Microbial Landscapes of the Gut–Biliary Axis: Implications for Benign and Malignant Biliary Tract Diseases
Abstract
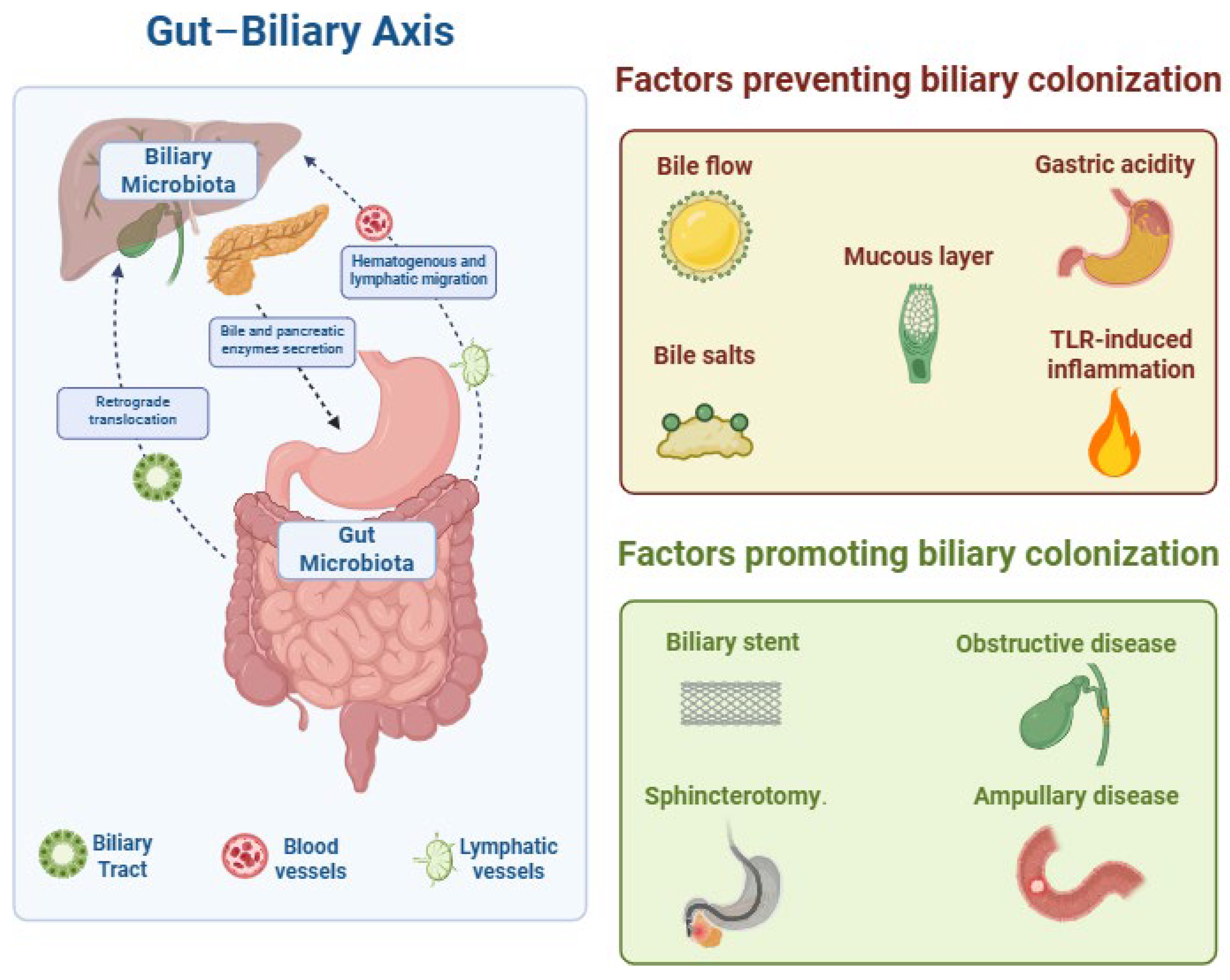
1. Introduction
2. The Microbiome and Primary Sclerosing Cholangitis
| Reference (Year) | Cohort/Sample Type | Sequencing or Culture Technique | Biodiversity vs. Controls | Dominant Genera in Controls | Alterations in PSC | Clinical Correlations |
|---|---|---|---|---|---|---|
| Pereira et al., 2017 [64] | 10 early-stage PSC vs. 9 gallstone controls; ERC bile | 16S rRNA NGS | No significant difference | Prevotella, Streptococcus, Veillonella, Fusobacterium, Haemophilus | Increased concentration of Streptococcus in dysplasia/CCA | Early disease shows near-physiological biliary community |
| Liwinski et al., 2020 [36] | 43 PSC (no dysplasia or CCA) vs. 34 controls; ERC bile | 16S rRNA NGS + quantitative culture | Reduced richness; higher intra-patient variability | Streptococcus predominant | Proteobacteria 25% (vs. 12%); Enterococcus faecalis, Staphylococcus, Neisseria ↑ | Enterococcus burden strongly correlates with TLCA levels, metalloproteinase activity, and accelerated fibrosis |
| Tyc et al., 2021 [65] | 20 PSC; ERC bile | 16S rRNA NGS | Markedly reduced | Expansion of Proteobacteria pathobionts mirrored Liwinski’s results | Supports link between Enterococcaceae dominance, Th17 polarization, and duct injury | |
| Miyabe et al., 2024 [66] | 32 PSC vs. 20 choledocholithiasis controls; ERC bile | 16S rRNA NGS | Similar overall diversity | Streptococcus core genus | Species richness, especially Fusobacteria, rises with PSC duration and CCA | Suggests inflammation- driven microbiota maturation contributes to oncogenesis |
| Early Culture or PCR studies 1998–2010 [61,62,63] | 76 PSC; bile or brushings | Aerobic or anaerobic culture, species- specific PCR | Frequent isolation of Streptococcus, Enterococcus, and Staphylococcus | First evidence that bile is not sterile and harbors opportunistic Gram-positive cocci |
3. Primary Biliary Cholangitis
4. The Microbiome and Autoimmune Hepatitis
5. The Oncobiome and Hepato-Biliary Malignancies
5.1. The Microbiome and Hepatocellular Carcinoma
5.2. The Microbiome and Cholangiocarcinoma
5.3. The Microbiome and Gallbladder Cancer
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AIH | Autoimmune Hepatitis |
| BA | Bile Acid |
| BSH | Bile Salt Hydrolase |
| CCA | Cholangiocarcinoma |
| CDT | Cytolethal Distending Toxin |
| dCCA | Distal Cholangiocarcinoma |
| dMBO | Distal Malignant Biliary Obstruction |
| HCC | Hepatocellular Carcinoma |
| iCCA | Intrahepatic Cholangiocarcinoma |
| NGS | Next-Generation Sequencing |
| PSC | Primary Sclerosing Cholangitis |
| PBC | Primary Biliary Cholangitis |
| SCFA | Short-Chain Fatty Acid |
| TGR5 | Takeda G-Protein-Coupled Receptor 5 |
| TLCA | Taurolithocholic Acid |
| UDCA | Ursodeoxycholic Acid |
References
- Nicoletti, A.; Ponziani, F.R.; Nardella, E.; Ianiro, G.; Gasbarrini, A.; Dal Verme, L.Z. Biliary tract microbiota: A new kid on the block of liver diseases? Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2750–2775. [Google Scholar] [CrossRef]
- Thomas, R.M.; Jobin, C. Microbiota in pancreatic health and disease: The next frontier in microbiome research. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 53–64. [Google Scholar] [CrossRef]
- Baquero, F.; Nombela, C. The Microbiome as a Human Organ. Clin. Microbiol. Infect. 2012, 18, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.V.; Pedersen, O. The human intestinal microbiome in health and disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef] [PubMed]
- Binda, C.; Gibiino, G.; Coluccio, C.; Sbrancia, M.; Dajti, E.; Sinagra, E.; Capurso, G.; Sambri, V.; Cucchetti, A.; Ercolani, G.; et al. Biliary Diseases from the Microbiome Perspective: How Microorganisms Could Change the Approach to Benign and Malignant Diseases. Microorganisms 2022, 10, 312. [Google Scholar] [CrossRef]
- Tözün, N.; Vardareli, E. Gut Microbiome and Gastrointestinal Cancer les liaisons Dangereuses. J. Clin. Gastroenterol. 2016, 50, S191–S196. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Fu, S.W.; Lu, L.; Zhao, H. A preliminary study of biliary microbiota in patients with bile duct stones or distal cholangiocarcinoma. BioMed Res. Int. 2019, 2019, 1092563. [Google Scholar] [CrossRef]
- Begley, M.; Gahan, C.G.M.; Hill, C. The interaction between bacteria and bile. FEMS Microbiol. Rev. 2005, 29, 625–651. [Google Scholar] [CrossRef]
- Schubert, K.; Olde Damink, S.W.M.; von Bergen, M.; Schaap, F.G. Interactions between bile salts, gut microbiota, and hepatic innate immunity. Immunol. Rev. 2017, 279, 23–35. [Google Scholar] [CrossRef]
- Long, S.L.; Gahan, C.G.M.; Joyce, S.A. Interactions between gut bacteria and bile in health and disease. Mol. Aspects. Med. 2017, 56, 54–65. [Google Scholar] [CrossRef]
- Duboc, H.; Rajca, S.; Rainteau, D.; Benarous, D.; Maubert, M.-A.; Quervain, E.; Thomas, G.; Barbu, V.; Humbert, L.; Despras, G.; et al. Connecting dysbiosis, bile-acid dysmetabolism and Gut inflammation in inflammatory bowel diseases. Gut 2013, 62, 531–539. [Google Scholar] [CrossRef]
- Li, Y.; Tang, R.; Leung, P.S.C.; Gershwin, M.E.; Ma, X. Bile acids and intestinal microbiota in autoimmune cholestatic liver diseases. Autoimmun. Rev. 2017, 16, 885–896. [Google Scholar] [CrossRef]
- Bernstein, H.; Bernstein, C.; Payne, C.M.; Dvorakova, K.; Garewal, H. Bile acids as carcinogens in human gastrointestinal cancers. Mutat. Res. Rev. Mutat. Res. 2005, 589, 47–65. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.Y.; Costerton, J.W.; Shaffer, E.A. Defense system in the biliary tract against bacterial infection. Dig. Dis. Sci. 1992, 37, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Ohba, K.; Ozaki, S.; Isse, K.; Hirayama, T.; Wada, A.; Nakanuma, Y. Peptide antibiotic human beta-defensin-1 and -2 contribute to antimicrobial defense of the intrahepatic biliary tree. Hepatology 2004, 40, 925–932. [Google Scholar] [CrossRef] [PubMed]
- Scheufele, F.; Aichinger, L.; Jäger, C.; Demir, I.E.; Schorn, S.; Sargut, M.; Erkan, M.; Kleeff, J.; Friess, H.; Ceyhan, G.O. Effect of preoperative biliary drainage on bacterial flora in bile of patients with periampullary cancer. Br. J. Surg. 2017, 104, e182–e188. [Google Scholar] [CrossRef]
- Gregg, J.A.; De Girolami, P.; Carr-Locke, D.L. Effects of sphincteroplasty and endoscopic sphincterotomy on the bacteriologic characteristics of the common bile duct. Am. J. Surg. 1985, 149, 668–671. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Hara, S.P.O.; Nelson, J.B.; Splinter, P.L.; Small, A.J.; Tietz, P.S.; Limper, A.H.; LaRusso, N.F. Multiple TLRs Are Expressed in Human Cholangiocytes and Mediate Host Epithelial Defense Responses to Cryptosporidium parvum via Activation of NF-κB 1. J. Immunol. 2005, 175, 7447–7456. [Google Scholar] [CrossRef]
- Sutherland, D.B.; Suzuki, K.; Fagarasan, S. Fostering of advanced mutualism with gut microbiota by Immunoglobulin A. Immunol. Rev. 2016, 270, 20–31. [Google Scholar] [CrossRef]
- De Martel, C.; Plummer, M.; Parsonnet, J.; van Doorn, L.-J.; Franceschi, S. Helicobacter species in cancers of the gallbladder and extrahepatic biliary tract. Br. J. Cancer 2009, 100, 194–199. [Google Scholar] [CrossRef]
- Brook, I. Aerobic and Anaerobic Microbiology of Biliary Tract Disease. J. Clin. Microbiol. 1989, 27, 2373–2375. [Google Scholar] [CrossRef]
- Van Velkinburgh, J.C.; Gunn, J.S. PhoP-PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect. Immun. 1999, 67, 1614–1622. [Google Scholar] [CrossRef][Green Version]
- Sakaguchi, Y.; Murata, K.; Kimura, M. Clostridium perfringens and other anaerobes isolated from bile. J. Clin. Pathol. 1983, 36, 345–349. [Google Scholar] [CrossRef]
- Flores, C.; Maguilnik, I.; Hadlich, E.; Goldani, L.Z. Microbiology of choledochal bile in patients with choledocholithiasis admitted to a tertiary hospital. J. Gastroenterol. Hepatol. 2003, 18, 333–336. [Google Scholar] [CrossRef]
- Hardy, J.; Francis, K.P.; Deboer, M.; Chu, P.; Gibbs, K.; Contag, C.H. Extracellular Replication of Listeria monocytogenes in the Murine Gall Bladder. Science 2004, 303, 851–853. [Google Scholar] [CrossRef] [PubMed]
- Flemma, R.J.; Flint, L.M.; Osterhout, S.; Shingleton, W.W. Bacteriologic studies of biliary tract infection. Ann. Surg. 1967, 166, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, E.; Sánchez, B.; Farina, A.; Margolles, A.; Rodríguez, J.M. Characterization of the bile and gall bladder microbiota of healthy pigs. Microbiologyopen 2014, 3, 937–949. [Google Scholar] [CrossRef] [PubMed]
- Molinero, N.; Ruiz, L.; Milani, C.; Gutiérrez-Díaz, I.; Sánchez, B.; Mangifesta, M.; Segura, J.; Cambero, I.; Campelo, A.B.; García-Bernardo, C.M.; et al. The human gallbladder microbiome is related to the physiological state and the biliary metabolic profile. Microbiome 2019, 7, 100. [Google Scholar] [CrossRef]
- Smruti, P.; Mautin, H.; Donnele, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E.; et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2019, 8, 403–416. [Google Scholar] [CrossRef]
- Ley, R.E.; Hamady, M.; Lozupone, C.; Turnbaugh, P.J.; Ramey, R.R.; Bircher, J.S.; Schlegel, M.L.; Tucker, T.A.; Schrenzel, M.D.; Knight, R.; et al. Evolution of mammals and their gut microbes. Science 2008, 320, 20144. [Google Scholar] [CrossRef]
- Thomas, R.M.; Gharaibeh, R.Z.; Gauthier, J.; Beveridge, M.; Pope, J.L.; Guijarro, M.V.; Yu, Q.; He, Z.; Ohland, C.; Newsome, R.; et al. Intestinal microbiota enhances pancreatic carcinogenesis in preclinical models. Carcinogenesis 2018, 39, 1068–1078. [Google Scholar] [CrossRef] [PubMed]
- Bravo-Blas, A.; Utriainen, L.; Clay, S.L.; Kästele, V.; Cerovic, V.; Cunningham, A.F.; Henderson, I.R.; Wall, D.M.; Milling, S.W.F. Salmonella enterica Serovar Typhimurium Travels to Mesenteric Lymph Nodes Both with Host Cells and Autonomously. J. Immunol. 2019, 202, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Diehl, G.E.; Longman, R.S.; Zhang, J.X.; Breart, B.; Galan, C.; Cuesta, A.; Schwab, S.R.; Littman, D.R. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX 3 CR1 hi cells. Nature 2013, 494, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Bowlus, C.L.; Arrivé, L.; Bergquist, A.; Deneau, M.; Forman, L.; Ilyas, S.I.; Lunsford, K.E.; Martinez, M.; Sapisochin, G.; Shroff, R.; et al. AASLD practice guidance on primary sclerosing cholangitis and cholangiocarcinoma. Hepatology 2023, 77, 659–702. [Google Scholar] [CrossRef]
- Karlsen, T.H.; Folseraas, T.; Thorburn, D.; Vesterhus, M. Primary sclerosing cholangitis—A comprehensive review. J. Hepatol. 2017, 67, 1298–1323. [Google Scholar] [CrossRef]
- Liwinski, T.; Zenouzi, R.; John, C.; Ehlken, H.; Rühlemann, M.C.; Bang, C.; Groth, S.; Lieb, W.; Kantowski, M.; Andersen, N.; et al. Alterations of the bile microbiome in primary sclerosing cholangitis. Gut 2020, 69, 665–672. [Google Scholar] [CrossRef]
- Shah, A.; Macdonald, G.A.; Morrison, M.; Holtmann, G. Targeting the Gut Microbiome as a Treatment for Primary Sclerosing Cholangitis: A Conceptional Framework. Am. J. Gastroenterol. 2020, 115, 814–822. [Google Scholar] [CrossRef]
- Sabino, J.; Vieira-Silva, S.; Machiels, K.; Joossens, M.; Falony, G.; Ballet, V.; Ferrante, M.; Van Assche, G.; Van der Merwe, S.; Vermeire, S.; et al. Primary sclerosing cholangitis is characterised by intestinal dysbiosis independent from IBD. Gut 2016, 65, 1681–1689. [Google Scholar] [CrossRef]
- Bajer, L.; Kverka, M.; Kostovcik, M.; Macinga, P.; Dvorak, J.; Stehlikova, Z.; Brezina, J.; Wohl, P.; Spicak, J.; Drastich, P. Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World J. Gastroenterol. 2017, 23, 4548–4558. [Google Scholar] [CrossRef]
- Kummen, M.; Holm, K.; Anmarkrud, J.A.; Nygård, S.; Vesterhus, M.; Høivik, M.L.; Trøseid, M.; Marschall, H.-U.; Schrumpf, E.; Moum, B.; et al. The gut microbial profile in patients with primary sclerosing cholangitis is distinct from patients with ulcerative colitis without biliary disease and healthy controls. Gut 2017, 66, 611–619. [Google Scholar] [CrossRef]
- Kummen, M.; Thingholm, L.B.; Rühlemann, M.C.; Holm, K.; Hansen, S.H.; Moitinho-Silva, L.; Liwinski, T.; Zenouzi, R.; Storm-Larsen, C.; Midttun, Ø.; et al. Altered Gut Microbial Metabolism of Essential Nutrients in Primary Sclerosing Cholangitis. Gastroenterology 2021, 160, 1784–1798.e0. [Google Scholar] [CrossRef]
- Rühlemann, M.C.; Heinsen, F.-A.; Zenouzi, R.; Lieb, W.; Franke, A.; Schramm, C. Faecal microbiota profiles as diagnostic biomarkers in primary sclerosing cholangitis. Gut 2017, 66, 753–754. [Google Scholar] [CrossRef]
- Rühlemann, M.; Liwinski, T.; Heinsen, F.-A.; Bang, C.; Zenouzi, R.; Kummen, M.; Thingholm, L.; Tempel, M.; Lieb, W.; Karlsen, T.; et al. Consistent alterations in faecal microbiomes of patients with primary sclerosing cholangitis independent of associated colitis. Aliment. Pharmacol. Ther. 2019, 50, 580–589. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Li, B.; Li, Y.; Wei, Y.; Huang, B.; Liang, J.; You, Z.; Li, Y.; Qian, Q.; Wang, R.; et al. Altered faecal microbiome and metabolome in IgG4-related sclerosing cholangitis and primary sclerosing cholangitis. Gut 2022, 71, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Rossen, N.G.; Fuentes, S.; Boonstra, K.; D’hAens, G.R.; Heilig, H.G.; Zoetendal, E.G.; de Vos, W.M.; Ponsioen, C.Y. The mucosa-associated microbiota of PSC patients is characterized by low diversity and low abundance of uncultured Clostridiales II. J. Crohns Colitis 2015, 9, 342–348. [Google Scholar] [CrossRef]
- Kevans, D.; Tyler, A.D.; Holm, K.; Jørgensen, K.K.; Vatn, M.H.; Karlsen, T.H.; Kaplan, G.G.; Eksteen, B.; Gevers, D.; Hov, J.; et al. Characterization of Intestinal Microbiota in Ulcerative Colitis Patients with and without Primary Sclerosing Cholangitis. J. Crohns Colitis 2016, 10, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Torres, J.; Bao, X.; Goel, A.; Colombel, J.; Pekow, J.; Jabri, B.; Williams, K.; Castillo, A.; Odin, J.; Meckel, K.; et al. The features of mucosa-associated microbiota in primary sclerosing cholangitis. Aliment. Pharmacol. Ther. 2016, 43, 790–801. [Google Scholar] [CrossRef]
- Quraishi, M.N.; Sergeant, M.; Kay, G.; Iqbal, T.; Chan, J.; Constantinidou, C.; Trivedi, P.; Ferguson, J.; Adams, D.H.; Pallen, M.; et al. The gut-adherent microbiota of PSC-IBD is distinct to that of IBD. Gut 2017, 66, 386–388. [Google Scholar] [CrossRef]
- Al-Shakhshir, S.; Quraishi, M.N.; Mullish, B.; Patel, A.; Vince, A.; Rowe, A.; Homer, V.; Jackson, N.; Gyimah, D.; Shabir, S.; et al. FAecal micRobiota transplantation in primary sclerosinG chOlangitis (FARGO): Study protocol for a randomised, multicentre, phase IIa, placebo-controlled trial. BMJ Open 2025, 15, e095392. [Google Scholar] [CrossRef]
- Liwinski, T.; Heinemann, M.; Schramm, C. The intestinal and biliary microbiome in autoimmune liver disease—Current evidence and concepts. Semin. Immunopathol. 2022, 44, 485–507. [Google Scholar] [CrossRef]
- Little, R.; Wine, E.; Kamath, B.M.; Griffiths, A.M.; Ricciuto, A. Gut microbiome in primary sclerosing cholangitis: A review. World J. Gastroenterol. 2020, 26, 2768–2780. [Google Scholar] [CrossRef]
- Wei, X.; Yan, X.; Zou, D.; Yang, Z.; Wang, X.; Liu, W.; Wang, S.; Li, X.; Han, J.; Huang, L.; et al. Abnormal fecal microbiota community and functions in patients with hepatitis B liver cirrhosis as revealed by a metagenomic approach. BMC Gastroenterol. 2013, 13, 175. [Google Scholar] [CrossRef]
- Qin, N.; Yang, F.; Li, A.; Prifti, E.; Chen, Y.; Shao, L.; Guo, J.; Le Chatelier, E.; Yao, J.; Wu, L.; et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014, 513, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.J.; Hirschfield, G.M.; Adams, D.H.; Vierling, J.M. Immunopathogenesis of Primary Biliary Cholangitis, Primary Sclerosing Cholangitis and Autoimmune Hepatitis: Themes and Concepts. Gastroenterology 2024, 166, 995–1019. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.J.; Tickle, J.; Vesterhus, M.N.; Eddowes, P.J.; Bruns, T.; Vainio, J.; Parker, R.; Smith, D.; Liaskou, E.; Thorbjørnsen, L.W.; et al. Vascular adhesion protein-1 is elevated in primary sclerosing cholangitis, is predictive of clinical outcome and facilitates recruitment of gut-tropic lymphocytes to liver in a substrate-dependent manner. Gut 2018, 67, 1135–1145. [Google Scholar] [CrossRef] [PubMed]
- Lemoinne, S.; Kemgang, A.; Ben Belkacem, K.; Straube, M.; Jegou, S.; Corpechot, C.; Network, S.-A.I.; Chazouillères, O.; Housset, C.; Sokol, H. Fungi participate in the dysbiosis of gut microbiota in patients with primary sclerosing cholangitis. Gut 2020, 69, 92–102. [Google Scholar] [CrossRef]
- Rühlemann, M.C.; Solovjeva, M.E.L.; Zenouzi, R.; Liwinski, T.; Kummen, M.; Lieb, W.; Hov, J.R.; Schramm, C.; Franke, A.; Bang, C. Gut mycobiome of primary sclerosing cholangitis patients is characterised by an increase of Trichocladium griseum and Candida species. Gut 2020, 69, 1890–1892. [Google Scholar] [CrossRef]
- Kulaksiz, H.; Rudolph, G.; Kloeters-Plachky, P.; Sauer, P.; Geiss, H.; Stiehl, A. Biliary candida infections in primary sclerosing cholangitis. J. Hepatol. 2006, 45, 711–716. [Google Scholar] [CrossRef]
- Allegretti, J.R.; Kassam, Z.; Carrellas, M.; Mullish, B.H.; Marchesi, J.R.; Pechlivanis, A.; Smith, M.; Gerardin, Y.; Timberlake, S.; Pratt, D.S.; et al. Fecal Microbiota Transplantation in Patients with Primary Sclerosing Cholangitis: A Pilot Clinical Trial. Am. J. Gastroenterol. 2019, 114, 1071–1079. [Google Scholar] [CrossRef]
- Vrieze, A.; Out, C.; Fuentes, S.; Jonker, L.; Reuling, I.; Kootte, R.S.; van Nood, E.; Holleman, F.; Knaapen, M.; Romijn, J.A.; et al. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J. Hepatol. 2014, 60, 824–831. [Google Scholar] [CrossRef]
- Olsson, R.; Björnsson, E.; Bäckman, L.; Friman, S.; Höckerstedt, K.; Kaijser, B.; Olausson, M. Bile duct bacterial isolates in primary sclerosing cholangitis: A study of explanted livers. J. Hepatol. 1998, 28, 426–432. [Google Scholar] [CrossRef]
- Katt, J.; Schwinge, D.; Schoknecht, T.; Quaas, A.; Sobottka, I.; Burandt, E.; Becker, C.; Neurath, M.F.; Lohse, A.W.; Herkel, J.; et al. Increased T helper type 17 response to pathogen stimulation in patients with primary sclerosing cholangitis. Hepatology 2013, 58, 1084–1093. [Google Scholar] [CrossRef] [PubMed]
- Björnsson, E.S.; Kilander, A.F.; Olsson, R.G. Bile duct bacterial isolates in primary sclerosing cholangitis and certain other forms of cholestasis--a study of bile cultures from ERCP. Hepatogastroenterology 2000, 47, 1504–1508. [Google Scholar] [PubMed]
- Pereira, P.; Aho, V.; Arola, J.; Boyd, S.; Jokelainen, K.; Paulin, L.; Auvinen, P.; Färkkilä, M.; Alpini, G.D. Bile microbiota in primary sclerosing cholangitis: Impact on disease progression and development of biliary dysplasia. PLoS ONE 2017, 12, e0182924. [Google Scholar] [CrossRef] [PubMed]
- Tyc, O.; Jansen, C.; Schierwagen, R.; Uschner, F.E.; Israelsen, M.; Klein, S.; Ortiz, C.; Strassburg, C.P.; Zeuzem, S.; Gu, W.; et al. Variation in Bile Microbiome by the Etiology of Cholestatic Liver Disease. Liver Transplant. 2020, 26, 1652–1657. [Google Scholar] [CrossRef]
- Miyabe, K.; Chandrasekhara, V.; Wongjarupong, N.; Chen, J.; Yang, L.; Johnson, S.; Chia, N.; Walther-Antonio, M.; Yao, J.Z.; Harrington, S.C.; et al. Potential Role of Inflammation-Promoting Biliary Microbiome in Primary Sclerosing Cholangitis and Cholangiocarcinoma. Cancers 2022, 14, 2120. [Google Scholar] [CrossRef]
- Steck, N.; Hoffmann, M.; Sava, I.G.; Kim, S.C.; Hahne, H.; Tonkonogy, S.L.; Mair, K.; Krueger, D.; Pruteanu, M.; Shanahan, F.; et al. Enterococcus faecalis metalloprotease compromises epithelial barrier and contributes to intestinal inflammation. Gastroenterology 2011, 141, 959–971. [Google Scholar] [CrossRef]
- Zigmond, E.; Zecher, B.F.; Bartels, A.-L.; Ziv-Baran, T.; Rösch, T.; Schachschal, G.; Lohse, A.W.; Ehlken, H.; Schramm, C. Bile Duct Colonization with Enterococcus sp. Associates with Disease Progression in Primary Sclerosing Cholangitis. Clin. Gastroenterol. Hepatol. 2023, 21, 1223–1232.e3. [Google Scholar] [CrossRef]
- Minich, J.J.; Zhu, Q.; Janssen, S.; Hendrickson, R.; Amir, A.; Vetter, R.; Hyde, J.; Doty, M.M.; Stillwell, K.; Benardini, J.; et al. KatharoSeq Enables High-Throughput Microbiome Analysis from Low-Biomass Samples. mSystems 2018, 3, e00218-17. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: The diagnosis and management of patients with primary biliary cholangitis. J. Hepatol. 2017, 67, 145–172. [Google Scholar] [CrossRef]
- Lleo, A.; Leung, P.S.C.; Hirschfield, G.M.; Gershwin, E.M. The Pathogenesis of Primary Biliary Cholangitis: A Comprehensive Review. Semin. Liver Dis. 2020, 40, 34–48. [Google Scholar] [CrossRef]
- Zhou, Y.-J.; Ying, G.-X.; Dong, S.-L.; Xiang, B.; Jin, Q.-F. Gut microbial profile of treatment-naive patients with primary biliary cholangitis. Front. Immunol. 2023, 14, 1126117. [Google Scholar] [CrossRef]
- Lv, L.; Fang, D.; Shi, D.; Chen, D.; Yan, R.; Zhu, Y.; Chen, Y.; Shao, L.; Guo, F.; Wu, W.; et al. Alterations and correlations of the gut microbiome, metabolism and immunity in patients with primary biliary cirrhosis. Environ. Microbiol. 2016, 18, 2272–2286. [Google Scholar] [CrossRef]
- Tang, R.; Wei, Y.; Li, Y.; Chen, W.; Chen, H.; Wang, Q.; Yang, F.; Miao, Q.; Xiao, X.; Zhang, H.; et al. Gut microbial profile is altered in primary biliary cholangitis and partially restored after UDCA therapy. Gut 2018, 67, 534–541. [Google Scholar] [CrossRef]
- Furukawa, M.; Moriya, K.; Nakayama, J.; Inoue, T.; Momoda, R.; Kawaratani, H.; Namisaki, T.; Sato, S.; Douhara, A.; Kaji, K.; et al. Gut dysbiosis associated with clinical prognosis of patients with primary biliary cholangitis. Hepatol. Res. 2020, 50, 840–852. [Google Scholar] [CrossRef]
- Chen, W.; Wei, Y.; Xiong, A.; Li, Y.; Guan, H.; Wang, Q.; Miao, Q.; Bian, Z.; Xiao, X.; Lian, M.; et al. Comprehensive Analysis of Serum and Fecal Bile Acid Profiles and Interaction with Gut Microbiota in Primary Biliary Cholangitis. Clin. Rev. Allergy Immunol. 2020, 58, 25–38. [Google Scholar] [CrossRef]
- Han, W.; Song, T.; Huang, Z.; Liu, Y.; Xu, B.; Huang, C. Distinct signatures of gut microbiota and metabolites in primary biliary cholangitis with poor biochemical response after ursodeoxycholic acid treatment. Cell Biosci. 2024, 14, 80. [Google Scholar] [CrossRef] [PubMed]
- Hiramatsu, K.; Harada, K.; Tsuneyama, K.; Sasaki, M.; Fujita, S.; Hashimoto, T.; Kaneko, S.; Kobayashi, K.; Nakanuma, Y. Amplification and sequence analysis of partial bacterial 16S ribosomal RNA gene in gallbladder bile from patients with primary biliary cirrhosis. J. Hepatol. 2000, 33, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Zachou, K.; Muratori, P.; Koukoulis, G.K.; Granito, A.; Gatselis, N.; Fabbri, A.; Dalekos, G.N.; Muratori, L. Review article: Autoimmune hepatitis—Current management and challenges. Aliment. Pharmacol. Ther. 2013, 38, 887–913. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Kang, Y. The Gut Microbiome and Autoimmune Hepatitis: Implications for Early Diagnostic Biomarkers and Novel Therapies. Mol. Nutr. Food Res. 2023, 67, 2300043. [Google Scholar] [CrossRef]
- Lou, J.; Jiang, Y.; Rao, B.; Li, A.; Ding, S.; Yan, H.; Zhou, H.; Liu, Z.; Shi, Q.; Cui, G.; et al. Fecal Microbiomes Distinguish Patients with Autoimmune Hepatitis from Healthy Individuals. Front. Cell Infect. Microbiol. 2020, 10, 342. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Li, Y.; Yan, L.; Sun, C.; Miao, Q.; Wang, Q.; Xiao, X.; Lian, M.; Li, B.; Chen, Y.; et al. Alterations of gut microbiome in autoimmune hepatitis. Gut 2020, 69, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Liwinski, T.; Casar, C.; Ruehlemann, M.C.; Bang, C.; Sebode, M.; Hohenester, S.; Denk, G.; Lieb, W.; Lohse, A.W.; Franke, A.; et al. A disease-specific decline of the relative abundance of Bifidobacterium in patients with autoimmune hepatitis. Aliment. Pharmacol. Ther. 2020, 51, 1417–1428. [Google Scholar] [CrossRef] [PubMed]
- Elsherbiny, N.M.; Rammadan, M.; Hassan, E.A.; Ali, M.E.; El-Rehim, A.S.A.; Abbas, W.A.; Abozaid, M.A.A.; Hassanin, E.; Hetta, H.F. Autoimmune Hepatitis: Shifts in Gut Microbiota and Metabolic Pathways among Egyptian Patients. Microorganisms 2020, 8, 1011. [Google Scholar] [CrossRef]
- Abe, K.; Takahashi, A.; Fujita, M.; Imaizumi, H.; Hayashi, M.; Okai, K.; Ohira, H.; Barbier, O. Dysbiosis of oral microbiota and its association with salivary immunological biomarkers in autoimmune liver disease. PLoS ONE 2018, 13, e0198757. [Google Scholar] [CrossRef]
- Abbas, M.; Tangney, M. The oncobiome; what, so what, now what? Microbiome Res. Rep. 2025, 4, 16. [Google Scholar] [CrossRef]
- Fu, A.; Yao, B.; Dong, T.; Chen, Y.; Yao, J.; Liu, Y.; Li, H.; Bai, H.; Liu, X.; Zhang, Y.; et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell 2022, 185, 1356–1372.e26. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, L.; Li, M.; Zhang, Y.; Sun, M.; Wang, L.; Lin, J.; Cui, Y.; Chen, Q.; Jin, C.; et al. Fusobacterium nucleatum reduces METTL3-mediated m6A modification and contributes to colorectal cancer metastasis. Nat. Commun. 2022, 13, 1248. [Google Scholar] [CrossRef]
- Gur, C.; Ibrahim, Y.; Isaacson, B.; Yamin, R.; Abed, J.; Gamliel, M.; Enk, J.; Bar-On, Y.; Stanietsky-Kaynan, N.; Coppenhagen-Glazer, S.; et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 2015, 42, 344–355. [Google Scholar] [CrossRef]
- Jinadasa, R.N.; Bloom, S.E.; Weiss, R.S.; Duhamel, G.E. Cytolethal distending toxin: A conserved bacterial genotoxin that blocks cell cycle progression, leading to apoptosis of a broad range of mammalian cell lineages. Microbiology 2011, 157, 1851–1875. [Google Scholar] [CrossRef]
- Ji, J.; Ji, F.; Bayarsaikhan, E. Intratumoral microbiota in HCC: A new kid on the block? Hepatology 2023, 78, 1012–1014. [Google Scholar] [CrossRef]
- Poudel, S.K.; Padmanabhan, R.; Dave, H.; Guinta, K.; Stevens, T.; Sanaka, M.R.; Chahal, P.; Sohal, D.P.S.; Khorana, A.A.; Eng, C.; et al. Microbiomic profiles of bile in patients with benign and malignant pancreaticobiliary disease. PLoS ONE 2023, 18, e0283021. [Google Scholar] [CrossRef] [PubMed]
- Dangtakot, R.; Intuyod, K.; Ahooja, A.; Wongwiwatchai, J.; Hanpanich, P.; Lulitanond, A.; Chamgramol, Y.; Pinlaor, S.; Pinlaor, P. Profiling of Bile Microbiome Identifies District Microbial Population between Choledocholithiasis and Cholangiocarcinoma Patients. Asian Pac. J. Cancer Prev. 2021, 22, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.K.; Tso, D.K.; Harris, A.C.; Malfair, D.; Chang, S.D. Extrahepatic Metastases of Hepatocellular Carcinoma: A Spectrum of Imaging Findings. Can. Assoc. Radiol. J. 2014, 65, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Rashidian, N.; Alseidi, A.; Kirks, R.C. Cancers Metastatic to the Liver. Surg. Clin. N. Am. 2020, 100, 551–563. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Fattovich, G.; Stroffolini, T.; Zagni, I.; Donato, F. Hepatocellular carcinoma in cirrhosis: Incidence and risk factors. Gastroenterology 2004, 127, S35–S50. [Google Scholar] [CrossRef]
- Song, Y.; Tian, S.; Li, Z.; Miao, J.; Wu, M.; Xu, T.; Wu, X.; Qiao, J.; Zhang, X.; Zhao, H.; et al. Progress in the Study of Intratumoral Microorganisms in Hepatocellular Carcinoma. J. Hepatocell. Carcinoma 2025, 12, 59–76. [Google Scholar] [CrossRef]
- Komiyama, S.; Yamada, T.; Takemura, N.; Kokudo, N.; Hase, K.; Kawamura, Y.I. Profiling of tumour-associated microbiota in human hepatocellular carcinoma. Sci. Rep. 2021, 11, 10589. [Google Scholar] [CrossRef]
- Yu, K.L.; Shen, S. Could intratumoural microbiota be key to unlocking treatment responses in hepatocellular carcinoma? Eur. J. Cancer 2025, 216, 115195. [Google Scholar] [CrossRef]
- He, Y.; Zhang, Q.; Yu, X.; Zhang, S.; Guo, W. Overview of microbial profiles in human hepatocellular carcinoma and adjacent nontumor tissues. J. Transl. Med. 2023, 21, 68. [Google Scholar] [CrossRef]
- Li, S.; Xia, H.; Wang, Z.; Zhang, X.; Song, T.; Li, J.; Xu, L.; Zhang, N.; Fan, S.; Li, Q.; et al. Intratumoral microbial heterogeneity affected tumor immune microenvironment and determined clinical outcome of HBV-related HCC. Hepatology 2023, 78, 1079–1091. [Google Scholar] [CrossRef]
- Nesić, D.; Hsu, Y.; Stebbins, C.E. Assembly and function of a bacterial genotoxin. Nature 2004, 429, 429–433. [Google Scholar] [CrossRef]
- Zhu, H.; Li, M.; Bi, D.; Yang, H.; Gao, Y.; Song, F.; Zheng, J.; Xie, R.; Zhang, Y.; Liu, H.; et al. Fusobacterium nucleatum promotes tumor progression in KRAS p.G12D-mutant colorectal cancer by binding to DHX15. Nat. Commun. 2024, 15, 1688. [Google Scholar] [CrossRef]
- Liu, B.; Zhou, Z.; Jin, Y.; Lu, J.; Feng, D.; Peng, R.; Sun, H.; Mu, X.; Li, C.; Chen, Y. Hepatic stellate cell activation and senescence induced by intrahepatic microbiota disturbances drive progression of liver cirrhosis toward hepatocellular carcinoma. J. Immunother. Cancer 2022, 10, e003069. [Google Scholar] [CrossRef] [PubMed]
- Qu, D.; Wang, Y.; Xia, Q.; Chang, J.; Jiang, X.; Zhang, H. Intratumoral Microbiome of Human Primary Liver Cancer. Hepatol. Commun. 2022, 6, 1741–1752. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.-H.; Wang, J.; Chai, X.-Q.; Li, Z.-C.; Jiang, Y.-H.; Li, J.; Liu, X.; Fan, J.; Cai, J.-B.; Liu, F.; et al. The Intratumoral Bacterial Metataxonomic Signature of Hepatocellular Carcinoma. Microbiol. Spectr. 2022, 10, e00983-22. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Ke, X.; Guan, A.; Jin, B.; Qu, J.; Wang, Y.; Xu, X.; Li, C.; Sun, H.; Xu, H.; et al. Intratumoural microbiome can predict the prognosis of hepatocellular carcinoma after surgery. Clin. Transl. Med. 2023, 13, e1331. [Google Scholar] [CrossRef]
- Jiang, F.; Dang, Y.; Zhang, Z.; Yan, Y.; Wang, Y.; Chen, Y.; Chen, L.; Zhang, J.; Liu, J.; Wang, J.; et al. Association of intratumoral microbiome diversity with hepatocellular carcinoma prognosis. mSystems 2025, 10, e00765-24. [Google Scholar] [CrossRef]
- Everhart, J.E.; Ruhl, C.E. Burden of Digestive Diseases in the United States Part III: Liver, Biliary Tract, and Pancreas. Gastroenterology 2009, 136, 1134–1144. [Google Scholar] [CrossRef]
- Ilyas, S.I.; Khan, S.A.; Hallemeier, C.L.; Kelley, R.K.; Gores, G.J.; Rizvi, S. Cholangiocarcinoma—Evolving concepts and therapeutic strategies. Nat. Rev. Clin. Oncol. 2018, 15, 95–111. [Google Scholar] [CrossRef]
- Liu, R.; Zhao, R.; Zhou, X.; Liang, X.; Campbell, D.J.; Zhang, X.; Zhang, L.; Shi, R.; Wang, G.; Pandak, W.M.; et al. Conjugated bile acids promote cholangiocarcinoma cell invasive growth through activation of sphingosine 1-phosphate receptor 2. Hepatology 2014, 60, 908–918. [Google Scholar] [CrossRef]
- Wadsworth, C.A.; Dixon, P.H.; Wong, J.H.; Chapman, M.H.; McKay, S.C.; Sharif, A.; Spalding, D.R.; Pereira, S.P.; Thomas, H.C.; Taylor-Robinson, S.D.; et al. Genetic factors in the pathogenesis of cholangiocarcinoma. Dig. Dis. 2011, 29, 93–97. [Google Scholar] [CrossRef]
- Sripa, B.; Kaewkes, S.; Sithithaworn, P.; Mairiang, E.; Laha, T.; Smout, M.; Pairojkul, C.; Bhudhisawasdi, V.; Tesana, S.; Thinkamrop, B.; et al. Liver fluke induces cholangiocarcinoma. PLoS Med. 2007, 4, e201. [Google Scholar] [CrossRef]
- Ito, Z.; Koido, S.; Kato, K.; Odamaki, T.; Horiuchi, S.; Akasu, T.; Saruta, M.; Hata, T.; Kumagai, Y.; Fujioka, S.; et al. Dysbiosis of the Fecal and Biliary Microbiota in Biliary Tract Cancer. Cancers 2022, 14, 5379. [Google Scholar] [CrossRef]
- Higuchi, R.; Takada, T.; Strasberg, S.M.; Pitt, H.A.; Gouma, D.J.; Garden, O.J.; Büchler, M.W.; Windsor, J.A.; Mayumi, T.; Yoshida, M.; et al. TG13 miscellaneous etiology of cholangitis and cholecystitis. J. Hepatobiliary Pancreat. Sci. 2013, 20, 97–105. [Google Scholar] [CrossRef]
- Di Carlo, P.; Serra, N.; Fasciana, T.M.A.; Giammanco, A.; D’aRpa, F.; Rea, T.; Napolitano, M.S.; Lucchesi, A.; Cascio, A.; Sergi, C.M.; et al. Microbial profile in bile from pancreatic and extra-pancreatic biliary tract cancer. PLoS ONE 2024, 19, e0294049. [Google Scholar] [CrossRef]
- Li, Z.; Chu, J.; Su, F.; Ding, X.; Zhang, Y.; Dou, L.; Liu, Y.; Ke, Y.; Liu, X.; Liu, Y.; et al. Characteristics of bile microbiota in cholelithiasis, perihilar cholangiocarcinoma, distal cholangiocarcinoma, and pancreatic cancer. Am. J. Transl. Res. 2022, 14, 2962–2971. [Google Scholar] [PubMed]
- Avilés-Jiménez, F.; Guitron, A.; Segura-López, F.; Méndez-Tenorio, A.; Iwai, S.; Hernández-Guerrero, A.; Torres, J. Microbiota studies in the bile duct strongly suggest a role for Helicobacter pylori in extrahepatic cholangiocarcinoma. Clin. Microbiol. Infect. 2016, 22, 178.e11–178.e22. [Google Scholar] [CrossRef] [PubMed]
- Sitthirak, S.; Suksawat, M.; Phetcharaburanin, J.; Wangwiwatsin, A.; Klanrit, P.; Namwat, N.; Khuntikeo, N.; Titapun, A.; Jarearnrat, A.; Sangkhamanon, S.; et al. Chemotherapeutic resistant cholangiocarcinoma displayed distinct intratumoral microbial composition and metabolic profiles. PeerJ 2022, 10, e13876. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Sunkara, T.; Thandra, K.C.; Barsouk, A. Epidemiology of gallbladder cancer. Clin. Exp. Hepatol. 2019, 5, 93–102. [Google Scholar] [CrossRef]
- Misra, S.P.; Dwivedi, M. Pancreaticobiliary ductal union. Gut 1990, 31, 1144–1149. [Google Scholar] [CrossRef]
- Takayama, S.; Takahashi, H.; Matsuo, Y.; Okada, Y.; Takeyama, H. Effect of Helicobacter bilis infection on human bile duct cancer cells. Dig. Dis. Sci. 2010, 55, 1905–1910. [Google Scholar] [CrossRef]
- Murata, H.; Tsuji, S.; Tsujii, M.; Fu, H.Y.; Tanimura, H.; Tsujimoto, M.; Matsuura, N.; Kawano, S.; Hori, M. Helicobacter bilis infection in biliary tract cancer. Aliment. Pharmacol. Ther. 2004, 20 (Suppl. S1), 90–94. [Google Scholar] [CrossRef]
- Caygill, C.P.; Hill, M.J.; Braddick, M.; Sharp, J. Cancer mortality in chronic typhoid and paratyphoid carriers. Lancet 1994, 343, 83–84. [Google Scholar] [CrossRef] [PubMed]
- Lara-Tejero, M.; Galán, J.E. A bacterial toxin that controls cell cycle progression as a deoxyribonuclease I-like protein. Science 2000, 290, 354–357. [Google Scholar] [CrossRef]
- Haghjoo, E.; Galán, J.E. Salmonella typhi encodes a functional cytolethal distending toxin that is delivered into host cells by a bacterial-internalization pathway. Proc. Natl. Acad. Sci. USA 2004, 101, 4614–4619. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, Y.; Loza, E.; Villa-Gomez, G.; Trujillo, C.C.; Baez, S.; Asai, T.; Ikoma, T.; Endoh, K.; Nakamura, K. Metagenomics of Microbial Communities in Gallbladder Bile from Patients with Gallbladder Cancer or Cholelithiasis. Asian Pac. J. Cancer Prev. 2018, 19, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.J.; Kim, Y.; Jeon, J.; Gwak, H.-J.; Kim, M.; Kang, K.; Kim, Y.; Jeong, J.; Jung, Y.K.; Lee, K.G.; et al. Association of Microbial Dysbiosis with Gallbladder Diseases Identified by Bile Microbiome Profiling. J. Korean Med. Sci. 2021, 36, e189. [Google Scholar] [CrossRef]
- Song, X.; Wang, X.; Hu, Y.; Li, H.; Ren, T.; Li, Y.; Liu, L.; Li, L.; Li, X.; Wang, Z.; et al. A metagenomic study of biliary microbiome change along the cholecystitis-carcinoma sequence. Clin. Transl. Med. 2020, 10, e97. [Google Scholar] [CrossRef]
- Dell’Anna, G.; Ogura, T.; Vanella, G.; Nishikawa, H.; Lakhtakia, S.; Arcidiacono, P.G. Endoscopic ultrasound guided biliary interventions. Best Pract. Res. Clin. Gastroenterol. 2022, 60–61, 101810. [Google Scholar] [CrossRef]
- Dell’Anna, G.; Nunziata, R.; Delogu, C.; Porta, P.; Grassini, M.V.; Dhar, J.; Barà, R.; Bencardino, S.; Fanizza, J.; Mandarino, F.V.; et al. The Role of Therapeutic Endoscopic Ultrasound in Management of Malignant Double Obstruction (Biliary and Gastric Outlet): A Comprehensive Review with Clinical Scenarios. J. Clin. Med. 2024, 13, 7731. [Google Scholar] [CrossRef]
| Reference (Year) | Study Population | Sampling Method and Sequencing Approach | Principal Taxa Increased in CCA (vs. Benign Disease) | Reported Diagnostic or Mechanistic Significance |
|---|---|---|---|---|
| Ito et al., 2020 [115] | 41 malignant biliary strictures (mixed iCCA + pCCA + dCCA) vs. 33 benign strictures | ERC-aspirated bile; 16S rRNA metagenomics | Enterobacteriaceae family; colibactin-positive E. coli | DNA damage-mediated carcinogenesis; Enterobacteriaceae abundance as malignant biomarker |
| Chen et al., 2021 [7] | 30 distal CCA vs. 30 choledocholithiasis | Intraoperative bile; 16S rRNA NGS | Enterobacteriaceae, Staphylococcus, Klebsiella, Faecalibacterium, Corynebacterium | Enterobacteriaceae enrichment plus higher α-diversity differentiate dCCA from benign obstructive disease |
| Di Carlo et al., 2022 [117] | 50 extra-pancreatic biliary cancers (34 CCA, 16 ampullary) vs. 58 pancreatic cancers | ERC bile; 16S rRNA NGS | Alcaligenes faecalis ↑, Acinetobacter spp. ↓ in CCA | Microbial signature improves discrimination between CCA and pancreatic ductal adenocarcinoma |
| Li et al., 2023 [118] | 47 perihilar CCA, 29 distal CCA, 38 pancreatic cancer | ERC bile; 16S rRNA NGS | pCCA: Pseudomonas, Sphingomonas, Halomonas dCCA: Streptococcus, Prevotella, Halomonas | Distinct taxa panels permit molecular subclassification of malignant biliary strictures |
| Avilés-Jiménez et al., 2023 [119] | 100 CCA (mixed sites) vs. 100 benign biliary disease | ERC brushings; 16S rRNA + qPCR for Helicobacter | Helicobacter, Campylobacter, Prevotella, Fusobacterium, Methylophilaceae | Chronic H. pylori colonization with virulence factors may drive progression from benign inflammation to CCA |
| Sitthirak et al. [120] | 62 CCA tumor specimens pre-chemotherapy | Shotgun metagenomics of tumor tissue | Gammaproteobacteria over-represented in gemcitabine/cisplatin-refractory tumors | Bacterial metabolic inactivation of nucleoside analogs underlies primary chemoresistance |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meacci, D.; Bruni, A.; Cocquio, A.; Dell’Anna, G.; Mandarino, F.V.; Marasco, G.; Cecinato, P.; Barbara, G.; Zagari, R.M. Microbial Landscapes of the Gut–Biliary Axis: Implications for Benign and Malignant Biliary Tract Diseases. Microorganisms 2025, 13, 1980. https://doi.org/10.3390/microorganisms13091980
Meacci D, Bruni A, Cocquio A, Dell’Anna G, Mandarino FV, Marasco G, Cecinato P, Barbara G, Zagari RM. Microbial Landscapes of the Gut–Biliary Axis: Implications for Benign and Malignant Biliary Tract Diseases. Microorganisms. 2025; 13(9):1980. https://doi.org/10.3390/microorganisms13091980
Chicago/Turabian StyleMeacci, David, Angelo Bruni, Alice Cocquio, Giuseppe Dell’Anna, Francesco Vito Mandarino, Giovanni Marasco, Paolo Cecinato, Giovanni Barbara, and Rocco Maurizio Zagari. 2025. "Microbial Landscapes of the Gut–Biliary Axis: Implications for Benign and Malignant Biliary Tract Diseases" Microorganisms 13, no. 9: 1980. https://doi.org/10.3390/microorganisms13091980
APA StyleMeacci, D., Bruni, A., Cocquio, A., Dell’Anna, G., Mandarino, F. V., Marasco, G., Cecinato, P., Barbara, G., & Zagari, R. M. (2025). Microbial Landscapes of the Gut–Biliary Axis: Implications for Benign and Malignant Biliary Tract Diseases. Microorganisms, 13(9), 1980. https://doi.org/10.3390/microorganisms13091980








