Comparative Analysis of Compound Probiotics, Seasonal Variation, and Age on Gut Microbial Composition and Function in Endangered Forest Musk Deer
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Processing
2.2. Extraction of Musk Deer Fecal DNA, Amplification of 16S rRNA Gene, and Purification
2.3. Construction of Sequencing Libraries and Sequencing on the Illumina-MiSeq Platform
2.4. Sequencing Data Processing and Bioinformatics Analysis
2.5. Statistical Analysis
3. Results
3.1. Analysis of Gut Microbiota Composition and α-Diversity in Captive Forest Musk Deer Under Different Influencing Factors
3.2. Analysis of Beta Diversity in Gut Microbial Composition of Musk Deer Under Different Influencing Factors
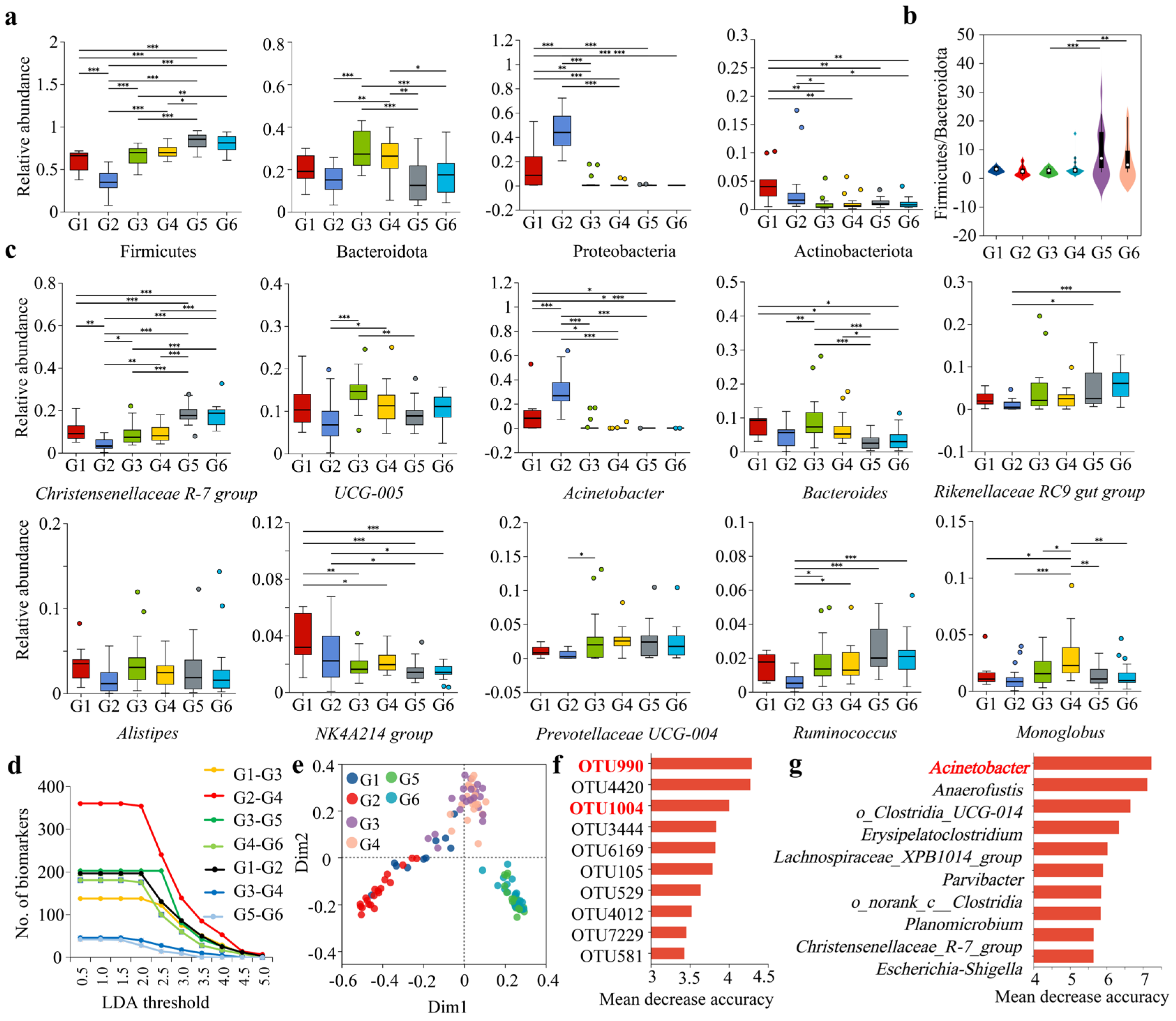
3.3. Analysis of Intergroup Differences in Dominant Gut Microbiota of Forest Musk Deer Under Different Influencing Factors
3.4. Combined Analysis of the Impact of Different Influencing Factors on the Composition and Diversity of the Gut Microbiota in Forest Musk Deer
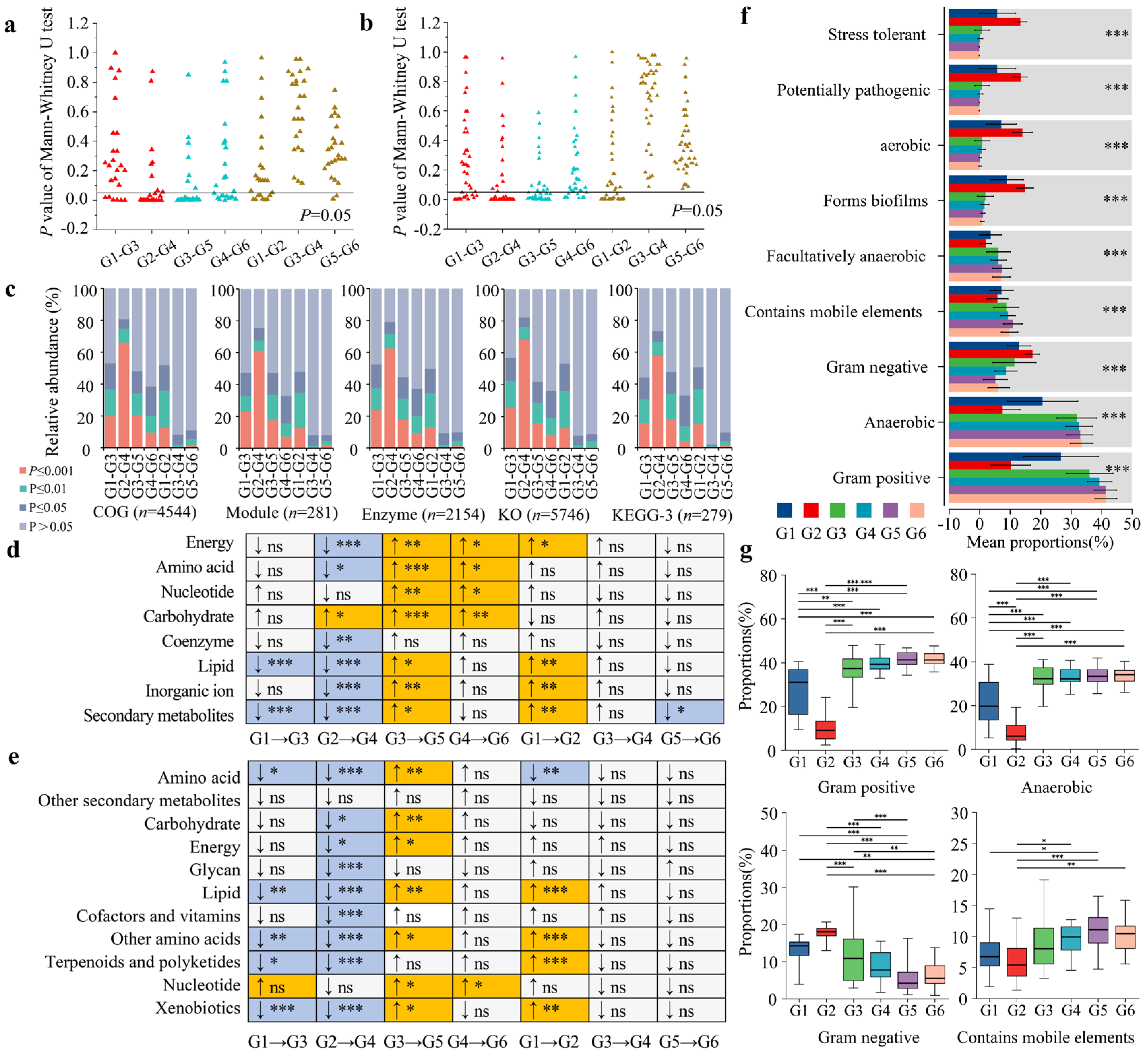
3.5. Analysis of Functional Differences in the Gut Microbiota of Forest Musk Deer Across Different Influencing Factors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, Q.; Meng, X.; Lin, X.; Feng, Z. Conservation status and causes of decline of musk deer (Moschus spp.) in China. Biol. Conserv. 2003, 109, 333–342. [Google Scholar] [CrossRef]
- Dhami, B.; Chhetri, N.B.; Neupane, B.; Adhikari, B.; Bashyal, B.; Maraseni, T.; Thapamagar, T.; Dhakal, Y.; Tripathi, A.; Koju, N.P. Predicting the current habitat refugia of Himalayan Musk deer (Moschus chrysogaster) across Nepal. Ecol. Evol. 2024, 4, e10949. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.S.; Liu, Y.R.; Lv, Y.; Duan, J.A.; Chen, S.Z.; Sun, J.; Song, Z.X.; Wu, X.M.; Liu, L. Quality markers of animal medicinal materials: Correlative analysis of musk reveals distinct metabolic changes induced by multiple factors. Phytomedicine 2018, 44, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Cai, R.; Jin, X.; Shafer, A.B.A.; Hu, X.; Yang, S.; Li, Y.; Qi, L.; Liu, S.; Hu, D. Blood transcriptomics of captive forest musk deer (Moschus berezovskii) and possible associations with the immune response to abscesses. Sci. Rep. 2018, 8, 599. [Google Scholar] [CrossRef]
- Wu, J.; Wang, W. The Musk Deer of China; The China Forestry Publishing House: Beijing, China, 2006. [Google Scholar]
- National Forestry Administration. China Key Terrestrial Wildlife Resources Survey; China Forestry Publishing House: Beijing, China, 2009. [Google Scholar]
- Li, Y.; Hu, X.; Yang, S.; Zhou, J.; Qi, L.; Sun, X.; Fan, M.; Xu, S.; Cha, M.; Zhang, M.; et al. Comparison between the fecal bacterial microbiota of healthy and diarrheic captive musk deer. Front. Microbiol. 2018, 9, 300. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hu, X.; Yang, S.; Zhou, J.; Zhang, T.; Qi, L.; Sun, X.; Fan, M.; Xu, S.; Cha, M.; et al. Comparative Analysis of the Gut Microbiota Composition between Captive and Wild Forest Musk Deer. Front. Microbiol. 2017, 8, 1705. [Google Scholar] [CrossRef]
- Zhao, K.L.; Liu, Y.; Zhang, X.Y.; Palahati, P.; Wang, H.N.; Yue, B.S. Detection and characterization of antibiotic-resistance genes in Arcanobacterium pyogenes strains from abscesses of forest musk deer. J. Med. Microbiol. 2011, 60, 1820–1826. [Google Scholar] [CrossRef]
- Zhang, L.; Jie, H.; Xiao, Y.; Zhou, C.; Lyu, W.; Bai, W. Genomic identification and expression analysis of the cathelicidin gene family of the forest musk deer. Animals 2019, 9, 481. [Google Scholar] [CrossRef]
- Ding, Y.; Wu, Q.; Hu, Y.; Wang, X.; Nie, Y.; Wu, X.; Wei, F. Advances and prospects of gut microbiome in wild mammals. Acta Theriol. Sin. 2017, 37, 399–406. [Google Scholar]
- Liu, X.; Yu, J.; Huan, Z.; Xu, M.; Song, T.; Yang, R.; Zhu, W.; Jiang, J. Comparing the gut microbiota of Sichuan golden monkeys across multiple captive and wild settings: Roles of anthropogenic activities and host factors. BMC Genom. 2024, 25, 148. [Google Scholar] [CrossRef]
- Aguirre, M.; Venema, K. The use of fecal samples for studying human obesity. Eur. J. Epidemiol. 2015, 30, 1067–1069. [Google Scholar] [CrossRef]
- Rounge, T.B.; Meisal, R.; Nordby, J.I.; Ambur, O.H.; de Lange, T.; Hoff, G. Evaluating gut microbiota profiles from archived fecal samples. BMC Gastroenterol. 2018, 18, 171. [Google Scholar] [CrossRef]
- Knutie, S.A.; Gotanda, K.M. A Non-invasive Method to Collect Fecal Samples from Wild Birds for Microbiome Studies. Microb. Ecol. 2018, 76, 851–855. [Google Scholar] [CrossRef]
- Ning, Y.; Qi, J.; Dobbins, M.T.; Liang, X.; Wang, J.; Chen, S.; Ma, J.; Jiang, G. Comparative analysis of microbial community structure and function in the gut of wild and captive amur tiger. Front. Microbiol. 2020, 11, 1665. [Google Scholar] [CrossRef]
- Wei, F.; Huang, G.; Fan, H.; Hu, Y. Research advances and perspectives of conservation genomics and meta-genomics of threatened mammals in China. Acta Theriol. Sin. 2021, 41, 581–590. [Google Scholar]
- Guo, R.; Zhang, W.; Shen, W.; Zhang, G.; Xie, T.; Li, L.; Jinmei, J.; Liu, Y.; Kong, F.; Guo, B.; et al. Analysis of gut microbiota in chinese donkey in different regions using metagenomic sequencing. BMC Genom. 2023, 24, 524. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef]
- Yoo, J.Y.; Groer, M.; Dutra, S.V.O.; Sarkar, A.; McSkimming, D.I. Gut microbiota and immune system interactions. Microorganisms 2020, 8, 1587, Erratum in Microorganisms 2020, 8, 2046. [Google Scholar] [CrossRef]
- Enjalbert, F.; Combes, S.; Zened, A.; Meynadier, A. Rumen microbiota and dietary fat: A mutual shaping. J. Appl. Microbiol. 2017, 123, 782–797. [Google Scholar] [CrossRef]
- Ghaisas, S.; Maher, J.; Kanthasamy, A. Gut microbiome in health and disease: Linking the microbiome-gut-brain axis and environmental factors in the pathogenesis of systemic and neurodegenerative diseases. Pharmacol. Ther. 2016, 158, 52–62. [Google Scholar] [CrossRef]
- Sassone-Corsi, M.; Nuccio, S.P.; Liu, H.; Hernandez, D.; Vu, C.T.; Takahashi, A.A.; Edwards, R.A.; Raffatellu, M. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 2016, 540, 280–283. [Google Scholar] [CrossRef]
- Auffret, M.D.; Dewhurst, R.J.; Duthie, C.A.; Rooke, J.A.; John Wallace, R.; Freeman, T.C.; Stewart, R.; Watson, M.; Roehe, R. The rumen microbiome as a reservoir of antimicrobial resistance and pathogenicity genes is directly affected by diet in beef cattle. Microbiome 2017, 5, 159, Erratum in Microbiome 2017, 5, 159. [Google Scholar] [CrossRef]
- O’Toole, P.W.; Jeffery, I.B. Gut microbiota and aging. Science 2015, 350, 1214–1215. [Google Scholar] [CrossRef]
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’Connor, E.M.; Cusack, S.; Harris, H.M.; Coakley, M.; Lakshminarayanan, B.; O’Sullivan, O.; et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012, 488, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, M.; Wang, S.; Han, R.; Cao, Y.; Hua, W.; Mao, Y.; Zhang, X.; Pang, X.; Wei, C.; et al. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010, 4, 232–241, Erratum in ISME J. 2010, 4, 232–241. [Google Scholar] [CrossRef]
- Zmora, N.; Suez, J.; Elinav, E. You are what you eat: Diet, health and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 35–56. [Google Scholar] [CrossRef]
- Kim, S.K.; Guevarra, R.B.; Kim, Y.T.; Kwon, J.; Kim, H.; Cho, J.H.; Kim, H.B.; Lee, J.H. Role of probiotics in human gut microbiome-associated diseases. J. Microbiol. Biotechnol. 2019, 29, 1335–1340. [Google Scholar] [CrossRef]
- Xue, Z.; Zhang, W.; Wang, L.; Hou, R.; Zhang, M.; Fei, L.; Zhang, X.; Huang, H.; Bridgewater, L.C.; Jiang, Y.; et al. The bamboo-eating giant panda harbors a carnivore-like gut microbiota, with excessive seasonal variations. mBio 2015, 6, e00022-15. [Google Scholar] [CrossRef]
- Amato, K.R.; Leigh, S.R.; Kent, A.; Mackie, R.I.; Yeoman, C.J.; Stumpf, R.M.; Wilson, B.A.; Nelson, K.E.; White, B.A.; Garber, P.A. The gut microbiota appears to compensate for seasonal diet variation in the wild black howler monkey (Alouatta pigra). Microb. Ecol. 2015, 69, 434–443. [Google Scholar] [CrossRef]
- Buford, T.W. (Dis)Trust your gut: The gut microbiome in age-related inflammation, health, and disease. Microbiome 2017, 5, 80. [Google Scholar] [CrossRef]
- Jiang, F.; Gao, H.; Qin, W.; Song, P.; Wang, H.; Zhang, J.; Liu, D.; Wang, D.; Zhang, T. Marked seasonal variation in structure and function of gut microbiota in forest and alpine musk deer. Front. Microbiol. 2021, 12, 699797. [Google Scholar] [CrossRef]
- Jiang, F.; Song, P.; Zhang, J.; Gao, H.; Wang, H.; Cai, Z.; Liu, D.; Zhang, T. Comparative analysis of gut microbial composition and functions of forest musk deer in different breeding centres. Acta Theriol. Sin. 2023, 43, 129–140. [Google Scholar]
- Robeson, M.S., II; O’Rourke, D.R.; Kaehler, B.D.; Ziemski, M.; Dillon, M.R.; Foster, J.T.; Bokulich, N.A. RESCRIPt: Reproducible sequence taxonomy reference database management. PLoS Comput. Biol. 2021, 17, e1009581. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534, Erratum in Mol. Biol. Evol. 2020, 37, 2461. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Community Ecology Package, Version 2.5-6. 2019.
- Zhao, X.; Zhang, X.; Chen, Z.; Wang, Z.; Lu, Y.; Cheng, D. The divergence in bacterial components associated with bactrocera dorsalis across developmental stages. Front. Microbiol. 2018, 9, 114. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Jiang, F.; Song, P.; Wang, H.; Zhang, J.; Liu, D.; Cai, Z.; Gao, H.; Chi, X.; Zhang, T. Comparative analysis of gut microbial composition and potential functions in captive forest and alpine musk deer. Appl. Microbiol. Biot. 2022, 106, 1325–1339. [Google Scholar] [CrossRef]
- Moustafa, M.A.M.; Chel, H.M.; Thu, M.J.; Bawm, S.; Htun, L.L.; Win, M.M.; Oo, Z.M.; Ohsawa, N.; Lahdenperä, M.; Mohamed, W.M.A.; et al. Anthropogenic interferences lead to gut microbiome dysbiosis in Asian elephants and may alter adaptation processes to surrounding environments. Sci. Rep. 2021, 11, 741. [Google Scholar] [CrossRef]
- Han, S.; Guan, Y.; Dou, H.; Yang, H.; Yao, M.; Ge, J.; Feng, L. Comparison of the fecal microbiota of two free-ranging Chinese subspecies of the leopard (Panthera pardus) using high-throughput sequencing. PeerJ 2019, 7, e6684. [Google Scholar] [CrossRef]
- Nelson, T.M.; Rogers, T.L.; Carlini, A.R.; Brown, M.V. Diet and phylogeny shape the gut microbiota of Antarctic seals: A comparison of wild and captive animals. Environ. Microbiol. 2013, 15, 1132–1145. [Google Scholar] [CrossRef]
- Barelli, C.; Albanese, D.; Stumpf, R.M.; Asangba, A.; Donati, C.; Rovero, F.; Hauffe, H.C. The gut microbiota communities of wild arboreal and ground-feeding tropical primates are affected differently by habitat disturbance. mSystems 2020, 5, e00061-20. [Google Scholar] [CrossRef]
- Hale, V.L.; Tan, C.L.; Niu, K.; Yang, Y.; Zhang, Q.; Knight, R.; Amato, K.R. Gut microbiota in wild and captive Guizhou snub-nosed monkeys, Rhinopithecus brelichi. Am. J. Primatol. 2019, 81, e22989. [Google Scholar] [CrossRef]
- Wang, W.; Wang, F.; Li, L.; Wang, A.; Sharshov, K.; Druzyaka, A.; Lancuo, Z.; Wang, S.; Shi, Y. Characterization of the gut microbiome of black-necked cranes (Grus nigricollis) in six wintering areas in China. Arch. Microbiol. 2020, 202, 983–993. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, Y.; Zhang, M.; Yao, Y.; Tian, H.; Sang, Z.; Wang, L.; Xu, H. Structural changes in the gut microbiota community of the black-necked crane (Grus nigricollis) in the wintering period. Arch. Microbiol. 2021, 203, 6203–6214. [Google Scholar] [CrossRef]
- Maynard, C.; Weinkove, D. The gut microbiota and ageing. Subcell. Biochem. 2018, 90, 351–371. [Google Scholar]
- Li, Y.; Chen, T.; Li, Y.; Tang, Y.; Huang, Z. Gut microbiota are associated with sex and age of host: Evidence from semi-provisioned rhesus macaques in southwest Guangxi, China. Ecol. Evol. 2021, 11, 8096–8122. [Google Scholar] [CrossRef]
- Walker, R.W.; Clemente, J.C.; Peter, I.; Loos, R.J.F. The prenatal gut microbiome: Are we colonized with bacteria in utero? Pediatr. Obes. 2017, 12 (Suppl. S1), 3–17. [Google Scholar] [CrossRef]
- McCoy, K.D.; Thomson, C.A. The Impact of Maternal Microbes and Microbial Colonization in Early Life on Hematopoiesis. J. Immunol. 2018, 200, 2519–2526. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, K.; Zhang, C.; Feng, Y.; Zhang, X.; Wang, X.; Wu, G. Dynamics and stabilization of the rumen microbiome in yearling Tibetan sheep. Sci. Rep. 2019, 9, 19620. [Google Scholar] [CrossRef]
- Stoffel, M.A.; Acevedo-Whitehouse, K.; Morales-Durán, N.; Grosser, S.; Chakarov, N.; Krüger, O.; Nichols, H.J.; Elorriaga-Verplancken, F.R.; Hoffman, J.I. Early sexual dimorphism in the developing gut microbiome of northern elephant seals. Mol. Ecol. 2020, 29, 2109–2122. [Google Scholar] [CrossRef]
- Su, K.; Lei, Y.; Jin, X.; Li, J. Study on gut microbiota of captive Bengal white tiger of different ages. Chin. J. Wildl. 2021, 2021, 737–746. [Google Scholar]
- Sandhu, K.V.; Sherwin, E.; Schellekens, H.; Stanton, C.; Dinan, T.G.; Cryan, J.F. Feeding the microbiota-gut-brain axis: Diet, microbiome, and neuropsychiatry. Transl. Res. 2017, 179, 223–244. [Google Scholar] [CrossRef]
- Liu, X.; Ma, Z.; Wang, Y.; Li, L.; Jia, H.; Zhang, L. Compound probiotics can improve intestinal health by affecting the gut microbiota of broilers. J. Anim. Sci. 2023, 101, skad388. [Google Scholar] [CrossRef]
- Zaman, S.; Gohar, M.; Kanwal, H.; Chaudhary, A.; Imran, M. Impact of probiotic geotrichum candidum QAUGC01 on health, productivity, and gut microbial diversity of dairy cattle. Curr. Microbiol. 2022, 79, 376. [Google Scholar] [CrossRef]
- Huang, W.; Ma, T.; Liu, Y.; Kwok, L.Y.; Li, Y.; Jin, H.; Zhao, F.; Shen, X.; Shi, X.; Sun, Z.; et al. Spraying compound probiotics improves growth performance and immunity and modulates gut microbiota and blood metabolites of suckling piglets. Sci. China Life Sci. 2023, 66, 1092–1107. [Google Scholar] [CrossRef]
- Wu, Q.; Wang, X.; Ding, Y.; Hu, Y.; Nie, Y.; Wei, W.; Ma, S.; Yan, L.; Zhu, L.; Wei, F. Seasonal variation in nutrient utilization shapes gut microbiome structure and function in wild giant pandas. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2017, 284, 20170955. [Google Scholar] [CrossRef]
- Tang, K.Y.; Wang, Z.W.; Wan, Q.H.; Fang, S.G. Metagenomics reveals seasonal functional adaptation of the gut microbiome to host feeding and fasting in the Chinese alligator. Front. Microbiol. 2019, 10, 2409. [Google Scholar] [CrossRef]
- Kartzinel, T.R.; Hsing, J.C.; Musili, P.M.; Brown, B.R.P.; Pringle, R.M. Covariation of diet and gut microbiome in African megafauna. Proc. Natl. Acad. Sci. USA 2019, 116, 23588–23593. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, F.; Song, P.; Cai, Z.; Wu, G.; He, S.; Gu, H.; Gao, H.; Zhang, T. Comparative Analysis of Compound Probiotics, Seasonal Variation, and Age on Gut Microbial Composition and Function in Endangered Forest Musk Deer. Microorganisms 2025, 13, 1991. https://doi.org/10.3390/microorganisms13091991
Jiang F, Song P, Cai Z, Wu G, He S, Gu H, Gao H, Zhang T. Comparative Analysis of Compound Probiotics, Seasonal Variation, and Age on Gut Microbial Composition and Function in Endangered Forest Musk Deer. Microorganisms. 2025; 13(9):1991. https://doi.org/10.3390/microorganisms13091991
Chicago/Turabian StyleJiang, Feng, Pengfei Song, Zhenyuan Cai, Guosheng Wu, Shunfu He, Haifeng Gu, Hongmei Gao, and Tongzuo Zhang. 2025. "Comparative Analysis of Compound Probiotics, Seasonal Variation, and Age on Gut Microbial Composition and Function in Endangered Forest Musk Deer" Microorganisms 13, no. 9: 1991. https://doi.org/10.3390/microorganisms13091991
APA StyleJiang, F., Song, P., Cai, Z., Wu, G., He, S., Gu, H., Gao, H., & Zhang, T. (2025). Comparative Analysis of Compound Probiotics, Seasonal Variation, and Age on Gut Microbial Composition and Function in Endangered Forest Musk Deer. Microorganisms, 13(9), 1991. https://doi.org/10.3390/microorganisms13091991





