Characterization of Drug Resistance Mutations in Mycobacterium tuberculosis Isolates from Moroccan Patients Using Deeplex Targeted Next-Generation Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Institutional Review Board Statement
2.2. Informed Consent Statement
2.3. Samples Processing and LJ Culture
2.4. Phenotypic Drug Susceptibility Testing (pDST):
2.5. Genomic DNA Extraction
2.6. Targeted Sequencing and Library Preparation
2.7. Bioinformatic Analysis and Interpretation of Drug Resistance
3. Results
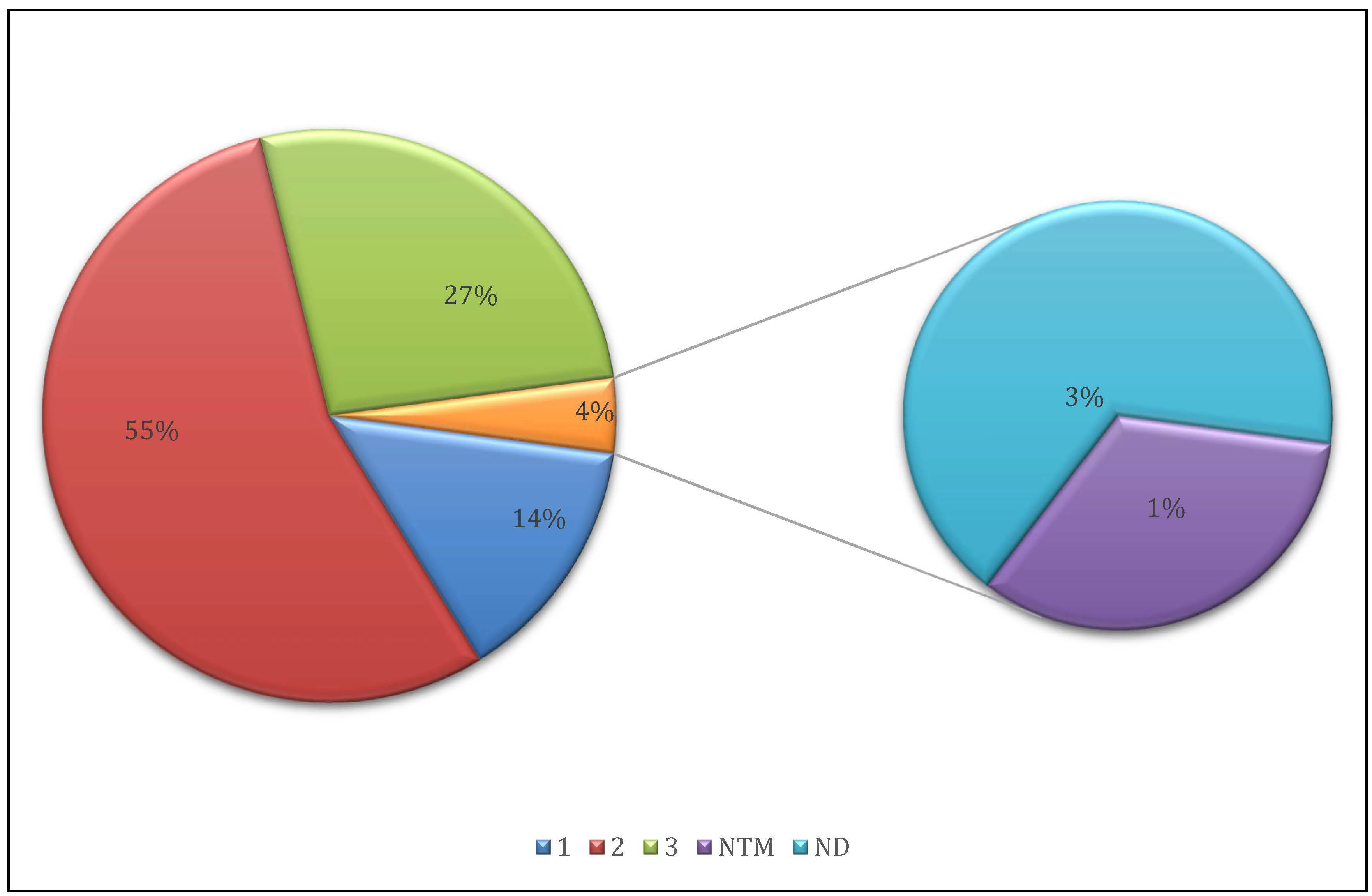
3.1. Sequencing Quality and Sample Selection
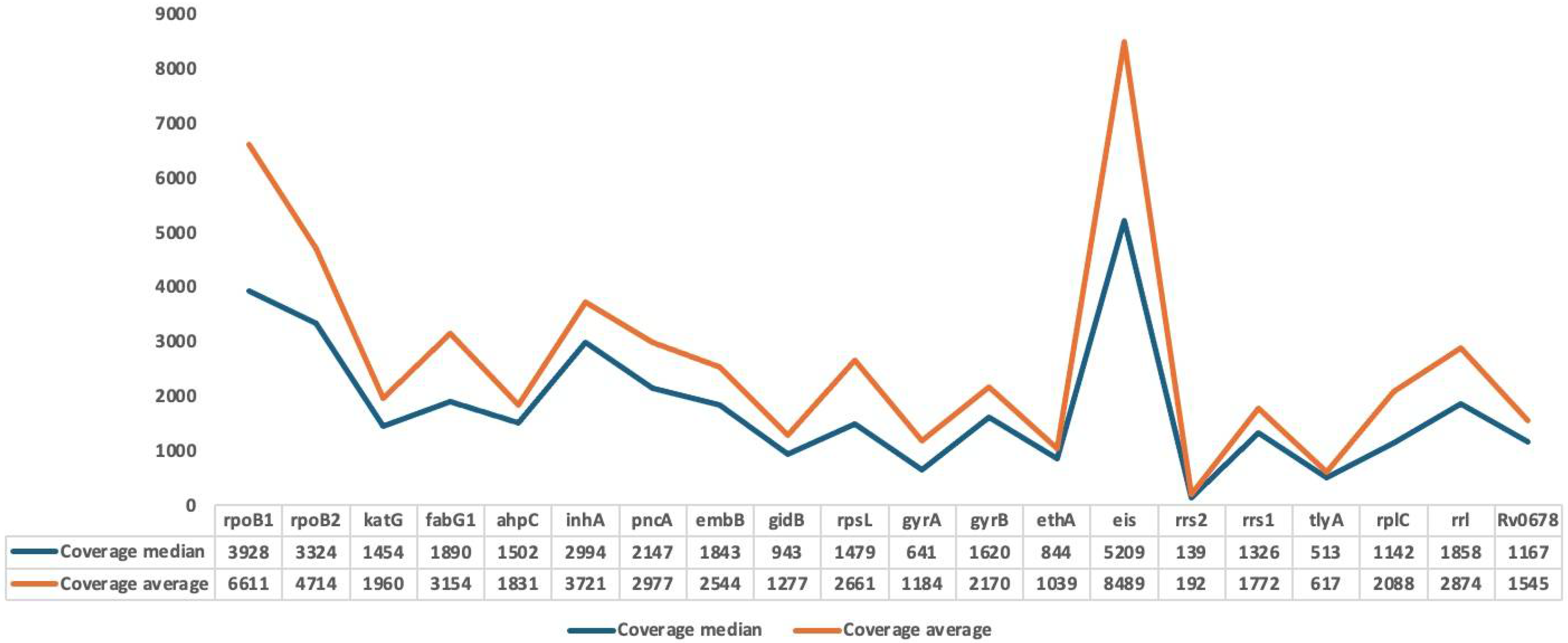
3.2. Mapping and Coverage Statistics
3.3. Drug Resistance Profiles
3.4. Resistance by Drug Class
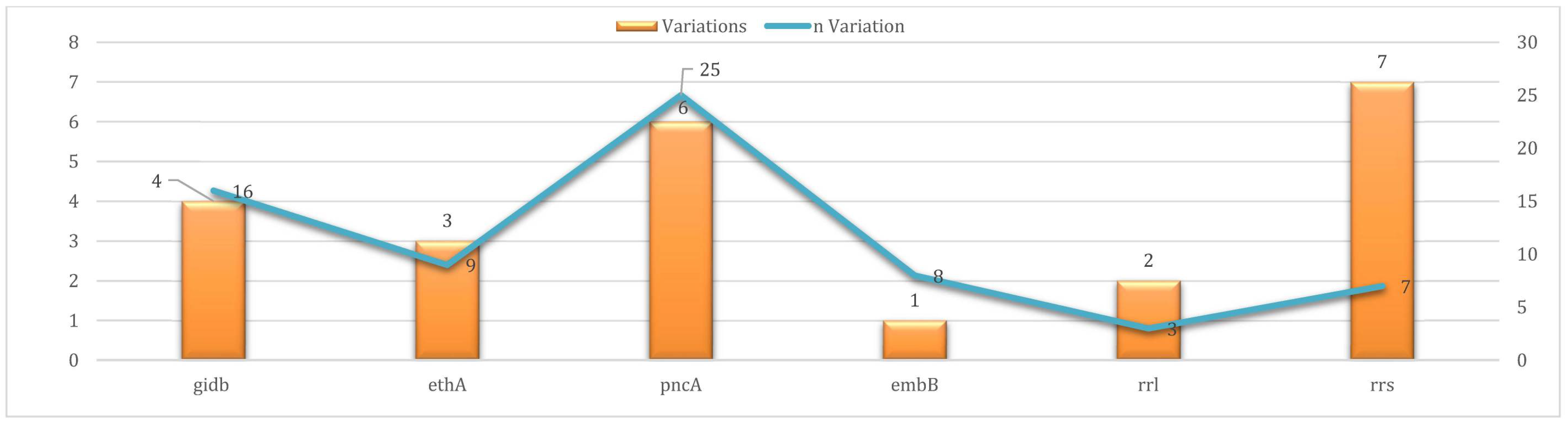
3.5. Uncharacterized Variants (UVs)
3.6. Concordance Between Phenotypic and Genotypic Testing
3.7. Statistical Analysis of Variant Distribution
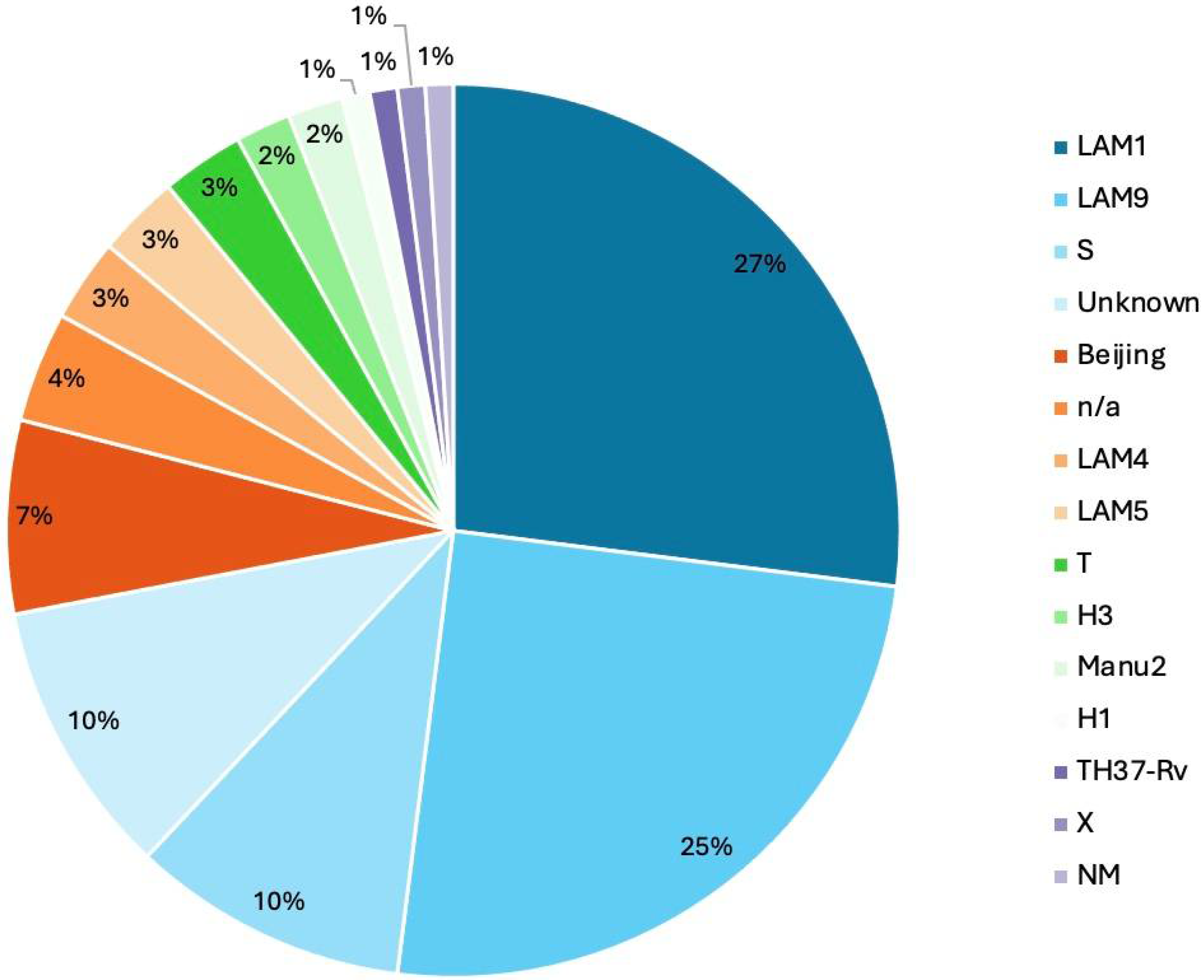
3.8. Phylogenetic Analysis and Lineage Distribution
3.9. Clade–Lineage Correlation in Drug-Resistant Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TB | Tuberculosis |
| WHO | World Health Organization |
| NGS | Next-Generation Sequencing |
| HRM | High-Resolution Melting Curve |
| LJ | Löwenstein–Jensen |
| RRDR | Rifampicin Resistance-Determining Region |
| MTBC | Mycobacterium Tuberculosis Complex |
| XDR-TB | Extensively Drug-Resistant TB |
| Pre- XD | Pre-extensively Drug-Resistant TB |
| MDR-TB | Multidrug-Resistant |
| RIF | Rifampicin |
| AA | Amino Acid |
| INH | Isoniazid |
| PZA | Pyrazinamide |
| EMB | Ethambutol |
| SM | Streptomycin |
| FQ | Fluoroquinolones |
| ETH | Ethionamide |
| PTH | Prothionamide |
| KAN | Kanamycin |
| AMI | Amikacin |
| CAP | Capreomycin |
| LIN | Linezolid |
| BDQ | Bedaquiline |
| CF | Clofazimine |
| NTM | Non-tuberculous Mycobacteria |
References
- World Health Organization. Drug-Resistant TB. Available online: https://www.who.int/teams/global-programme-on-tuberculosis-and-lung-health/tb-reports/global-tuberculosis-report-2024/tb-disease-burden/ (accessed on 25 June 2025).
- Centers for Disease Control and Prevention. (n.d.); Clinical Overview of Drug-Resistant Tuberculosis Disease. Available online: https://www.cdc.gov/tb/hcp/clinical-overview/drug-resistant-tuberculosis-disease.html/ (accessed on 25 June 2025).
- Cho, E.H.; Bae, H.K.; Kang, S.K.; Lee, E.H. Detection of isoniazid and rifampicin resistance by sequencing of katG, inhA, and rpoB genes in Korea. Korean J. Lab. Med. 2009, 29, 455–460. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, W.-L.; Chen, H.-Y.; Kuo, Y.-M.; Jou, R. Performance assessment of the GenoType MTBDRplus test and DNA sequencing in detection of multidrug-resistant Mycobacterium tuberculosis. J. Clin. Microbiol. 2009, 47, 2520–2524. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.; Kalantri, S.; Flores, L.; Pai, M. A commercial line probe assay for the rapid detection of rifampicin resistance in Mycobacterium tuberculosis: A systematic review and meta-analysis. BMC Infect. Dis. 2005, 5, 62. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Morris, S.L.; Langone, J.J.; Bockstahler, L.E. Microarray and allele specific PCR detection of point mutations in Mycobacterium tuberculosis genes associated with drug resistance. J. Microbiol. Methods 2005, 63, 318–330. [Google Scholar] [CrossRef]
- World Health Organization. The Use of Molecular Line Probe Assay for the Detection of Resistance to Isoniazid and Rifampicin. Available online: https://www.who.int/publications/i/item/9789241511261/ (accessed on 31 July 2025).
- World Health Organization. Xpert MTB/RIF Assay for the Diagnosis of Pulmonary and Extrapulmonary TB in Adults and Children. Available online: https://www.who.int/publications/i/item/9789241506335/ (accessed on 25 June 2025).
- World Health Organization. Next-Generation Xpert® MTB/RIF Ultra Assay Recommended by WHO. Available online: https://www.who.int/publications/i/item/9789240040090// (accessed on 25 June 2025).
- Chaudhary, R.; Bhatta, S.; Singh, A.; Pradhan, M.; Srivastava, B.; Singh, Y.I.; Sah, R.; Fathah, Z.; Mehta, R.; Rabaan, A.A.; et al. Diagnostic performance of GeneXpert MTB/RIF assay compared to conventional Mycobacterium tuberculosis culture for diagnosis of pulmonary and extrapulmonary tuberculosis, Nepal. Narra J. 2021, 1, e33. [Google Scholar] [CrossRef]
- Terzi, H.; Aydemir, Ö.; Karakeçe, E.; Demiray, T.; Köroğlu, M. Evaluation of the Xpert MTB/RIF test performance in the diagnosis of suspected Mycobacterium tuberculosis in pulmonary and extrapulmonary clinical specimens. Mediterr. J. Infect. Microbes Antimicrob. 2024, 13, 13. [Google Scholar] [CrossRef]
- El Achkar, S.; Demanche, C.; Osman, M.; Rafei, R.; Ismail, M.B.; Gaudin, C.; Duthoy, S.; De Matos, F.; Yaacoub, H.; Pinçon, C.; et al. Zoonotic tuberculosis in humans assessed by next-generation sequencing: An 18-month nationwide study in Lebanon. Eur. Respir. J. 2020, 55, 1900513. [Google Scholar] [CrossRef]
- Tagliani, E.; Hassan, M.O.; Waberi, Y.; De Filippo, M.R.; Falzon, D.; Dean, A.; Zignol, M.; Supply, P.; Abdoulkader, M.A.; Hassangue, H.; et al. Culture and next-generation sequencing-based drug susceptibility testing unveil high levels of drug-resistant TB in Djibouti: Results from the first national survey. Sci. Rep. 2017, 7, 17672. [Google Scholar] [CrossRef]
- Jouet, A.; Gaudin, C.; Badalato, N.; Allix-Béguec, C.; Duthoy, S.; Ferré, A.; Diels, M.; Laurent, Y.; Contreras, S.; Feuerriegel, S.; et al. Deep amplicon sequencing for culture-free prediction of susceptibility or resistance to 13 anti-tuberculous drugs. Eur. Respir. J. 2021, 57, 2002338. [Google Scholar] [CrossRef]
- Cabibbe, A.M.; Spitaleri, A.; Battaglia, S.; Colman, R.E.; Suresh, A.; Uplekar, S.; Rodwell, T.C.; Cirillo, D.M. Application of Targeted Next-Generation Sequencing Assay on a Portable Sequencing Platform for Culture-Free Detection of Drug-Resistant Tuberculosis from Clinical Samples. J. Clin. Microbiol. 2020, 58, e00632-20. [Google Scholar] [CrossRef]
- Meehan, C.J.; Goig, G.A.; Kohl, T.A.; Verboven, L.; Dippenaar, A.; Ezewudo, M.; Farhat, M.R.; Guthrie, J.L.; Laukens, K.; Miotto, P.; et al. Whole-genome sequencing of Mycobacterium tuberculosis: Current standards and open issues. Nat. Rev. Microbiol. 2019, 17, 533–545. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. (n.d.); Next Generation Sequencing (NGS) Quality Initiative. Available online: https://www.cdc.gov/lab-quality/php/ngs-quality-initiative/index.html (accessed on 31 July 2025).
- Bentaleb, E.M.; El Messaoudi, M.D.; Abid, M.; Messaoudi, M.; Yetisen, A.K.; Sefrioui, H.; Amzazi, S.; Benhassou, H.A. Plasmid-based high-resolution melting analysis for accurate detection of rpoB mutations in Mycobacterium tuberculosis isolates from Moroccan patients. BMC Infect. Dis. 2017, 17, 548. [Google Scholar] [CrossRef]
- Solari, L.; Santos-Lazaro, D.; Puyen, Z.M. Mutations in Mycobacterium tuberculosis isolates with discordant results for drug-susceptibility testing in Peru. Int. J. Microbiol. 2020, 2020, 8253546. [Google Scholar] [CrossRef]
- Merker, M.; Egbe, N.F.; Ngangue, Y.R.; Vuchas, C.; Kohl, T.A.; Dreyer, V.; Kuaban, C.; Noeske, J.; Niemann, S.; Sander, M.S. Transmission patterns of rifampicin resistant Mycobacterium tuberculosis complex strains in Cameroon: A genomic epidemiological study. BMC Infect. Dis. 2021, 21, 891. [Google Scholar] [CrossRef]
- Sandgren, A.; Strong, M.; Muthukrishnan, P.; Weiner, B.K.; Church, G.M.; Murray, M.B. Tuberculosis drug resistance mutation database. PLoS Med. 2009, 6, e1000002. [Google Scholar] [CrossRef] [PubMed]
- Zimenkov, D.V.; Kulagina, E.V.; Antonova, O.V.; Zhuravlev, V.Y.; Gryadunov, D.A. Simultaneous drug resistance detection and genotyping of Mycobacterium tuberculosis using a low-density hydrogel microarray. J. Antimicrob. Chemother. 2016, 71, 1520–1531. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Catalogue of Mutations in Mycobacterium Tuberculosis Complex and Their Association with Drug Resistance (2nd ed.). Available online: https://www.who.int/publications/i/item/9789240082410 (accessed on 31 July 2025).
- Yadon, A.N.; Maharaj, K.; Adamson, J.H.; Lai, Y.-P.; Sacchettini, J.C.; Ioerger, T.R.; Rubin, E.J.; Pym, A.S. A comprehensive characterization of pncA polymorphisms that confer resistance to pyrazinamide. Nat. Commun. 2017, 8, 588. [Google Scholar] [CrossRef]
- Zignol, M.; Dean, A.S.; Alikhanova, N.; Andres, S.; Cabibbe, A.M.; Cirillo, D.M.; Dadu, A.; Dreyer, A.; Driesen, M.; Gilpin, C.; et al. Population-based resistance of Mycobacterium tuberculosis isolates to pyrazinamide and fluoroquinolones: Results from a multicountry surveillance project. Lancet Infect. Dis. 2016, 16, 1185–1192. [Google Scholar] [CrossRef]
- Ndawula, C.; Petra, N.P.; Wasswa, F.B.; Bazira, J. Prevalence and clinical implications of pyrazinamide resistance in newly diagnosed TB patients in Uganda. Infect. Drug Resist. 2025, 18, 1629–1635. [Google Scholar] [CrossRef]
- Khan, M.T.; Malik, S.I.; Ali, S.; Masood, N.; Nadeem, T.; Khan, A.S.; Afzal, M.T. Pyrazinamide resistance and mutations in pncA among isolates of Mycobacterium tuberculosis from Khyber Pakhtunkhwa, Pakistan. BMC Infect. Dis. 2019, 19, 116. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Chen, J.; Zhang, S.; Zhang, W.; Zhang, Y. Identification of novel mutations in LprG (rv1411c), rv0521, rv3630, rv0010c, ppsC, and cyp128 associated with pyrazinoic acid/pyrazinamide resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2018, 62, e00430-18. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Zhang, X.; Jiang, X.; Yuan, H.; Lee, J.S.; Barry, C.E., 3rd; Wang, H.; Zhang, W.; Zhang, Y. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science 2011, 333, 1630–1632. [Google Scholar] [CrossRef] [PubMed]
- Morlock, G.P.; Metchock, B.; Sikes, D.; Crawford, J.T.; Cooksey, R.C. ethA, inhA, and katG loci of ethionamide-resistant clinical Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 2003, 47, 3799–3805. [Google Scholar] [CrossRef]
- Wong, S.Y.; Lee, J.S.; Kwak, H.K.; Via, L.E.; Boshoff, H.I.M.; Barry, C.E., III. Mutations in gidB confer low-level streptomycin resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2011, 55, 2515–2522. [Google Scholar] [CrossRef]
- Zetola, N.M.; Shin, S.S.; Tumedi, K.A.; Moeti, K.; Ncube, R.; Nicol, M.; Collman, R.G.; Klausner, J.D.; Modongo, C.; Land, G.A. Mixed Mycobacterium tuberculosis complex infections and false-negative results for rifampin resistance by genexpert MTB/RIF are associated with poor clinical outcomes. J. Clin. Microbiol. 2014, 52, 2422–2429. [Google Scholar] [CrossRef]
- Cohen, T.; van Helden, P.D.; Wilson, D.; Colijn, C.; McLaughlin, M.M.; Abubakar, I.; Warren, R.M. Mixed-strain Mycobacterium tuberculosis infections and the implications for tuberculosis treatment and control. Clin. Microbiol. Rev. 2012, 25, 708–719. [Google Scholar] [CrossRef]
- Van Rie, A.; Victor, T.C.; Richardson, M.; Johnson, R.; Van Der Spuy, G.D.; Murray, E.J.; Beyers, N.; Van Pittius, N.C.G.; Van Helden, P.D.; Warren, R.M. Reinfection and Mixed Infection Cause Changing Mycobacterium tuberculosis Drug-Resistance Patterns. Am. J. Respir. Crit. Care Med. 2005, 172, 636–642. [Google Scholar] [CrossRef]
- Komakech, K.; Nakiyingi, L.; Fred, A.; Achan, B.; Joloba, M.; Kirenga, B.J.; Ssengooba, W. Effect of mixed Mycobacterium tuberculosis infection on rapid molecular diagnostics among patients starting MDR-TB treatment in Uganda. BMC Infect Dis. 2024, 24, 70. [Google Scholar] [CrossRef] [PubMed]
- Warren, R.M.; Victor, T.C.; Streicher, E.M.; Richardson, M.; van der Spuy, G.D.; Johnson, R.; Chihota, V.N.; Locht, C.; Supply, P.; van Helden, P.D.; et al. Clonal expansion of a globally disseminated lineage of Mycobacterium tuberculosis with low IS6110 copy numbers. J. Clin. Microbiol. 2004, 42, 5774–5782. [Google Scholar] [CrossRef]
- Thwaites, G.E.; Chau, T.T.H.; Stepniewska, K.; Phu, N.H.; Chuong, L.V.; Sinh, D.X.; White, N.J.; Parry, C.M.; Farrar, J.J. Diagnosis of adult tuberculous meningitis by use of clinical and laboratory features. Lancet 2002, 360, 1287–1292. [Google Scholar] [CrossRef]
- Ghosh, S.; Moonan, P.K.; Cowan, L.; Grant, J.; Kammerer, S.; Navin, T.R. Tuberculosis Genotyping Information Management System: Enhancing Tuberculosis Surveillance in the United States. Infect. Genet. Evol. 2012, 12, 782–788. [Google Scholar] [CrossRef] [PubMed]
- Lukoye, D.; Adatu, F.; Musisi, K.; Kasule, G.W.; Were, W.; Odeke, R.; Kalamya, J.N.; Awor, A.; Date, A.; Joloba, M.L.; et al. Anti-tuberculosis drug resistance among new and previously treated sputum smear-positive tuberculosis patients in Uganda: Results of the first national survey. PLoS ONE 2013, 8, e70763. [Google Scholar] [CrossRef]
- Nimmo, C.; Shaw, L.P.; Doyle, R.; Williams, R.; Brien, K.; Burgess, C.; Breuer, J.; Balloux, F.; Pym, A.S. Whole genome sequencing Mycobacterium tuberculosis directly from sputum identifies more genetic diversity than sequencing from culture. BMC Genom. 2019, 20, 389. [Google Scholar] [CrossRef]
- Cabibbe, A.M.; Walker, T.M.; Niemann, S.; Cirillo, D.M. Whole genome sequencing of Mycobacterium tuberculosis. Eur. Respir. J. 2018, 52, 1801163. [Google Scholar] [CrossRef]
- Lahlou, O.; Millet, J.; Chaoui, I.; Sabouni, R.; Filali-Maltouf, A.; Akrim, M.; El Mzibri, M.; Rastogi, N.; El Aouad, R.; Mokrousov, I. The genotypic population structure of Mycobacterium tuberculosis complex from Moroccan patients reveals a predominance of Euro-American lineages. PLoS ONE 2012, 7, e47113. [Google Scholar] [CrossRef] [PubMed]
- Chaoui, I.; Zozio, T.; Lahlou, O.; Sabouni, R.; Abid., M.; El Aouad, R.; Akrim, M.; Amzazi, S.; Rastogi, N.; El Mzibri, M.; et al. Contribution of spoligotyping and MIRU-VNTRs to characterize prevalent Mycobacterium tuberculosis genotypes infecting tuberculosis patients in Morocco. Infect. Genet. Evol. 2014, 21, 463–471. [Google Scholar] [CrossRef]
- Oudghiri, A.; Momen, G.; Aainouss, A.; Laglaoui, A.; El Messaoudi, M.D.; El Mzibri, M.; Chaoui, I. Genotypic diversity of multi- and pre-extensively drug-resistant Mycobacterium tuberculosis isolates from Morocco. PLoS ONE 2021, 16, e0253826. [Google Scholar] [CrossRef] [PubMed]
- WHO. WHO Consolidated Guidelines on Tuberculosis: Module 3: Diagnosis: Rapid Diagnostics for Tuberculosis Detection, 3rd ed. Available online: https://www.who.int/publications/i/item/9789240089488 (accessed on 19 July 2025).
- WHO. Use of Targeted Next-Generation Sequencing to Detect Drug-Resistant Tuberculosis: Rapid Communication, July 2023. Available online: https://www.who.int/publications/i/item/9789240076372 (accessed on 19 July 2025).

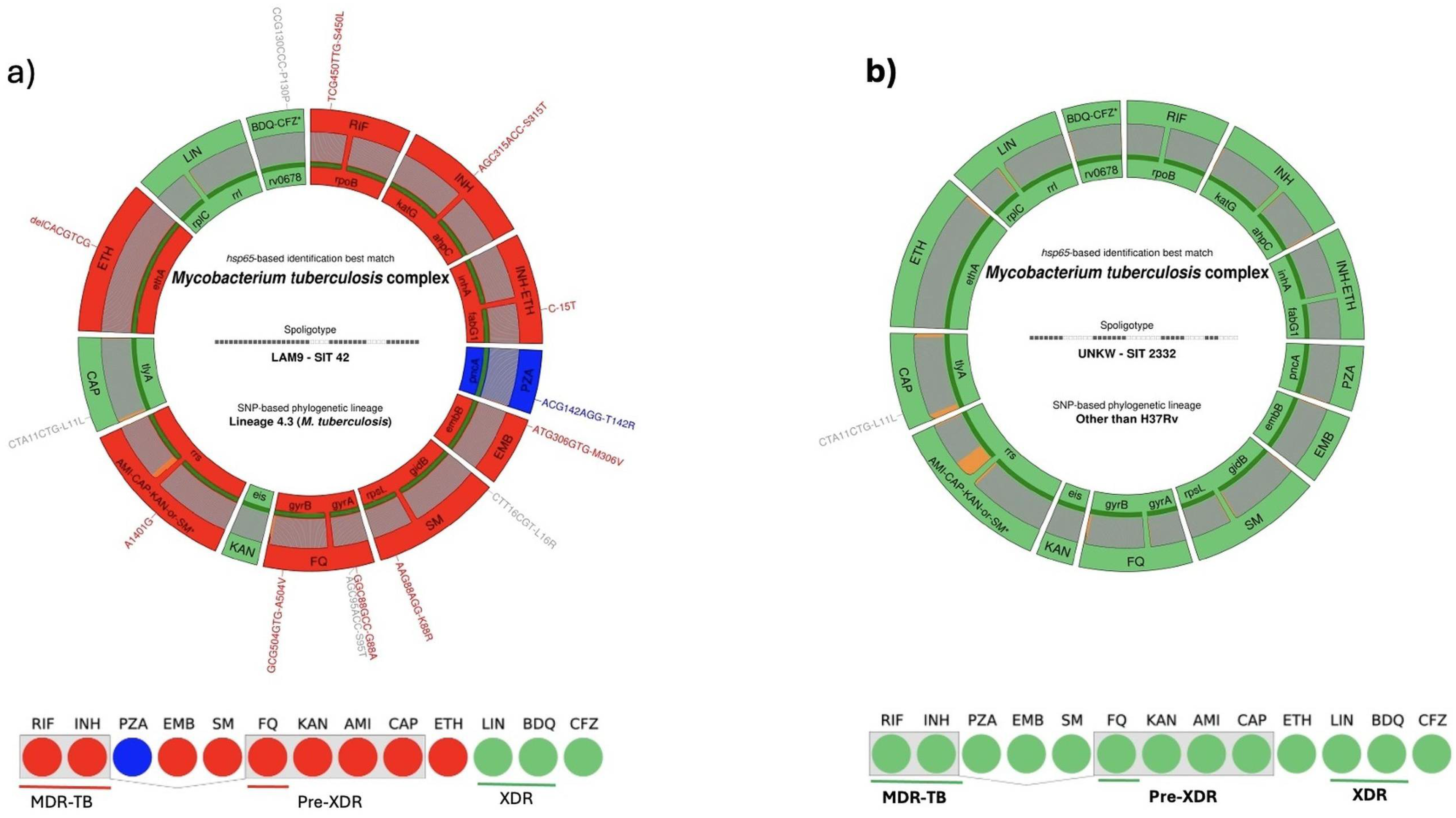
| Deeplex®-MycTB Assay | ||||
|---|---|---|---|---|
| Drug Type | Drug | Locus | Number of Samples with RVs | Number of Samples with UVs |
| FIRST-LINE | RIF | rpoB | 40 | 0 |
| INH | katG | 42 | 0 | |
| fabG1 | 16 | 0 | ||
| ahpC | 0 | 0 | ||
| inhA | 0 | 0 | ||
| PZA | pncA | 18 | 25 | |
| EMB | embB | 29 | 8 | |
| GROUP A/B (XDR) | FQ | gyrA | 14 | 0 |
| gyrB | 5 | 0 | ||
| LIN | rplC | 0 | 0 | |
| rrl | 0 | 3 | ||
| BDQ/CFZ | Rv0678 | 0 | 0 | |
| GROUP C | AMI | rrs | 11 | 0 |
| SM | gidB | 0 | 16 | |
| rpsL | 21 | 0 | ||
| Rrs | 11 | 1 | ||
| ETH | ethA | 32 | 9 | |
| inhA | 0 | 0 | ||
| fabG1 | 16 | 0 | ||
| OTHERS | KAN | Eis | 1 | 0 |
| KAN/CAP | rrs | 11 | 1 | |
| CAP | tlyA | 0 | 0 | |
| Gene | Genomic Position | Codon Change | AA Change | Number of Samples | Drug |
|---|---|---|---|---|---|
| embB | 4247429 | ATG306GTG | M306V | 10 | EMB |
| 4248002 | CAG497AAG | Q497K | 9 | ||
| 4247431 | ATG306GATT | M306I | 2 | ||
| 4247431 | ATG306ATA | M306I | 4 | ||
| 4247730 | GGC406GCC | G406A | 3 | ||
| 4247402 | TCG297GCG | S297A | 1 | ||
| gyrA | 7582 | GAC94GGC | D94G | 4 | FQ |
| 7570 | GCG90GTG | A90V | 3 | ||
| 7572 | TCG91CCG | S91P | 1 | ||
| 7581 | GAC94AAC | D94N | 1 | ||
| 7581 | GAC94CAC | D94H | 1 | ||
| 7564 | GGC88GCC | G88A | 3 | ||
| 7581 | GAC94TAC | D94Y | 2 | ||
| katG | 2155168 | AGC315ACC | S315T | 42 | INH |
| rpoB | 761155 | TCG450TTG | S450L | 31 | RIF |
| 761155 | TCG450TGG | S450W | 2 | ||
| 761140 | CAC445CTC | H445L | 2 | ||
| 761139 | CAC445TAC | H445Y | 5 | ||
| 760314 | GTC170TTC | V170F | 2 | ||
| 761140 | CAC445CGC | H445R | 3 | ||
| ethA | 4326930 | inserC | FrSh | 1 | ETH |
| 4326210 | delG | FrSh | 6 | ||
| 4326640-6 | delCACGTCG | FrSh | 15 | ||
| 4327367 | delT | FrSh | 10 | ||
| pncA | 2288852 | inserC | FsSh | 1 | PZA |
| 2289218 | GAC8GAG | D8E | 5 | ||
| 2289010 | delC | FrSh | 6 | ||
| 2289252 | A-11G | n/a | 2 | ||
| 2288869 | GTC125TTC | V125F | 2 | ||
| 2288853-65 | delACATCGACCTCAT | FrSh | 1 | ||
| 2288794 | inserC | FrSh | 1 | ||
| 2289057 | CCG62CTG | P62L | 1 | ||
| Rrs | 1472362 | C517T | n/a | 3 | AMI, KAN, SM |
| 1472359 | A514C | n/a | 3 | ||
| 1473246 | A1401G | n/a | 4 | ||
| rpsL | 781822 | AAG88AAG | K88R | 13 | SM |
| 781687 | AAG43AGG | K43R | 8 | SM | |
| fabG1 | 1673425 | C-15T | n/a | 16 | INH, EMB |
| gyrB | 6620 | GAC461AAC | D461N | 2 | FQ |
| 6750 | GCG504GTG | A504V | 1 | FQ | |
| 6738 | ACC500AAC | T500N | 2 | FQ | |
| Eis | 2715346 | C-14T | n/a | 1 | KAN |
| Gene | Genomic Position | Codon Change | AA Change | Number of Samples | Drug |
|---|---|---|---|---|---|
| gidB | 4408072 | CTA44CCA | L44P | 1 | SM |
| 4408105 | delC | FrSh | 2 | ||
| 4408137 | TAC22TAA | Y22Stop | 7 | ||
| 4407856 | inserC | FrSh | 6 | ||
| ethA | 4326663 | CAG271TAG | Q271Stop | 5 | ETH |
| 4327314 | CGC54AGC | R54S | 2 | ||
| 4326284 | CTG397CCG | L397P | 2 | ||
| pncA | 2288754 | CTG163GG | V163G | 3 | PZA |
| 2288817 | ACG142AGG | T142R | 14 | ||
| 2289097 | GAC49TAC | D49Y | 2 | ||
| 2289071 | CAC57CAA | H57Q | 2 | ||
| 2289027 | TGC72TAC | C72Y | 3 | ||
| 2288731 | GCG171ACG | A171T | 1 | ||
| embB | 4247399 | AAT296CAT | N296H | 8 | EMB |
| rrl | 1476007 | T2350G | n/a | 2 | AMI, KAN, SM |
| 1476251 | T2594C | n/a | 1 | ||
| rrs | 1473123 | A1278T | n/a | 1 | AMI, KAN, SM |
| 1472850 | delT | deletion | 1 | ||
| 1472857 | delA | deletion | 1 | ||
| 1472845 | inserC | Insertion | 1 | ||
| 1472860 | inserG | Insertion | 1 | ||
| 1473120 | inserT | Insertion | 1 | ||
| 1473396 | inserG | Insertion | 1 |
| Resistance | Gene(s) with Mutations | Number of Samples with Mutations (tNGS) | Phenotypic DST Result | Concordance |
|---|---|---|---|---|
| Rifampicin (RIF) | rpoB | 40 | All 40 resistant | 100% |
| Isoniazid (INH) | katG and/or fabG1 | 42 | All 42 resistant | 100% |
| Sample ID | INH | RIF | FQ | LIN BDQ/CFZ | MDR | Pre-XDR | Spoligotype | SNP Phylogenetic Lineage |
|---|---|---|---|---|---|---|---|---|
| Clade | Lineage | |||||||
| 1 | LAM1 | L4.3 | ||||||
| 2 | n/a | L4.3 | ||||||
| 5 | T1 | Other than H37Rv | ||||||
| 6 | n/a | Other than H37Rv | ||||||
| 8 | LAM9 | L4.3 | ||||||
| 11 | LAM9 | L4.3 | ||||||
| 12 | LAM9 | L4.3 | ||||||
| 16 | S | Other than H37Rv | ||||||
| 17 | Beijing | L2 | ||||||
| 18 | Manu2 | Other than H37Rv | ||||||
| 19 | LAM9 | L4.3 | ||||||
| 20 | T1 | Other than H37Rv | ||||||
| 21 | Unknown | L2 | ||||||
| 22 | LAM9 | L4.3 | ||||||
| 24 | T1 | L4.3 | ||||||
| 26 | T1 | L4.3 | ||||||
| 27 | T1 | Other than H37Rv | ||||||
| 29 | LAM9 | L4.3 | ||||||
| 33 | T1 | Other than H37Rv | ||||||
| 34 | T1 | L4.3 | ||||||
| 35 | T1 | L4.3 | ||||||
| 37 | LAM9 | L4.3 | ||||||
| 40 | LAM9 | L4.3 | ||||||
| 41 | LAM9 | L4.3 | ||||||
| 43 | H1 | Other than H37Rv | ||||||
| 45 | Beijing | L2 | ||||||
| 46 | S | Other than H37Rv | ||||||
| 47 | T1 | Other than H37Rv | ||||||
| 48 | S | Other than H37Rv | ||||||
| 52 | Manu2 | L2 | ||||||
| 53 | Beijing | L2 | ||||||
| 54 | S | Other than H37Rv | ||||||
| 55 | T1 | L4.3 | ||||||
| 56 | T1 | Other than H37Rv | ||||||
| 57 | Beijing | L2 | ||||||
| 58 | S | Other than H37Rv | ||||||
| 60 | n/a | Other than H37Rv | ||||||
| 62 | LAM9 | L4.3 | ||||||
| 63 | Beijing | L2 | ||||||
| 64 | LAM9 | L4.3 | ||||||
| 65 | LAM9 | L4.3 | ||||||
| 66 | LAM9 | L4.3 | ||||||
| 42 | 40 | 40 | 5 |
| Spoligotype Clade | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LAM1 | LAM4 | LAM5 | LAM9 | T1 | Beijing | Manu2 | H1 | H3 | S | X | TH37-Rv | n/a | Unknown | NTM | ||
| Lineage | L4.3 | 1 | 1 | 1 | 18 | 8 | * | * | * | * | * | * | * | 2 | 1 | * |
| Other than H37rv | * | * | * | * | 11 | * | 1 | 2 | 2 | 7 | 1 | * | 4 | 2 | * | |
| L2 | * | * | * | * | * | 5 | 1 | * | * | * | * | * | * | * | * | |
| Not detected | * | * | * | * | * | * | * | * | * | * | * | * | 1 | * | 1 | |
| No specific | * | * | * | * | * | * | * | * | * | * | * | 1 | * | * | * | |
| Total | 1 | 1 | 1 | 18 | 19 | 5 | 1 | 2 | 2 | 7 | 1 | 1 | 7 | 3 | 1 | |
| Spoligotype Clade | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LAM1 | LAM9 | T1 | Beijing | Manu2 | S | H1 | n/a | Unknown | ||
| Lineage | % | 2.5 | 30 | 28 | 10 | 5 | 12.5 | 2.5 | 5 | 2.5 |
| L4.3 | 1 | 12 | 5 | * | * | * | * | 1 | 1 | |
| Other than H37rv | * | * | 6 | * | 1 | 5 | 1 | 1 | * | |
| L2 | * | * | * | 4 | 1 | * | * | * | * | |
| Total | 1 | 12 | 11 | 4 | 2 | 5 | 1 | 2 | 1 | |
| Mixed Infection | Number of Samples | % |
|---|---|---|
| Yes | 11 | 16.2 |
| No | 57 | 83.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laatri, S.; El Kassimi, S.; Bentaleb, E.M.; El Messaoudi, M.D.; Irobi, J.; Belkadi, B.; Filali-Maltouf, A.; Ait Benhassou, H. Characterization of Drug Resistance Mutations in Mycobacterium tuberculosis Isolates from Moroccan Patients Using Deeplex Targeted Next-Generation Sequencing. Microorganisms 2025, 13, 2163. https://doi.org/10.3390/microorganisms13092163
Laatri S, El Kassimi S, Bentaleb EM, El Messaoudi MD, Irobi J, Belkadi B, Filali-Maltouf A, Ait Benhassou H. Characterization of Drug Resistance Mutations in Mycobacterium tuberculosis Isolates from Moroccan Patients Using Deeplex Targeted Next-Generation Sequencing. Microorganisms. 2025; 13(9):2163. https://doi.org/10.3390/microorganisms13092163
Chicago/Turabian StyleLaatri, Said, Safaa El Kassimi, El Mehdi Bentaleb, My Driss El Messaoudi, Joy Irobi, Bouchra Belkadi, Abdelkarim Filali-Maltouf, and Hassan Ait Benhassou. 2025. "Characterization of Drug Resistance Mutations in Mycobacterium tuberculosis Isolates from Moroccan Patients Using Deeplex Targeted Next-Generation Sequencing" Microorganisms 13, no. 9: 2163. https://doi.org/10.3390/microorganisms13092163
APA StyleLaatri, S., El Kassimi, S., Bentaleb, E. M., El Messaoudi, M. D., Irobi, J., Belkadi, B., Filali-Maltouf, A., & Ait Benhassou, H. (2025). Characterization of Drug Resistance Mutations in Mycobacterium tuberculosis Isolates from Moroccan Patients Using Deeplex Targeted Next-Generation Sequencing. Microorganisms, 13(9), 2163. https://doi.org/10.3390/microorganisms13092163








