Helicobacter pylori Diagnostic Testing Accuracy in a High-Prevalence Native American Population of Northern Arizona
Abstract
1. Introduction
2. Materials and Methods
2.1. Setting and Subjects
2.2. Sample Testing
2.3. Medical Record Abstraction
2.4. Diagnostic Risk Factors
2.5. Data Analysis
2.6. Ethical Approvals
2.7. Patient and Public Involvement
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wroblewski, L.E.; Peek, R.M.; Wilson, K.T. Helicobacter pylori and gastric cancer: Factors that modulate disease risk. Clin. Microbiol. Rev. 2010, 23, 713–739. [Google Scholar] [CrossRef]
- Hooi, J.K.Y.; Lai, W.Y.; Ng, W.K.; Suen, M.M.Y.; Underwood, F.E.; Tanyingoh, D.; Malfertheiner, P.; Graham, D.Y.; Wong, V.W.S.; Wu, J.C.Y.; et al. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology 2017, 153, 420–429. [Google Scholar] [CrossRef]
- Harris, R.B.; Brown, H.E.; Begay, R.L.; Sanderson, P.R.; Chief, C.; Monroy, F.P.; Oren, E. Helicobacter pylori prevalence and risk factors in three rural Indigenous communities of Northern Arizona. Int. J. Environ. Res. Public Health 2022, 19, 797. [Google Scholar] [CrossRef]
- Navajo Epidemiology Center. Cancer Among the Navajo 2005–2013; Navajo Epidemiology Center: Window Rock, AZ, USA, 2018. [Google Scholar]
- Stancioiu, F.; Ahmed, S.H.; Buschor, R. Helicobacter pylori: Findings in a Native American Population. IHS Prim. Care Provid. 2005, 30, 59–63. [Google Scholar]
- Shirani, M.; Pakzad, R.; Haddadi, M.H.; Akrami, S.; Asadi, A.; Kazemian, H.; Moradi, M.; Kaviar, V.H.; Zomorodi, A.R.; Khoshnood, S.; et al. The global prevalence of gastric cancer in Helicobacter pylori-infected individuals: A systematic review and meta-analysis. BMC Infect. Dis. 2023, 23, 1–30. [Google Scholar] [CrossRef]
- Uotani, T.; Graham, D.Y. Diagnosis of Helicobacter pylori using the rapid urease test. Ann. Transl. Med. 2015, 3, 9. [Google Scholar]
- Lan, H.C.; Chen, T.S.; Li, A.F.Y.; Chang, F.Y.; Lin, H.C. Additional corpus biopsy enhances the detection of Helicobacter pylori infection in a background of gastritis with atrophy. BMC Gastroenterol. 2012, 12, 182. [Google Scholar] [CrossRef]
- Choi, Y.J.; Kim, N.; Lim, J.; Jo, S.Y.; Shin, C.M.; Lee, H.S.; Park, Y.S.; Hwang, J.-H.; Kim, J.-W.; Jeong, S.-H.; et al. Accuracy of diagnostic tests for Helicobacter pylori in patients with peptic ulcer bleeding. Helicobacter 2012, 17, 77–85. [Google Scholar] [CrossRef]
- Tu, T.C.; Lee, C.L.; Wu, C.H.; Chen, T.K.; Chan, C.C.; Huang, S.H.; Lee, S.C. Comparison of invasive and noninvasive tests for detecting Helicobacter pylori infection in bleeding peptic ulcers. Gastrointest. Endosc. 1999, 49 Pt 1, 302–306. [Google Scholar] [CrossRef]
- Gisbert, J.P.; Abraira, V. Accuracy of Helicobacter pylori diagnostic tests in patients with bleeding peptic ulcer: A systematic review and meta-analysis. Am. J. Gastroenterol. 2006, 101, 848–863. [Google Scholar] [CrossRef]
- Kokkola, A.; Rautelin, H.; Puolakkainen, P.; Sipponen, P.; Färkkilä, M.; Haapiainen, R.; Kosunen, T.U. Diagnosis of Helicobacter pylori infection in patients with atrophic gastritis: Comparison of histology, 13C-urea breath test, and serology. Scand. J. Gastroenterol. 2000, 35, 138–141. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kim, N. Diagnosis of Helicobacter pylori by invasive test: Histology. Ann. Transl. Med. 2015, 3, 10. [Google Scholar]
- Griñó, P.; Pascual, S.; Such, J.; Casellas, J.A.; Niveiro, M.; Andreu, M.; Pérez-Mateo, P. Comparison of diagnostic methods for Helicobacter pylori infection in patients with upper gastrointestinal bleeding. Scand. J. Gastroenterol. 2001, 36, 1254–1258. [Google Scholar] [CrossRef]
- Archimandritis, A.; Tzivras, M.; Sougioultzis, S.; Papaparaskevas, I.; Apostolopoulos, P.; Avlami, A.; Davaris, P. Rapid urease test is less sensitive than histology in diagnosing Helicobacter pylori infection in patients with non-variceal upper gastrointestinal bleeding. J. Gastroenterol. Hepatol. 2000, 15, 369–373. [Google Scholar] [CrossRef]
- Hoover, J.; Gonzales, M.; Shuey, C.; Barney, Y.; Lewis, J. Elevated arsenic and uranium concentrations in unregulated water sources on the Navajo Nation, USA. Expo. Health 2017, 9, 113–124. [Google Scholar] [CrossRef]
- Jeong, C.H.; Seok, J.S.; Petriello, M.C.; Han, S.G. Arsenic downregulates tight junction claudin proteins through p38 and NF-κB in intestinal epithelial cell line, HT-29. Toxicology 2017, 379, 31–39. [Google Scholar] [CrossRef]
- Rahman, M.M.; Chowdhury, U.K.; Mukherjee, S.C.; Mondal, B.K.; Paul, K.; Lodh, D.; Biswas, B.K.; Chanda, C.R.; Basu, G.K.; Saha, K.C.; et al. Chronic Arsenic Toxicity in Bangladesh and West Bengal, India—A Review and Commentary. J. Toxicol. Clin. Toxicol. 2001, 39, 683–700. [Google Scholar] [CrossRef]
- Weather. Navajo National Monument, AZ Climate Averages, Monthly Weather Conditions [Internet]. Available online: https://www.weatherworld.com/climate-averages/az/navajo+national+monument.html (accessed on 18 March 2024).
- Monroy, F.P.; Brown, H.E.; Sanderson, P.R.; Jarrin, G.; Mbegbu, M.; Kyman, S.; Harris, R.B. Helicobacter pylori in Native Americans in Northern Arizona. Diseases 2022, 10, 19. [Google Scholar] [CrossRef]
- WHO. A Healthy Lifestyle—WHO Recommendations [Internet]. 2024. Available online: https://www.who.int/europe/news-room/fact-sheets/item/a-healthy-lifestyle---who-recommendations (accessed on 2 May 2024).
- Patel, S.K.; Pratap, C.B.; Jain, A.K.; Gulati, A.K.; Nath, G. Diagnosis of Helicobacter pylori: What should be the gold standard? World J. Gastroenterol. WJG 2014, 20, 12847. [Google Scholar] [CrossRef]
- Hunt, R.; Xiao, S.; Megraud, F.; Leon-Barua, R.; Bazzoli, F.; van der Merwe, S.; Vaz Coelho, L.G.; Fock, M.; Fedail, S.; Cohen, H.; et al. World Gastroenterology Organisation Global Guideline Helicobacter pylori in Developing Countries. J. Dig. Dis. 2011, 12, 319–329. [Google Scholar] [CrossRef]
- Chey, W.D.; Wong, B.C.Y. American College of Gastroenterology guideline on the management of Helicobacter pylori infection. Am. J. Gastroenterol. 2007, 102, 1808–1825. [Google Scholar] [CrossRef]
- Miftahussurur, M.; Yamaoka, Y. Diagnostic Methods of Helicobacter pylori Infection for Epidemiological Studies: Critical Importance of Indirect Test Validation. BioMed Res. Int. 2016, 2016, 4819423. [Google Scholar] [CrossRef]
- Bruden, D.L.; Bruce, M.G.; Miernyk, K.M.; Morris, J.; Hurlburt, D.; Hennessy, T.W.; Peters, H.; Sacco, F.; Parkinson, A.J.; McMahon, B.J. Diagnostic accuracy of tests for Helicobacter pylori in an Alaska Native population. World J. Gastroenterol. 2011, 17, 4682–4688. [Google Scholar] [CrossRef]
- Mounsey, A.; Leonard, E.A. Noninvasive Diagnostic Tests for Helicobacter pylori Infection. Am. Fam. Physician 2019, 100, 16–17. [Google Scholar]
- Cardos, A.I.; Maghiar, A.; Zaha, D.C.; Pop, O.; Fritea, L.; Miere, F.; Cavalu, S. Evolution of Diagnostic Methods for Helicobacter pylori Infections: From Traditional Tests to High Technology, Advanced Sensitivity and Discrimination Tools. Diagnostics 2022, 12, 508. [Google Scholar] [CrossRef]
- Landis, J.R.; Koch, G.G. The measurement of observer agreement for categorical data. Biometrics 1977, 33, 159. [Google Scholar] [CrossRef]
- Nagy, P.; Johansson, S.; Molloy-Bland, M. Systematic review of time trends in the prevalence of Helicobacter pylori infection in China and the USA. Gut Pathog. 2016, 8, 8. [Google Scholar] [CrossRef]
- Ferro, A.; Morais, S.; Pelucchi, C.; Dierssen-Sotos, T.; Martín, V.; López-Carrillo, L.; Malekzadeh, R.; Tsugane, S.; Hamada, G.; Hidaka, A.; et al. Sex differences in the prevalence of Helicobacter pylori infection. Eur. J. Gastroenterol. Hepatol. 2019, 31, 593–598. [Google Scholar] [CrossRef]
- Chey, W.D.; Leontiadis, G.I.; Howden, C.W.; Moss, S.F. ACG Clinical Guideline: Treatment of Helicobacter pylori Infection. Am. J. Gastroenterol. 2017, 112, 212–238. [Google Scholar] [CrossRef]
- Bailey, K.S.; Brown, H.E.; Lekic, V.; Pradeep, K.; Merchant, J.L.; Harris, R.B. Helicobacter pylori treatment knowledge, access and barriers: A cross-sectional study. Helicobacter 2023, 28, e12954. [Google Scholar] [CrossRef]
- Hulten, K.G.; Lamberth, L.B.; Kalfus, I.N.; Graham, D.Y. National and Regional US Antibiotic Resistance to Helicobacter pylori: Lessons From a Clinical Trial. Gastroenterology 2021, 161, 342–344.e1. [Google Scholar] [CrossRef] [PubMed]
- Monroy, F.P.; Brown, H.E.; Acevedo-Solis, C.M.; Rodriguez-Galaviz, A.; Dholakia, R.; Pauli, L.; Harris, R.B. Antibiotic resistance rates for Helicobacter pylori in rural Arizona: A molecular-based study. Microorganisms 2023, 11, 2290. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.G.; Park, R.W.; Shin, S.J.; Yoon, D.; Kang, J.K.; Hwang, J.C.; Kim, S.S.; Kim, J.H.; Lee, K.M. The relationship between the failure to eradicate Helicobacter pylori and previous antibiotics use. Dig. Liver Dis. 2016, 48, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Dixon, M.F.; Genta, R.M.; Yardley, J.H.; Correa, P.; Batts, K.P.; Dahms, B.B.; Filipe, M.I.; Haggitt, R.C.; Haot, J.; Hui, P.K.; et al. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am. J. Surg. Pathol. 1996, 20, 1161–1181. [Google Scholar] [CrossRef] [PubMed]
- Malfertheiner, P.; Megraud, F.; Rokkas, T.; Gisbert, J.P.; Liou, J.M.; Schulz, C.; Gasbarrini, A.; Hunt, R.H.; Leja, M.; O’Morain, C.; et al. Management of Helicobacter pylori infection: The Maastricht VI/Florence consensus report. Gut 2022, 71, 1724–1762. [Google Scholar] [CrossRef]

| Total * | H. pylori + | H. pylori − | Statistics | |
|---|---|---|---|---|
| (N = 399) | (N = 108) | (N = 291) | ||
| Age (mean (sd)) | 56.2 (14.2) | 52.5 (15.6) | 57.6 (13.5) | t = 3.2, p = 0.001 |
| Missing (n = 2) | ||||
| Age Group, years (n (%)) | X2 = 14.6, p = 0.006 | |||
| 18–34 | 39 (9.8%) | 18 (16.7%) | 21 (7.3%) | |
| 35–44 | 44 (11.1%) | 18 (16.7%) | 26 (9.0%) | |
| 45–54 | 80 (20.2%) | 18 (16.7%) | 62 (21.5%) | |
| 55–64. | 103 (25.9%) | 26 (24.1%) | 77 (26.6%) | |
| 65+ | 131 (33.0%) | 28 (25.9%) | 103 (35.6%) | |
| Missing (n = 2) | ||||
| Tribal Affiliation (n (%)) | X2 = 3.5, p = 0.17 | |||
| Navajo | 377 (95.7%) | 102 (96.2%) | 275 (95.5%) | |
| Hopi | 14 (3.6%) | 2 (1.9%) | 12 (4.2%) | |
| Other | 3 (0.8%) | 2 (1.9%) | 1 (0.4%) | |
| Missing (n = 5) | ||||
| Sex (n (%)) | X2 = 7.3, p = 0.007 | |||
| Female | 290 (72.9%) | 68 (63.0%) | 222 (76.6%) | |
| Male | 108 (27.1%) | 40 (37.0%) | 68 (23.4%) | |
| Missing (n = 1) | ||||
| BMI (n (%)) | X2 = 5.3, p = 0.15 | |||
| Underweight (<18.5) | 6 (1.6%) | 3 (2.9%) | 3 (1.1%) | |
| Normal (18.5–24.9) | 42 (10.8%) | 13 (12.4%) | 29 (10.3%) | |
| Overweight (25–29.9) | 125 (32.3%) | 40 (38.1%) | 85 (30.1%) | |
| Obese (>30) | 214 (55.3%) | 49 (46.7%) | 165 (58.5%) | |
| Missing (n = 12) | ||||
| Current Tobacco Use (smoke &/or smokeless) (n (%)) | X2 = 0.91, p = 0.63 | |||
| Yes | 52 (13.1%) | 17 (15.7%) | 35 (12.2%) | |
| No | 278 (70.2%) | 73 (67.6%) | 205 (71.2%) | |
| Ceremonial Use only | 66 (16.7%) | 18 (16.7%) | 48 (16.7%) | |
| Missing (n = 3) | ||||
| GI Bleed Indicated (anemia, blood in vomit or stool) (n (%)) | X2 = 0.58, p = 0.44 | |||
| Yes | 71 (17.8%) | 22 (20.2%) | 49 (16.9%) | |
| No | 328 (82.2%) | 87 (79.8%) | 241 (83.1%) | |
| Missing (n = 0) | ||||
| History of Helicobacter pylori infection (n (%)) | X2 = 1.34, p = 0.25 | |||
| Yes | 165 (41.4%) | 40 (36.7%) | 125 (43.1%) | |
| No | 234 (58.7%) | 69 (63.3%) | 165 (56.9%) | |
| Missing (n = 0) | ||||
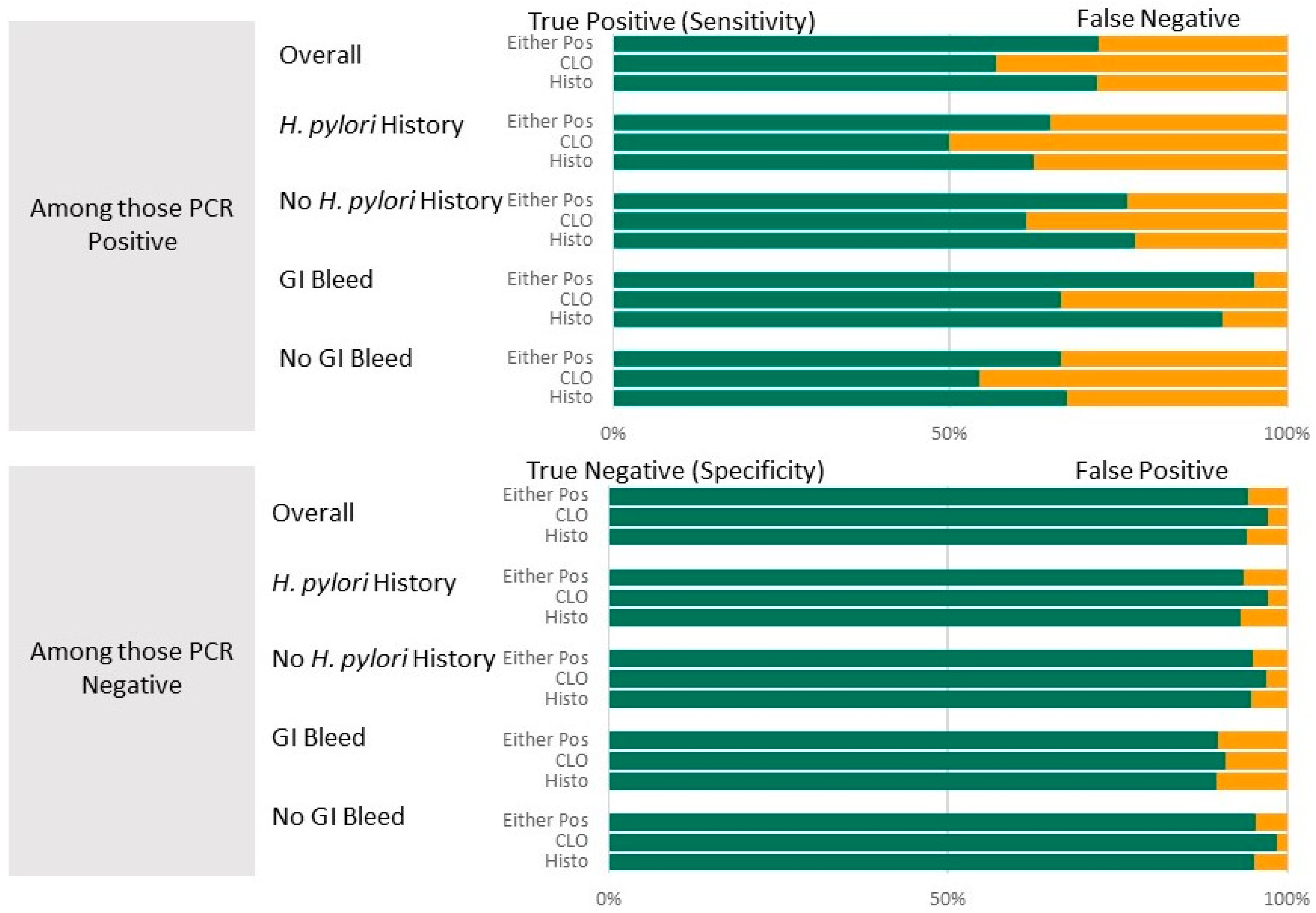
| Diagnostic Test | Sensitivity% (95% CI) | Specificity% (95% CI) | PPV% (95% CI) | NPV % (95% CI) | ROC Area Value (95% CI) | Agreement% | Kappa |
|---|---|---|---|---|---|---|---|
| Histopathology | |||||||
| Overall (n = 384) | 72.0 (62.5, 80.2) | 94.2 (90.8, 96.7) | 78.8 (69.5, 85.9) | 91.8 (89.2, 93.9) | 0.831 (0.786, 0.876) | 87.9 | 0.68, p < 0.001 |
| Hp HX * (n = 160) | 62.5 (45.8, 77.3) | 93.3 (87.3, 97.1) | 73.7 (57.9, 85.1) | 89.3 (84.8, 92.6) | 0.779 (0.700, 0.858) | 85.6 | 0.59, p < 0.001 |
| No Hp Hx (n = 224) | 77.6 (65.8, 86.9) | 94.9 (90.2, 97.8) | 82.0 (69.6, 90.0) | 93.4 (90.1, 95.7) | 0.863 (0.809, 0.916) | 89.7 | 0.77, p < 0.001 |
| GI Bleed * (n = 69) | 90.5 (69.6, 98.8) | 89.6 (77.3, 96.5) | 72.2 (52.8, 85.7) | 96.9 (89.4, 99.2) | 0.900 (0.823, 0.978) | 89.9 | 0.77, p < 0.001 |
| No GI Bleed (n = 315) | 67.4 (56.5, 77.2) | 95.2 (91.6, 97.6) | 80.7 (69.8, 88.4) | 90.7 (87.8, 93.0) | 0.813 (0.761, 0.865) | 87.6 | 0.67, p < 0.001 |
| CLO | |||||||
| Overall (n = 337) | 57.0 (45.8, 67.6) | 97.2 (94.3, 98.9) | 85.9 (74.2, 92.8) | 88.3 (85.6, 90.6) | 0.771 (0.717, 0.825) | 86.9 | 0.61, p < 0.001 |
| Hp HX (n = 146) | 50.0 (32.4, 67.6) | 97.3 (92.4, 99.4) | 84.8 (63.5, 94.7) | 86.7 (82.3, 90.1) | 0.737 (0.650, 0.823) | 86.3 | 0.55, p < 0.001 |
| No Hp Hx (n = 191) * | 61.5 (44.0, 74.7) | 97.1 (92.8, 99.2) | 86.5 (70.4, 94.5) | 89.4 (85.7, 92.3) | 0.793 (0.725, 0.862) | 87.3 | 0.65, p < 0.001 |
| GI Bleed * (n = 63) | 66.7 (41.0, 86.7) | 91.1 (78.8, 97.5) | 69.1 (45.4, 85.8) | 90.1 (82.6, 94.7) | 0.789 (0.669, 0.909) | 84.1 | 0.60, p < 0.001 |
| No GI Bleed * (n = 274) | 54.4 (41.9, 66.5) | 98.5 (95.8, 99.7) | 91.8 (78.0, 97.2) | 87.9 (84.9, 90.4) | 0.765 (0.705, 0.825) | 87.6 | 0.61, p < 0.001 |
| Combination (Histopathology or CLO Positive treated as positive) | |||||||
| Overall * (n = 397) | 72.2 (62.8, 80.4) | 94.5 (91.1; 96.8) | 79.6 (70.5, 86.4) | 91.9 (89.3, 93.9) | 0.833 (0.789, 0.878) | 88.4 | 0.70, p < 0.001 |
| Hp HX * (n = 165) | 65.0 (48.3, 79.4) | 93.6 (87.8, 97.4) | 75.2 (59.9, 86.0) | 90.0 (85.4, 93.2) | 0.793 (0.715, 0.871) | 86.7 | 0.62, p < 0.001 |
| No Hp Hx * (n = 232) | 76.5 (64.6, 85.9) | 95.1 (90.6, 97.9) | 82.4 (70.2, 90.3) | 93.1 (89.8, 95.4) | 0.858 (0.805, 0.911) | 89.7 | 0.74, p < 0.001 |
| GI Bleed * (n = 70) | 95.2 (76.2; 99.9) | 89.8 (77.8; 96.6) | 73.6 (54.7, 86.5) | 98.4 (90.3, 99.8) | 0.925 (0.862, 0.989) | 91.4 | 0.81, p < 0.001 |
| No GI Bleed * (n = 327) | 66.7 (55.7, 76.4) | 95.4 (91.9, 97.7) | 81.3 (70.5, 88.7) | 90.6 (87.7, 92.8) | 0.810 (0.759, 0.862) | 87.8 | 0.66, p < 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brown, H.E.; Pauli, L.; Dholakia, R.; Gunderson, J.; Jernberg, J.; Sanderson, P.R.; Harris, R.B.; Monroy, F.P. Helicobacter pylori Diagnostic Testing Accuracy in a High-Prevalence Native American Population of Northern Arizona. Microorganisms 2025, 13, 1920. https://doi.org/10.3390/microorganisms13081920
Brown HE, Pauli L, Dholakia R, Gunderson J, Jernberg J, Sanderson PR, Harris RB, Monroy FP. Helicobacter pylori Diagnostic Testing Accuracy in a High-Prevalence Native American Population of Northern Arizona. Microorganisms. 2025; 13(8):1920. https://doi.org/10.3390/microorganisms13081920
Chicago/Turabian StyleBrown, Heidi E., Laura Pauli, Rishi Dholakia, Joseph Gunderson, Julia Jernberg, Priscilla R. Sanderson, Robin B. Harris, and Fernando P. Monroy. 2025. "Helicobacter pylori Diagnostic Testing Accuracy in a High-Prevalence Native American Population of Northern Arizona" Microorganisms 13, no. 8: 1920. https://doi.org/10.3390/microorganisms13081920
APA StyleBrown, H. E., Pauli, L., Dholakia, R., Gunderson, J., Jernberg, J., Sanderson, P. R., Harris, R. B., & Monroy, F. P. (2025). Helicobacter pylori Diagnostic Testing Accuracy in a High-Prevalence Native American Population of Northern Arizona. Microorganisms, 13(8), 1920. https://doi.org/10.3390/microorganisms13081920






