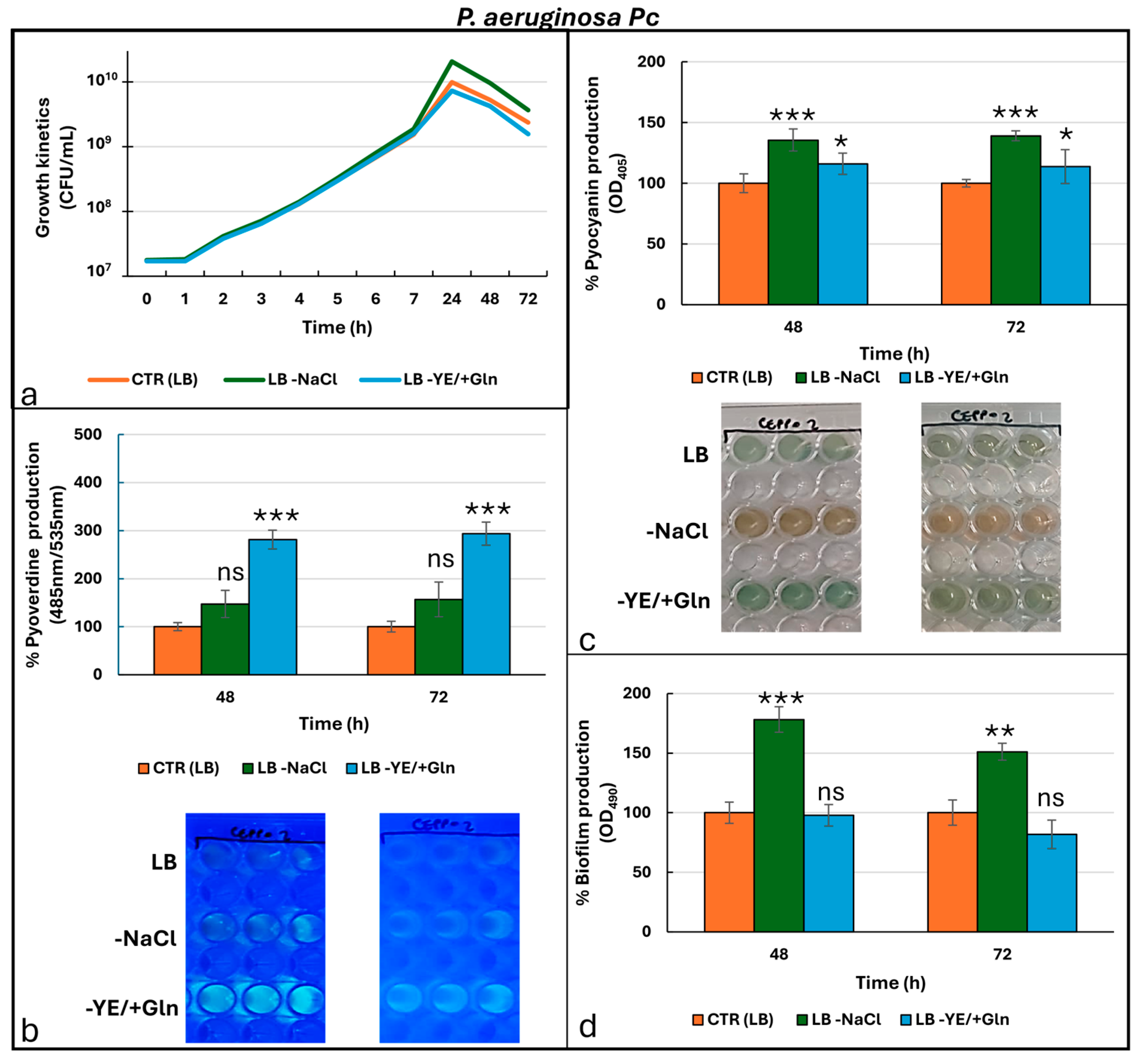
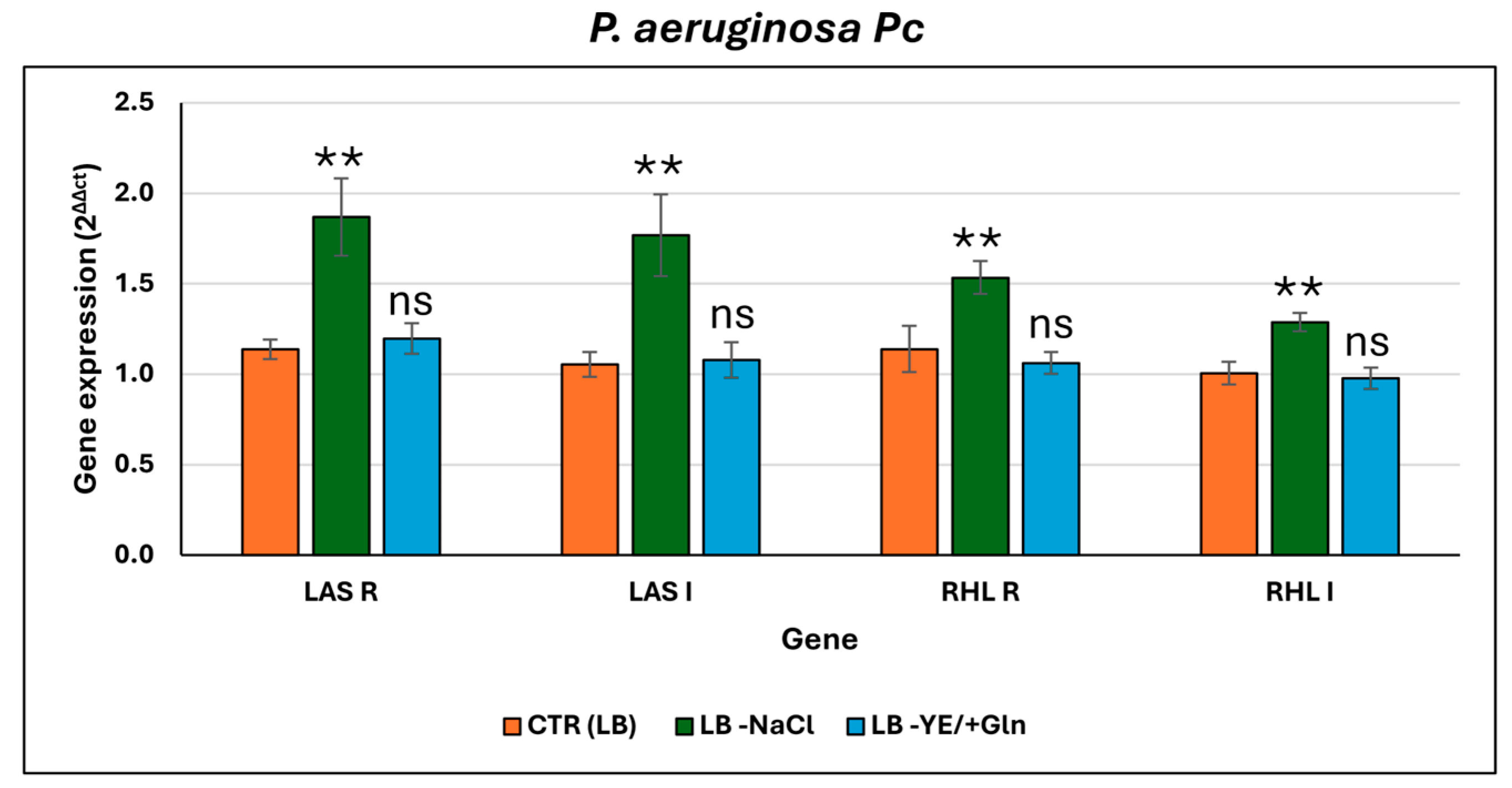
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fongang, H.; Mbaveng, A.T.; Kuete, V. Chapter One—Global Burden of Bacterial Infections and Drug Resistance. In Advances in Botanical Research; Kuete, V., Ed.; African Flora to Fight Bacterial Resistance, Part I: Standards for the Activity of Plant-Derived Products; Academic Press: Cambridge, MA, USA, 2023; Volume 106, pp. 1–20. [Google Scholar]
- La Rosa, R.; Johansen, H.K.; Molin, S. Persistent Bacterial Infections, Antibiotic Treatment Failure, and Microbial Adaptive Evolution. Antibiotics 2022, 11, 419. [Google Scholar] [CrossRef] [PubMed]
- Elfadadny, A.; Ragab, R.F.; AlHarbi, M.; Badshah, F.; Ibáñez-Arancibia, E.; Farag, A.; Hendawy, A.O.; De los Ríos-Escalante, P.R.; Aboubakr, M.; Zakai, S.A.; et al. Antimicrobial Resistance of Pseudomonas aeruginosa: Navigating Clinical Impacts, Current Resistance Trends, and Innovations in Breaking Therapies. Front. Microbiol. 2024, 15, 1374466. [Google Scholar] [CrossRef]
- Yin, R.; Cheng, J.; Wang, J.; Li, P.; Lin, J. Treatment of Pseudomonas aeruginosa Infectious Biofilms: Challenges and Strategies. Front. Microbiol. 2022, 13, 955286. [Google Scholar] [CrossRef]
- Zhen, X.; Lundborg, C.S.; Sun, X.; Hu, X.; Dong, H. Economic Burden of Antibiotic Resistance in ESKAPE Organisms: A Systematic Review. Antimicrob. Resist. Infect. Control 2019, 8, 137. [Google Scholar] [CrossRef]
- Di Fermo, P.; Diban, F.; Ancarani, E.; Yu, K.; D’Arcangelo, S.; D’Ercole, S.; Di Lodovico, S.; Di Giulio, M.; Cellini, L. New Commercial Wipes Inhibit the Dispersion and Adhesion of Staphylococcus Aureus and Pseudomonas aeruginosa Biofilms. J. Appl. Microbiol. 2024, 135, lxae234. [Google Scholar] [CrossRef] [PubMed]
- Labovská, S. Pseudomonas aeruginosa as a Cause of Nosocomial Infections. In Pseudomonas aeruginosa—Biofilm Formation, Infections and Treatments; IntechOpen: London, UK, 2021; ISBN 978-1-83968-648-1. [Google Scholar]
- Horcajada, J.P.; Montero, M.; Oliver, A.; Sorlí, L.; Luque, S.; Gómez-Zorrilla, S.; Benito, N.; Grau, S. Epidemiology and Treatment of Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa Infections. Clin. Microbiol. Rev. 2019, 32, e00031-19. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, S.; Hayes, D.; Wozniak, D.J. Cystic Fibrosis and Pseudomonas aeruginosa: The Host-Microbe Interface. Clin. Microbiol. Rev. 2019, 32, e00138-18. [Google Scholar] [CrossRef]
- Bhagirath, A.Y.; Li, Y.; Somayajula, D.; Dadashi, M.; Badr, S.; Duan, K. Cystic Fibrosis Lung Environment and Pseudomonas aeruginosa Infection. BMC Pulm. Med. 2016, 16, 174. [Google Scholar] [CrossRef]
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H.A. Pseudomonas aeruginosa Biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef]
- Vetrivel, A.; Ramasamy, M.; Vetrivel, P.; Natchimuthu, S.; Arunachalam, S.; Kim, G.-S.; Murugesan, R. Pseudomonas aeruginosa Biofilm Formation and Its Control. Biologics 2021, 1, 312–336. [Google Scholar] [CrossRef]
- Behzadi, P.; Gajdács, M.; Pallós, P.; Ónodi, B.; Stájer, A.; Matusovits, D.; Kárpáti, K.; Burián, K.; Battah, B.; Ferrari, M.; et al. Relationship between Biofilm-Formation, Phenotypic Virulence Factors and Antibiotic Resistance in Environmental Pseudomonas aeruginosa. Pathogens 2022, 11, 1015. [Google Scholar] [CrossRef]
- Kim, S.; Li, X.-H.; Hwang, H.-J.; Lee, J.-H. Thermoregulation of Pseudomonas aeruginosa Biofilm Formation. Appl. Environ. Microbiol. 2020, 86, e01584-20. [Google Scholar] [CrossRef]
- Li, X.; Gu, N.; Huang, T.Y.; Zhong, F.; Peng, G. Pseudomonas aeruginosa: A Typical Biofilm Forming Pathogen and an Emerging but Underestimated Pathogen in Food Processing. Front. Microbiol. 2023, 13, 1114199. [Google Scholar] [CrossRef]
- Sahoo, K.; Meshram, S. Biofilm Formation in Chronic Infections: A Comprehensive Review of Pathogenesis, Clinical Implications, and Novel Therapeutic Approaches. Cureus 2024, 16, e70629. [Google Scholar] [CrossRef] [PubMed]
- De Plano, L.M.; Caratozzolo, M.; Conoci, S.; Guglielmino, S.P.P.; Franco, D. Impact of Nutrient Starvation on Biofilm Formation in Pseudomonas aeruginosa: An Analysis of Growth, Adhesion, and Spatial Distribution. Antibiotics 2024, 13, 987. [Google Scholar] [CrossRef]
- Horak, R.D.; Ciemniecki, J.A.; Newman, D.K. Bioenergetic Suppression by Redox-Active Metabolites Promotes Antibiotic Tolerance in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2024, 121, e2406555121. [Google Scholar] [CrossRef] [PubMed]
- Tuon, F.F.; Dantas, L.R.; Suss, P.H.; Tasca Ribeiro, V.S. Pathogenesis of the Pseudomonas aeruginosa Biofilm: A Review. Pathogens 2022, 11, 300. [Google Scholar] [CrossRef]
- Cianciulli Sesso, A.; Resch, A.; Moll, I.; Bläsi, U.; Sonnleitner, E. The FinO/ProQ-like Protein PA2582 Impacts Antimicrobial Resistance in Pseudomonas aeruginosa. Front. Microbiol. 2024, 15, 1422742. [Google Scholar] [CrossRef]
- da Cruz Nizer, W.S.; Inkovskiy, V.; Versey, Z.; Strempel, N.; Cassol, E.; Overhage, J. Oxidative Stress Response in Pseudomonas aeruginosa. Pathogens 2021, 10, 1187. [Google Scholar] [CrossRef]
- Fernández, L.; Hancock, R.E.W. Adaptive and Mutational Resistance: Role of Porins and Efflux Pumps in Drug Resistance. Clin. Microbiol. Rev. 2012, 25, 661–681. [Google Scholar] [CrossRef]
- Zhao, A.; Sun, J.; Liu, Y. Understanding Bacterial Biofilms: From Definition to Treatment Strategies. Front. Cell. Infect. Microbiol. 2023, 13, 1137947. [Google Scholar] [CrossRef] [PubMed]
- Lundgren, B.R.; Shoytush, J.M.; Scheel, R.A.; Sain, S.; Sarwar, Z.; Nomura, C.T. Utilization of L-Glutamate as a Preferred or Sole Nutrient in Pseudomonas aeruginosa PAO1 Depends on Genes Encoding for the Enhancer-Binding Protein AauR, the Sigma Factor RpoN and the Transporter Complex AatJQMP. BMC Microbiol. 2021, 21, 83. [Google Scholar] [CrossRef]
- Delgado-Nungaray, J.A.; Figueroa-Yáñez, L.J.; Reynaga-Delgado, E.; García-Ramírez, M.A.; Aguilar-Corona, K.E.; Gonzalez-Reynoso, O. Influence of Amino Acids on Quorum Sensing-Related Pathways in Pseudomonas aeruginosa PAO1: Insights from the GEM iJD1249. Metabolites 2025, 15, 236. [Google Scholar] [CrossRef]
- Zhao, X.-L.; Chen, Z.-G.; Yang, T.-C.; Jiang, M.; Wang, J.; Cheng, Z.-X.; Yang, M.-J.; Zhu, J.-X.; Zhang, T.-T.; Li, H.; et al. Glutamine Promotes Antibiotic Uptake to Kill Multidrug-Resistant Uropathogenic Bacteria. Sci. Transl. Med. 2021, 13, eabj0716. [Google Scholar] [CrossRef]
- Hassanov, T.; Karunker, I.; Steinberg, N.; Erez, A.; Kolodkin-Gal, I. Novel Antibiofilm Chemotherapies Target Nitrogen from Glutamate and Glutamine. Sci. Rep. 2018, 8, 7097. [Google Scholar] [CrossRef]
- Sathe, N.; Beech, P.; Croft, L.; Suphioglu, C.; Kapat, A.; Athan, E. Pseudomonas aeruginosa: Infections and Novel Approaches to Treatment “Knowing the Enemy” the Threat of Pseudomonas aeruginosa and Exploring Novel Approaches to Treatment. Infect. Med. 2023, 2, 178–194. [Google Scholar] [CrossRef]
- Pai, L.; Patil, S.; Liu, S.; Wen, F. A Growing Battlefield in the War against Biofilm-Induced Antimicrobial Resistance: Insights from Reviews on Antibiotic Resistance. Front. Cell. Infect. Microbiol. 2023, 13, 1327069. [Google Scholar] [CrossRef] [PubMed]
- Mahamadou Harouna, B.; Bounou Issoufa, B.; Benkortbi, O.; Amrane, A. New Alternative of Enhancing Hospital Hygiene Facing Pseudomonas aeruginosa Drug Resistance Impact of Hypertonic Saline Solutions on the Behavior of P. aeruginosa. ACS Omega 2019, 4, 475–482. [Google Scholar] [CrossRef]
- Michon, A.-L.; Jumas-Bilak, E.; Chiron, R.; Lamy, B.; Marchandin, H. Advances toward the Elucidation of Hypertonic Saline Effects on Pseudomonas aeruginosa from Cystic Fibrosis Patients. PLoS ONE 2014, 9, e90164. [Google Scholar] [CrossRef] [PubMed]
- Camilios-Neto, D.; Nascimento, R.R.D.; Ratko, J.; Caldas Pan, N.; Casagrande, R.; Verri, W.A.; Vignoli, J.A. Pseudomonas aeruginosa: A Bacterial Platform for Biopharmaceutical Production. Future Pharmacol. 2024, 4, 892–918. [Google Scholar] [CrossRef]
- Leinweber, A.; Laffont, C.; Lardi, M.; Eberl, L.; Pessi, G.; Kümmerli, R. RNA-Seq Reveals That Pseudomonas aeruginosa Mounts Growth Medium-Dependent Competitive Responses When Sensing Diffusible Cues from Burkholderia Cenocepacia. Commun. Biol. 2024, 7, 995. [Google Scholar] [CrossRef] [PubMed]
- Fekete-Kertész, I.; Berkl, Z.; Buda, K.; Fenyvesi, É.; Szente, L.; Molnár, M. Quorum Quenching Effect of Cyclodextrins on the Pyocyanin and Pyoverdine Production of Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2024, 108, 271. [Google Scholar] [CrossRef]
- Zanni, E.; Bruni, E.; Chandraiahgari, C.R.; De Bellis, G.; Santangelo, M.G.; Leone, M.; Bregnocchi, A.; Mancini, P.; Sarto, M.S.; Uccelletti, D. Evaluation of the Antibacterial Power and Biocompatibility of Zinc Oxide Nanorods Decorated Graphene Nanoplatelets: New Perspectives for Antibiodeteriorative Approaches. J. Nanobiotechnol. 2017, 15, 57. [Google Scholar] [CrossRef]
- Coffey, B.M.; Anderson, G.G. Biofilm Formation in the 96-Well Microtiter Plate. In Pseudomonas Methods and Protocols; Filloux, A., Ramos, J.-L., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2014; pp. 631–641. ISBN 978-1-4939-0473-0. [Google Scholar]
- ImProm-IITM Reverse Transcription System. Available online: https://ita.promega.com/en/products/pcr/rt-pcr/improm_ii-reverse-transcription-system/ (accessed on 4 September 2025).
- SsoAdvanced Universal SYBR Green Supermix|Bio-Rad. Available online: https://www.bio-rad.com/it-it/product/ssoadvanced-universal-sybr-green-supermix?ID=MH5H1EE8Z (accessed on 4 September 2025).
- Tenover, F.C.; Nicolau, D.P.; Gill, C.M. Carbapenemase-Producing Pseudomonas aeruginosa—An Emerging Challenge. Emerg. Microbes Infect. 2022, 11, 811–814. [Google Scholar] [CrossRef]
- Theodorakis, N.; Feretzakis, G.; Hitas, C.; Kreouzi, M.; Kalantzi, S.; Spyridaki, A.; Boufeas, I.Z.; Sakagianni, A.; Paxinou, E.; Verykios, V.S.; et al. Antibiotic Resistance in the Elderly: Mechanisms, Risk Factors, and Solutions. Microorganisms 2024, 12, 1978. [Google Scholar] [CrossRef]
- Karlowsky, J.A.; Wise, M.G.; Hackel, M.A.; Six, D.A.; Uehara, T.; Daigle, D.M.; Pevear, D.C.; Moeck, G.; Sahm, D.F. Cefepime–Taniborbactam Activity against Antimicrobial-Resistant Clinical Isolates of Enterobacterales and Pseudomonas aeruginosa: GEARS Global Surveillance Programme 2018–22. J. Antimicrob. Chemother. 2024, 79, 3116–3131. [Google Scholar] [CrossRef]
- Alonso-García, I.; Vázquez-Ucha, J.C.; Martínez-Guitián, M.; Lasarte-Monterrubio, C.; Rodríguez-Pallares, S.; Camacho-Zamora, P.; Rumbo-Feal, S.; Aja-Macaya, P.; González-Pinto, L.; Outeda-García, M.; et al. Interplay between OXA-10 β-Lactamase Production and Low Outer-Membrane Permeability in Carbapenem Resistance in Enterobacterales. Antibiotics 2023, 12, 999. [Google Scholar] [CrossRef]
- Zakhour, J.; Sharara, S.L.; Hindy, J.-R.; Haddad, S.F.; Kanj, S.S. Antimicrobial Treatment of Pseudomonas aeruginosa Severe Sepsis. Antibiotics 2022, 11, 1432. [Google Scholar] [CrossRef]
- Pachori, P.; Gothalwal, R.; Gandhi, P. Emergence of Antibiotic Resistance Pseudomonas aeruginosa in Intensive Care Unit; a Critical Review. Genes Dis. 2019, 6, 109. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, S.; Bhatia, S.; Singh, S.; Franco, F., Jr. Growing Emergence of Drug-Resistant Pseudomonas aeruginosa and Attenuation of Its Virulence Using Quorum Sensing Inhibitors: A Critical Review. Iran. J. Basic Med. Sci. 2021, 24, 699. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Kirienko, D.R.; Webster, P.; Fisher, A.L.; Kirienko, N.V. Pyoverdine, a Siderophore from Pseudomonas aeruginosa, Translocates into C. Elegans, Removes Iron, and Activates a Distinct Host Response. Virulence 2018, 9, 804–817. [Google Scholar] [CrossRef] [PubMed]
- Meirelles, L.A.; Newman, D.K. Both Toxic and Beneficial Effects of Pyocyanin Contribute to the Lifecycle of Pseudomonas aeruginosa. Mol. Microbiol. 2018, 110, 995–1010. [Google Scholar] [CrossRef] [PubMed]
- Mould, D.L.; Stevanovic, M.; Ashare, A.; Schultz, D.; Hogan, D.A. Metabolic Basis for the Evolution of a Common Pathogenic Pseudomonas aeruginosa Variant. eLife 2022, 11, e76555. [Google Scholar] [CrossRef] [PubMed]
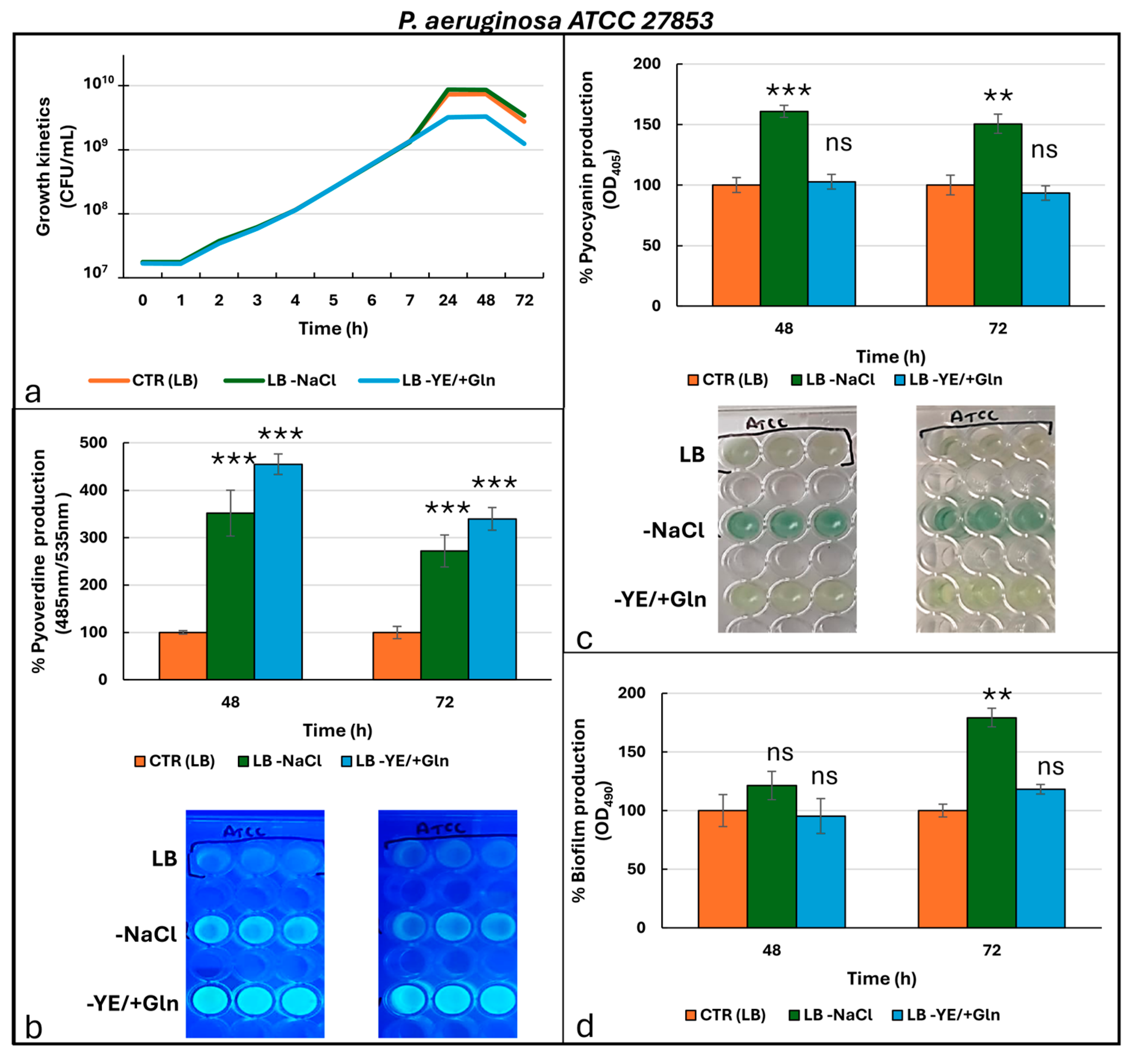
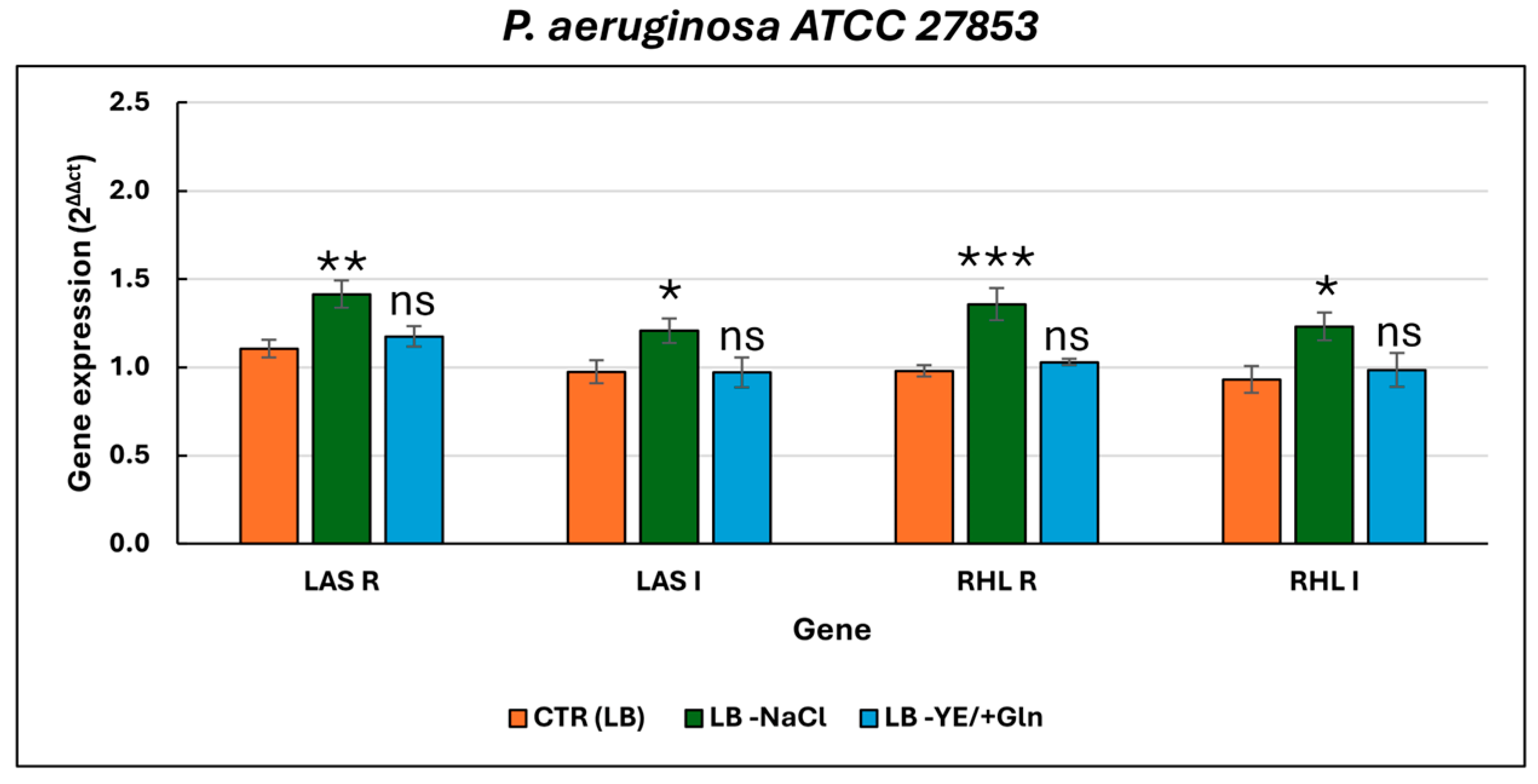
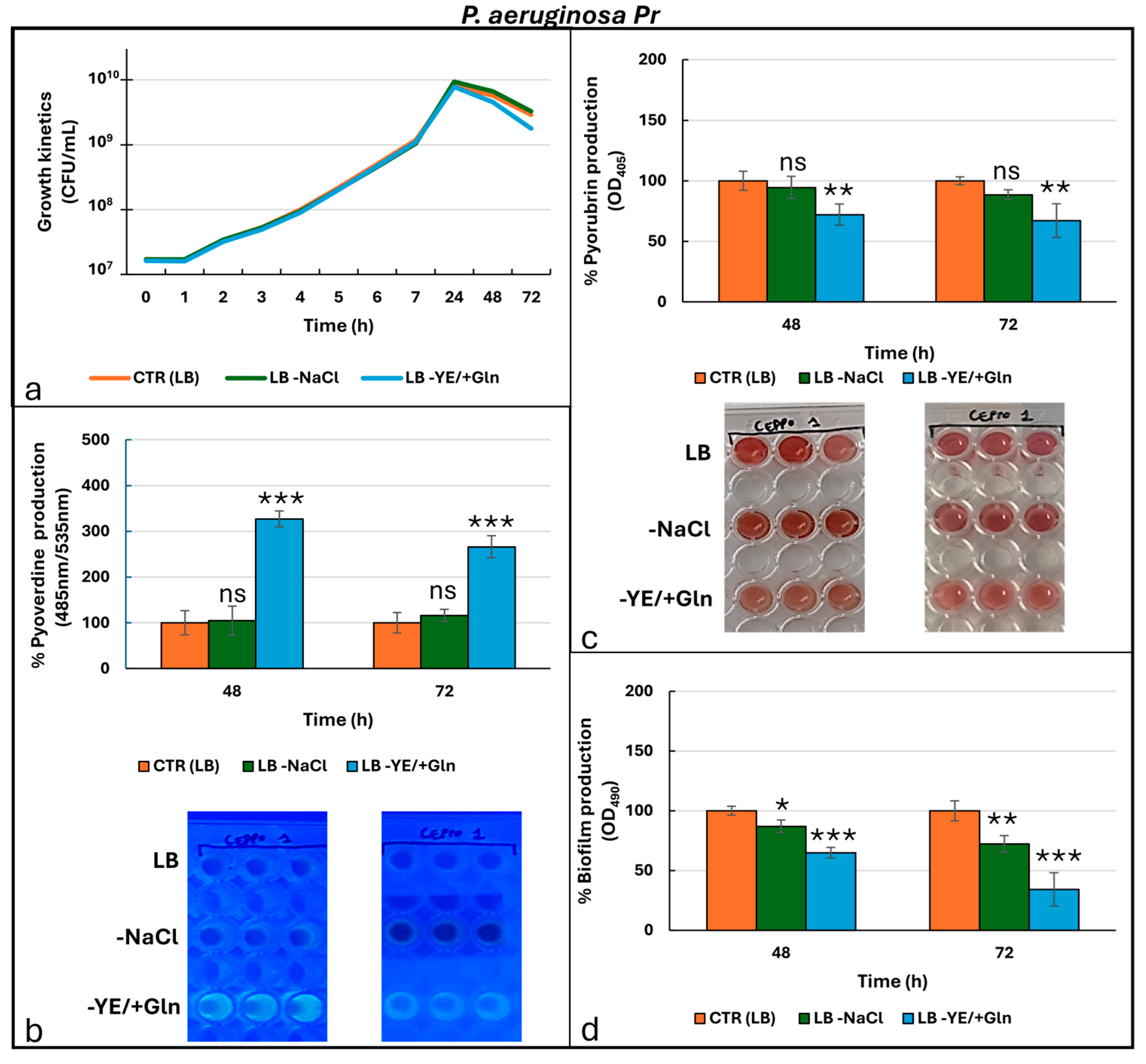
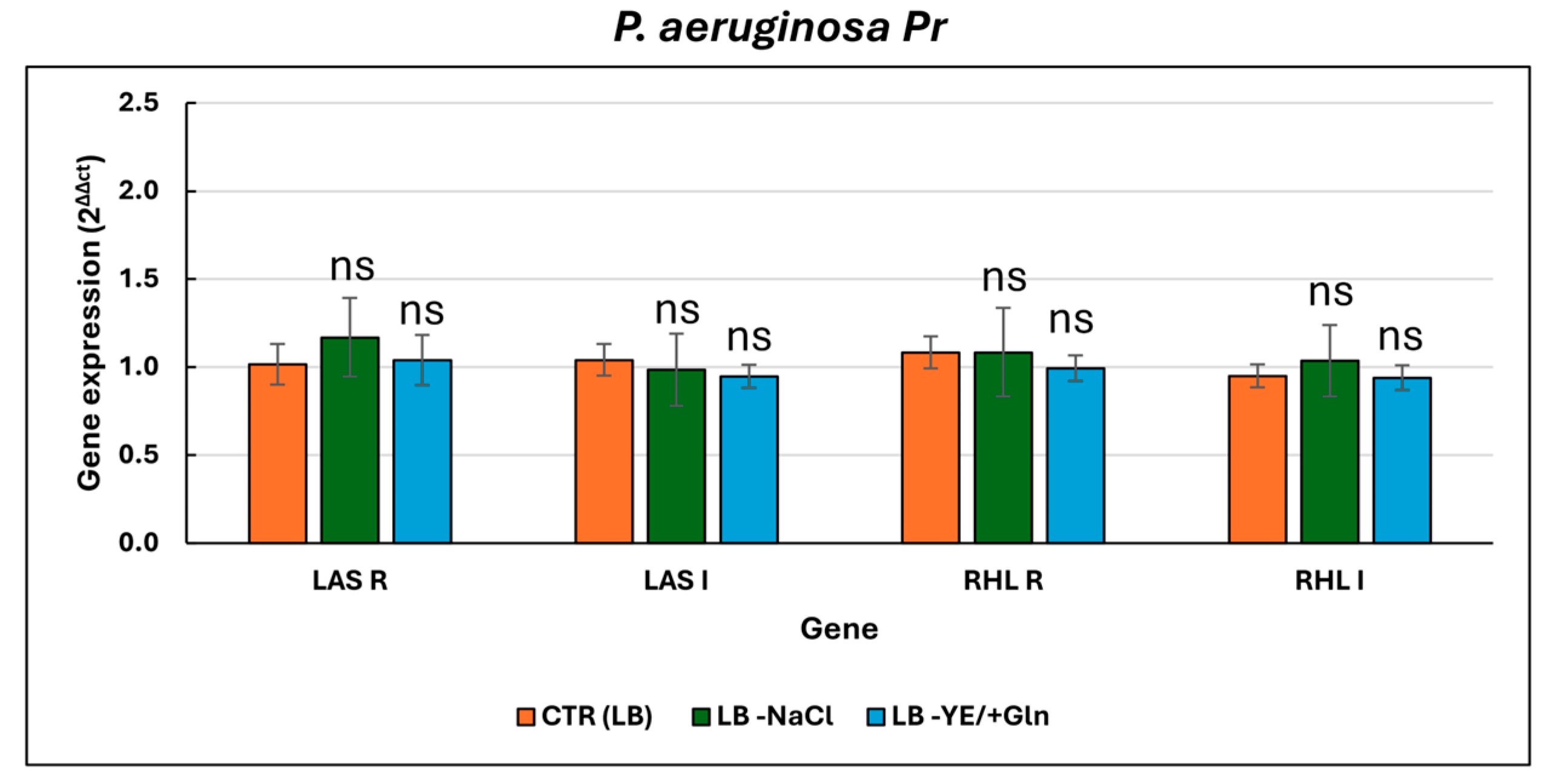
| ANTIBIOTICS | P. aeruginosa ATCC 27853 | P. aeruginosa Pr (Pyorubrin Producer) | P. aeruginosa Pc (Pyocyanin Producer) | |||
|---|---|---|---|---|---|---|
| MIC | Interpretation | MIC | Interpretation | MIC | Interpretation | |
| Amikacin (AMK) | 4 | S | 2 | S | 4 | S |
| Cefepime (FEP) | 0.5 | I | 8 | I | 16 | R |
| Ceftazidime (CAZ) | 2 | I | 2 | I | 32 | R |
| Ceftazidime/Avibactam (CAZ/AVI) | 2 | S | 8 | S | 2 | S |
| Ceftazidime/Tazobactam (CAZ/TZP) | 1 | S | 1 | S | 1 | S |
| Ciprofloxacin (CIP) | 0.5 | I | 0.5 | I | 0.12 | I |
| Colistin (COL) | 2 | S | 2 | S | ≤0.5 | S |
| Imipenem (IPM) | 2 | I | >8 | R | 2 | I |
| Meropem (MEM) | 0.5 | S | >8 | R | 0.5 | S |
| Piperacillina/Tazobactam (PIP/TZP) | ≤4 | I | 32 | R | >64 | R |
| Cell Viability (CFU/mL) | Time (h) | Cultural Condition | P. aeruginosa ATCC27853 | P. aeruginosa Pr | P. aeruginosa Pc |
| 24 | CTR (LB) | 6.4 × 109 ± 3 | 8.3 × 109 ± 0.9 | 1.3 × 1010 ± 0.7 | |
| 48 | CTR (LB) | 6.5 × 109 ± 2.9 | 5.6 × 109 ± 1 | 6.3 × 109 ± 2.7 | |
| 72 | CTR (LB) | 2.4 × 109 ± 1.1 | 2.7 × 109 ± 0.8 | 2.5 × 109 ± 1 | |
| 24 | LB -NaCl | 6.3 × 109 ± 2.7 | 8.2 × 109 ± 0.7 | 1.3 × 1010 ± 0.7 | |
| 48 | LB -NaCl | 2.4 × 109 ± 0.3 | 2.7 × 109 ± 1.2 | 2.5 × 109 ± 1.1 | |
| 72 | LB -NaCl | 6.4 × 109 ± 1.2 | 5.8 × 109 ± 0.7 | 6.3 × 109 ± 2.8 | |
| 24 | LB -YE/+Gln | 6.5 × 109 ± 1.1 | 8.5 × 109 ± 0.8 | 1.2 × 1010 ± 1.1 | |
| 48 | LB -YE/+Gln | 6.4 × 109 ± 2.7 | 5.6 × 109 ± 1.1 | 6.5 × 109 ± 2.9 | |
| 72 | LB -YE/+Gln | 2.5 × 109 ± 1.1 | 2.6 × 109 ± 0.8 | 2.5 × 109 ± 1.1 | |
| Pyoverdine (485 nm/535 nm) | Time (h) | Cultural condition | P. aeruginosa ATCC27853 | P. aeruginosa Pr | P. aeruginosa Pc |
| 48 h | CTR (LB) | 4.60 × 103 ± 0.16 | 1.42 × 103 ± 0.38 | 2.13 × 103 ± 0.18 | |
| 72 h | CTR (LB) | 7.28 × 103 ± 0.94 | 7.71 × 102 ± 1.73 | 1.57 × 103 ± 0.17 | |
| 48 h | LB -NaCl | 7.28 × 103 ± 2.23 | 7.71 × 102 ± 4.57 | 1.57 × 103 ± 0.6 | |
| 72 h | LB -NaCl | 1.62 × 104 ± 0.24 | 1.49 × 103 ± 0.1 | 3.14 × 103 ± 0.57 | |
| 48 h | LB -YE/+Gln | 2.09 × 104 ± 0.1 | 4.65 × 103 ± 0.25 | 6.00 × 103 ± 0.42 | |
| 72 h | LB -YE/+Gln | 2.47 × 104 ± 0.17 | 2.05 × 103 ± 0.19 | 4.60 × 103 ± 0.38 | |
| Phenazine (OD405) | Time (h) | Cultural condition | P. aeruginosa ATCC27853 | P. aeruginosa Pr | P. aeruginosa Pc |
| 48 h | CTR (LB) | 0.33 ± 0.02 | 0.53 ± 0.04 | 0.39 ± 0.02 | |
| 72 h | CTR (LB) | 0.32 ± 0.03 | 0.52 ± 0.02 | 0.38 ± 0.01 | |
| 48 h | LB -NaCl | 0.32 ± 0.03 | 0.52 ± 0.05 | 0.38 ± 0.02 | |
| 72 h | LB -NaCl | 0.53 ± 0.04 | 0.50 ± 0.02 | 0.53 ± 0.02 | |
| 48 h | LB -YE/+Gln | 0.34 ± 0.02 | 0.38 ± 0.03 | 0.45 ± 0.03 | |
| 72 h | LB -YE/+Gln | 0.30 ± 0.02 | 0.35 ± 0.05 | 0.43 ± 0.03 | |
| Biofilm (OD490) | Time (h) | Cultural condition | P. aeruginosa ATCC27853 | P. aeruginosa Pr | P. aeruginosa Pc |
| 48 h | CTR (LB) | 0.60 ± 0.09 | 0.49 ± 0.02 | 0.44 ± 0.04 | |
| 72 h | CTR (LB) | 0.47 ± 0.03 | 0.67 ± 0.06 | 0.54 ± 0.06 | |
| 48 h | LB -NaCl | 0.47 ± 0.09 | 0.67 ± 0.02 | 0.54 ± 0.09 | |
| 72 h | LB -NaCl | 0.73 ± 0.07 | 0.42 ± 0.04 | 0.78 ± 0.06 | |
| 48 h | LB -YE/+Gln | 0.57 ± 0.08 | 0.31 ± 0.02 | 0.43 ± 0.04 | |
| 72 h | LB -YE/+Gln | 0.55 ± 0.02 | 0.23 ± 0.03 | 0.44 ± 0.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Plano, L.M.; Iaconis, A.; Papasergi, S.; Mediati, F.; Caruso, D.; Guglielmino, S.P.P.; Franco, D. Role of NaCl and Glutamine on Biofilm Production from Pseudomonas aeruginosa. Microorganisms 2025, 13, 2198. https://doi.org/10.3390/microorganisms13092198
De Plano LM, Iaconis A, Papasergi S, Mediati F, Caruso D, Guglielmino SPP, Franco D. Role of NaCl and Glutamine on Biofilm Production from Pseudomonas aeruginosa. Microorganisms. 2025; 13(9):2198. https://doi.org/10.3390/microorganisms13092198
Chicago/Turabian StyleDe Plano, Laura Maria, Antonella Iaconis, Salvatore Papasergi, Francesco Mediati, Daniele Caruso, Salvatore Pietro Paolo Guglielmino, and Domenico Franco. 2025. "Role of NaCl and Glutamine on Biofilm Production from Pseudomonas aeruginosa" Microorganisms 13, no. 9: 2198. https://doi.org/10.3390/microorganisms13092198
APA StyleDe Plano, L. M., Iaconis, A., Papasergi, S., Mediati, F., Caruso, D., Guglielmino, S. P. P., & Franco, D. (2025). Role of NaCl and Glutamine on Biofilm Production from Pseudomonas aeruginosa. Microorganisms, 13(9), 2198. https://doi.org/10.3390/microorganisms13092198









