The Molecular Epidemiology of Hepatitis B Virus and Its Resistance-Associated Mutations in the Polymerase Gene in the Americas
Abstract
1. Introduction
2. Methods
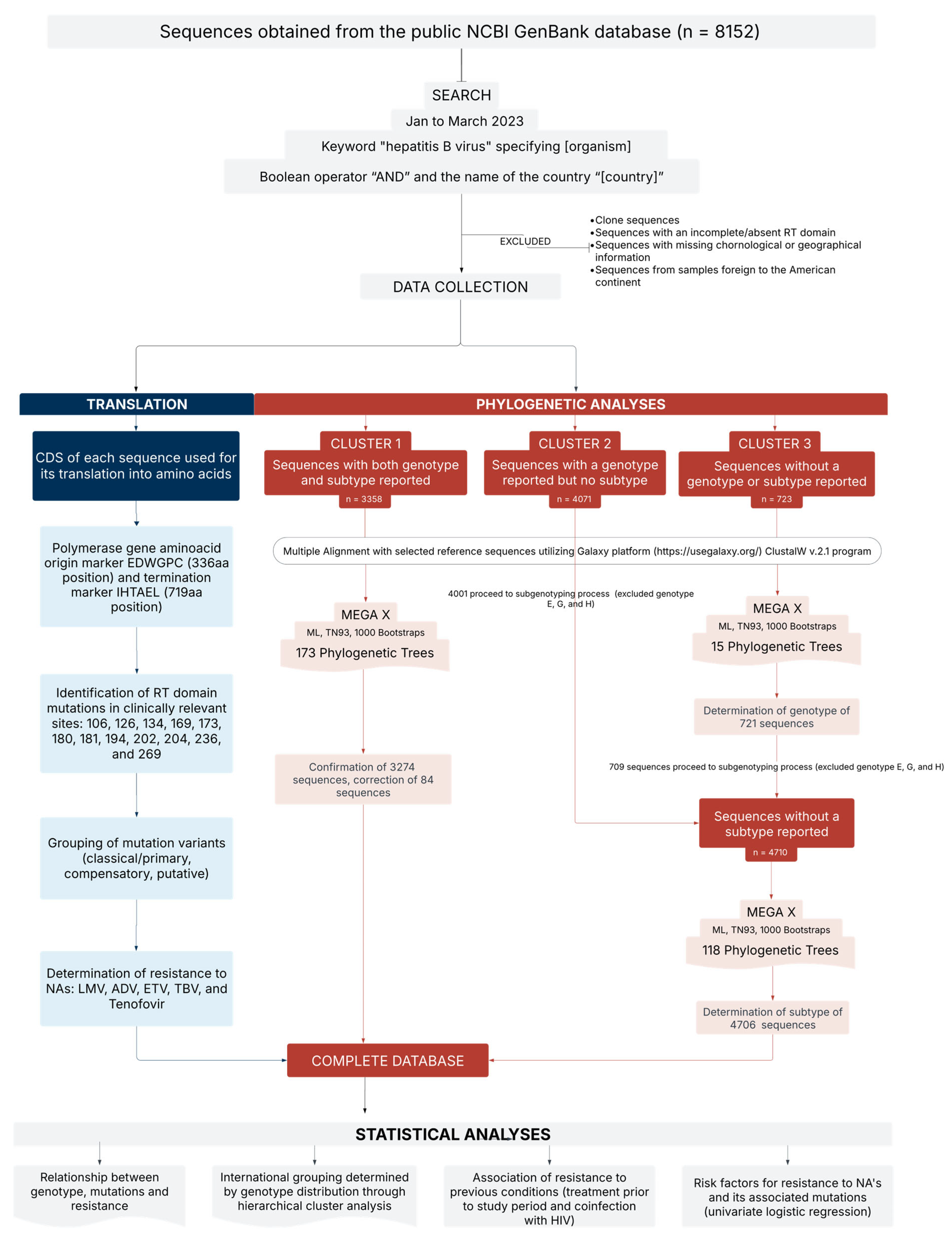
2.1. Study Design
2.2. Sequence Classification and Phylogenetic Analysis
- Cluster 1: Genotype and subtype identified in GenBank or associated publication (n = 3358).
- Cluster 2: Genotype provided; subtype not reported (n = 4071).
- Cluster 3: Neither genotype nor subtype reported (n = 723).
2.3. Analysis of Resistance-Associated Mutations
2.4. Statistical Analysis
3. Results
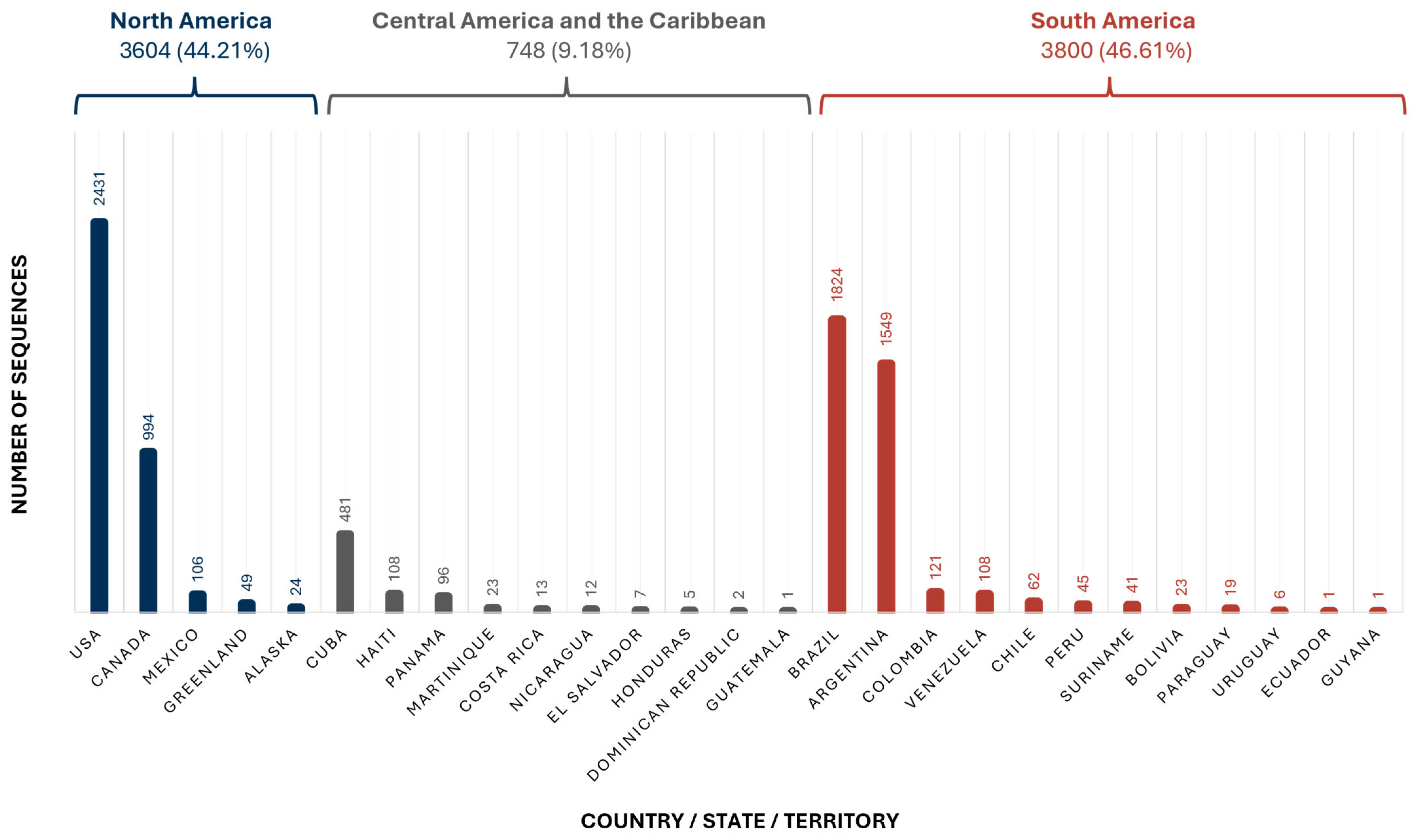
3.1. Geographical Distribution
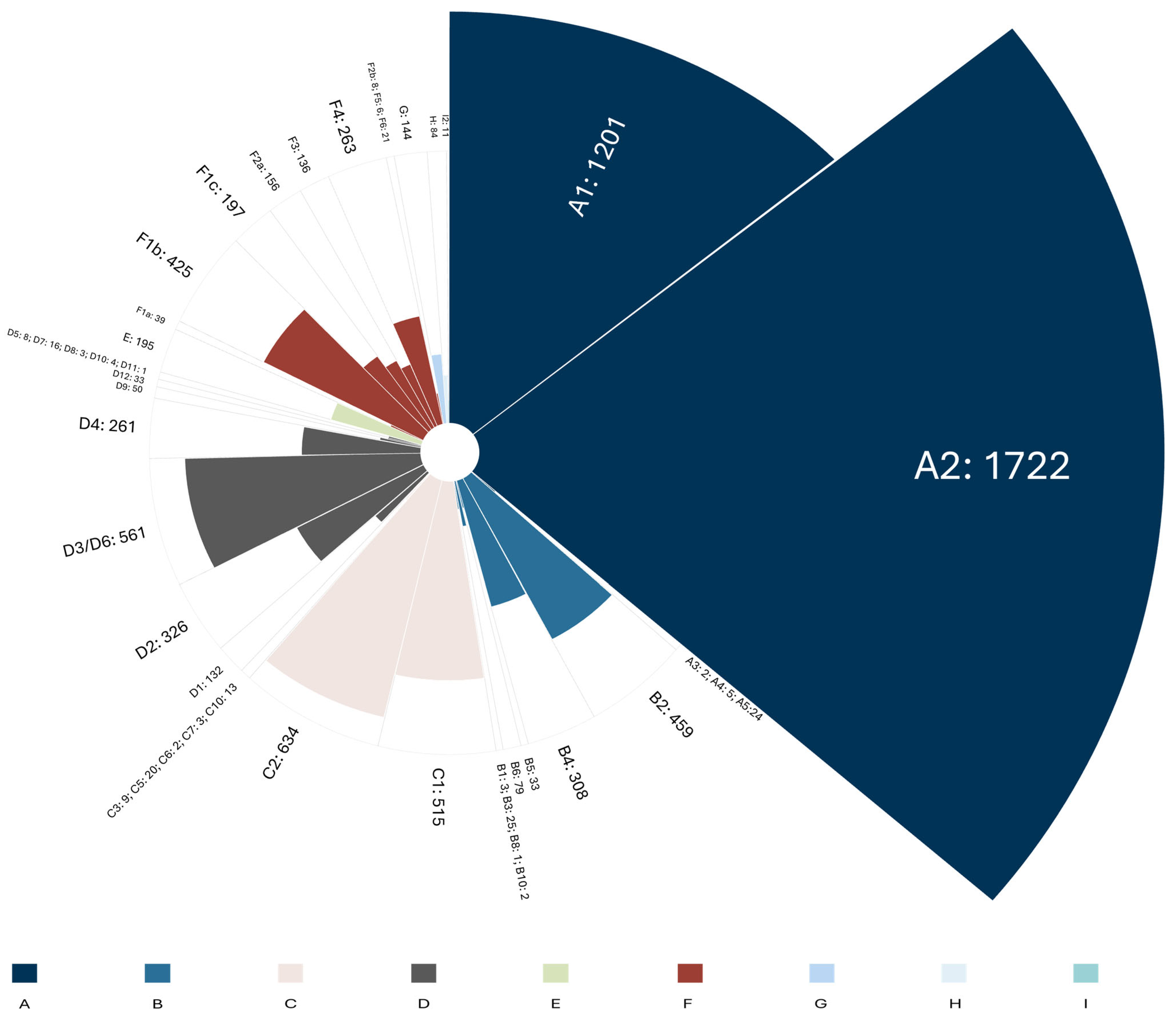
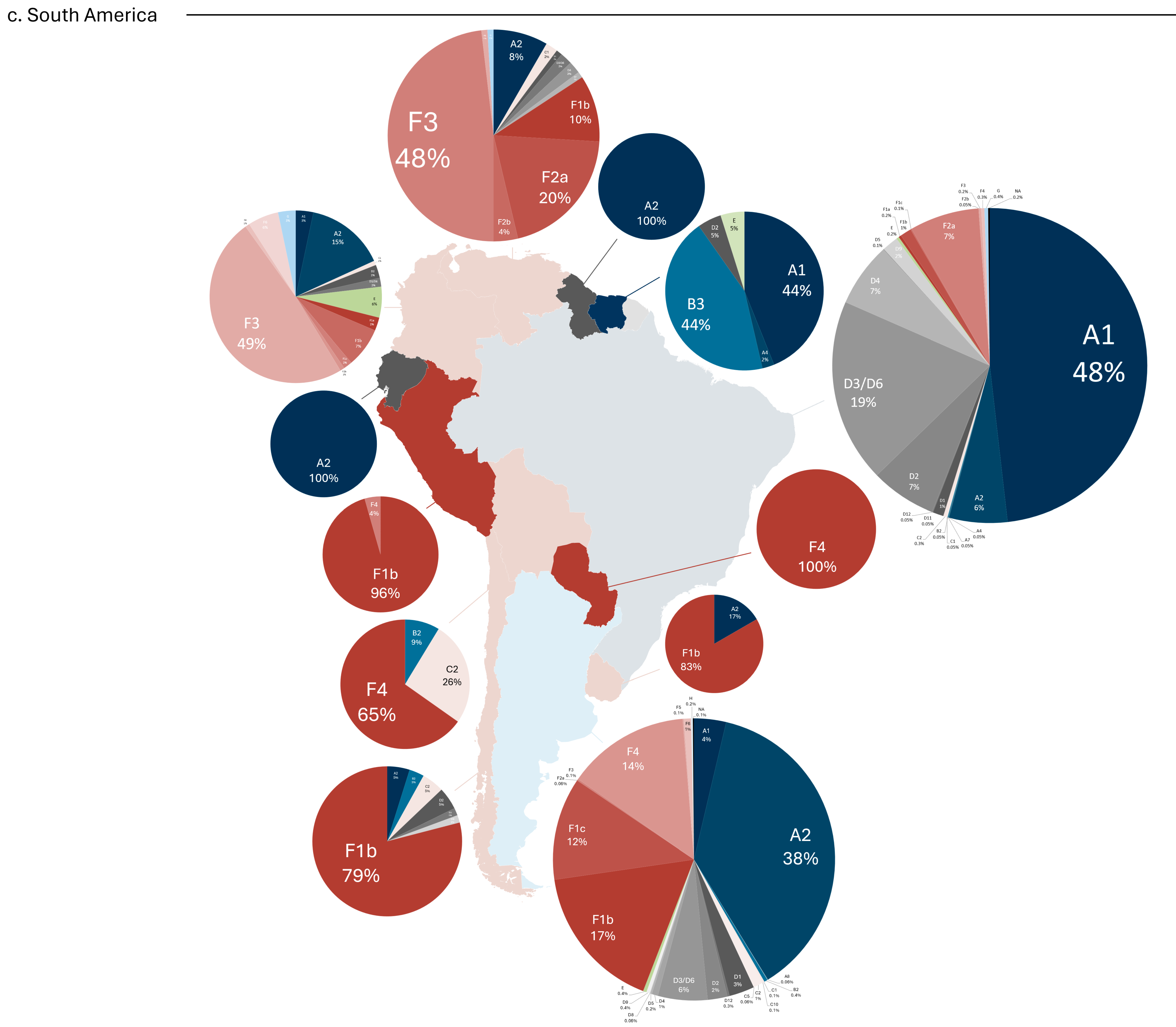
3.2. Molecular Epidemiology
3.3. Resistance-Associated Mutations
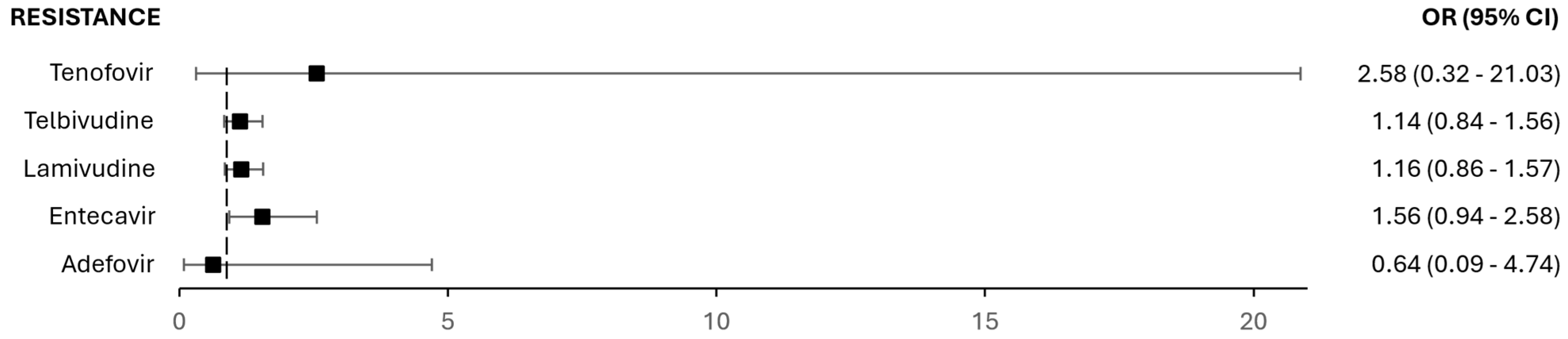
3.4. Resistance in Prior Treatments and Co-Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Global Hepatitis Report 2024: Action for Access in Low- and Middle-Income Countries. Available online: https://iris.who.int/handle/10665/376461 (accessed on 11 August 2025).
- Guidelines for the Prevention, Diagnosis, Care and Treatment for People with Chronic Hepatitis B Infection. Available online: https://iris.who.int/handle/10665/376353 (accessed on 11 August 2025).
- Campos-Valdez, M.; Monroy-Ramírez, H.C.; Armendáriz-Borunda, J.; Sánchez-Orozco, L.V. Molecular Mechanisms during Hepatitis B Infection and the Effects of the Virus Variability. Viruses 2021, 13, 1167. [Google Scholar] [CrossRef]
- Tsukuda, S.; Watashi, K. Hepatitis B virus biology and life cycle. Antivir. Res. 2020, 182, 104925. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Wu, C.C.; Chen, X.W.; Li, X.; Li, J.; Lu, M.J. Genetic variation of hepatitis B virus and its significance for pathogenesis. World J. Gastroenterol. 2016, 22, 126–144. [Google Scholar] [CrossRef]
- Chen, J.; Li, L.; Yin, Q.; Shen, T. A review of epidemiology and clinical relevance of Hepatitis B virus genotypes and subgenotypes. Clin. Res. Hepatol. Gastroenterol. 2023, 47, 102180. [Google Scholar] [CrossRef] [PubMed]
- Kocher, A.; Papac, L.; Barquera, R.; Key, F.M.; Spyrou, M.A.; Hübler, R.; Rohrlach, A.B.; Aron, F.; Stahl, R.; Wissgott, A.; et al. Ten millennia of hepatitis B virus evolution. Science 2021, 374, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Araujo, N.M.; Osiowy, C. Hepatitis B Virus Genotype G: The Odd Cousin of the Family. Front. Microbiol. 2022, 13, 872766. [Google Scholar] [CrossRef] [PubMed]
- Wolf, J.M.; Pereira, V.R.Z.B.; Simon, D.; Lunge, V.R. Evolutionary history of hepatitis B virus genotype H. J. Med. Virol. 2021, 93, 4004–4009. [Google Scholar] [CrossRef]
- Marchio, A.; Sitbounlang, P.; Deharo, E.; Paboriboune, P.; Pineau, P. Concealed for a Long Time on the Marches of Empires: Hepatitis B Virus Genotype I. Microorganisms 2023, 11, 2204. [Google Scholar] [CrossRef]
- Toyé, R.M.; Loureiro, C.L.; Jaspe, R.C.; Zoulim, F.; Pujol, F.H.; Chemin, I. The Hepatitis B Virus Genotypes E to J: The Overlooked Genotypes. Microorganisms 2023, 11, 1908. [Google Scholar] [CrossRef]
- Tatematsu, K.; Tanaka, Y.; Kurbanov, F.; Sugauchi, F.; Mano, S.; Maeshiro, T.; Nakayoshi, T.; Wakuta, M.; Miyakawa, Y.; Mizokami, M. A genetic variant of hepatitis B virus divergent from known human and ape genotypes isolated from a Japanese patient and provisionally assigned to new genotype J. J. Virol. 2009, 83, 10538–10547. [Google Scholar] [CrossRef]
- Sun, B.; Valtueña, A.A.; Kocher, A.; Gao, S.; Li, C.; Fu, S.; Zhang, F.; Ma, P.; Yang, X.; Qiu, Y.; et al. Origin and dispersal history of Hepatitis B virus in Eastern Eurasia. Nat. Commun. 2024, 15, 2951. [Google Scholar] [CrossRef]
- Araujo, N.M.; Teles, S.A.; Spitz, N. Comprehensive Analysis of Clinically Significant Hepatitis B Virus Mutations in Relation to Genotype, Subgenotype and Geographic Region. Front. Microbiol. 2020, 11, 616023. [Google Scholar] [CrossRef] [PubMed]
- Whalley, S.A.; Murray, J.M.; Brown, D.; Webster, G.J.; Emery, V.C.; Dusheiko, G.M.; Perelson, A.S. Kinetics of acute hepatitis B virus infection in humans. J. Exp. Med. 2001, 193, 847–854. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, X.; Wei, M.; Zhang, C.; Xu, T.; Liu, L.; Xu, Z. Potential resistant mutations within HBV reverse transcriptase sequences in nucleos(t)ide analogues-experienced patients with hepatitis B virus infection. Sci. Rep. 2019, 9, 8078. [Google Scholar] [CrossRef]
- Irwin, K.K.; Renzette, N.; Kowalik, T.F.; Jensen, J.D. Antiviral drug resistance as an adaptive process. Virus Evol. 2016, 2, vew014. [Google Scholar] [CrossRef] [PubMed]
- Boulard, Y.; Bressanelli, S. Recapitulating Trafficking of Nucleosides Into the Active Site of Polymerases of RNA Viruses: The Challenge and the Prize. Front. Med. Technol. 2021, 3, 705875. [Google Scholar] [CrossRef] [PubMed]
- Thames, J.E.; Seley-Radtke, K.L. Comparison of the Old and New-Novel Mechanisms of Action for Anti-coronavirus Nucleoside Analogues. Chimia 2022, 76, 409–417. [Google Scholar] [CrossRef]
- Tacke, F.; Kroy, D.C. Treatment for hepatitis B in patients with drug resistance. Ann. Transl. Med. 2016, 4, 334. [Google Scholar] [CrossRef]
- Hepatitis B Virus: Overview of Management-UpToDate. Available online: https://www.uptodate.com/contents/hepatitis-b-virus-overview-of-management (accessed on 11 August 2025).
- Kimberlin, D.W. 295—Antiviral Agents. In Principles and Practice of Pediatric Infectious Diseases, 6th ed.; Long, S.S., Ed.; Elsevier: Philadelphia, PA, USA, 2023; pp. 1583–1598.e6. Available online: https://www.sciencedirect.com/science/article/pii/B9780323756082002950 (accessed on 11 August 2025).
- Laoi, B.N.; Herra, C.; Norris, S.; Crowley, B. First report of genotypic resistance to adefovir in chronic HBV in the Republic of Ireland. J. Antimicrob. Chemother. 2006, 57, 1009–1010. [Google Scholar] [CrossRef]
- Quirós-Santana, C. Telbivudina. Acta Médica Costarric. 2008, 50, 55. [Google Scholar]
- Xia, Y.; Guo, H. Hepatitis B virus cccDNA: Formation, regulation and therapeutic potential. Antivir. Res. 2020, 180, 104824. [Google Scholar] [CrossRef]
- FDALabel. Available online: https://nctr-crs.fda.gov/fdalabel/ui/spl-summaries/criteria/391290 (accessed on 11 August 2025).
- Pourkarim, M.R.; Amini-Bavil-Olyaee, S.; Lemey, P.; Maes, P.; Van Ranst, M. HBV subgenotype misclassification expands quasi-subgenotype A3. Clin. Microbiol. Infect. 2011, 17, 947–949. [Google Scholar] [CrossRef] [PubMed]
- Meldal, B.H.M.; Bon, A.H.; Prati, D.; Ayob, Y.; Allain, J.P. Diversity of hepatitis B virus infecting Malaysian candidate blood donors is driven by viral and host factors. J. Viral Hepat. 2011, 18, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Sant’Anna, T.B.; Araujo, N.M. Hepatitis B Virus Genotype D: An Overview of Molecular Epidemiology, Evolutionary History, and Clinical Characteristics. Microorganisms 2023, 11, 1101. [Google Scholar] [CrossRef]
- Zhang, M.; Mouzannar, K.; Zhang, Z.; Teraoka, Y.; Piotrowski, J.; Ishida, Y.; Tateno-Mukaidani, C.; Saito, T.; Abe-Chayama, H.; Chayama, K.; et al. Hepatitis B virus genotypes A1 and A2 have distinct replication phenotypes due to polymorphisms in the HBx gene. PLoS Pathog. 2025, 21, e1012803. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Chen, H.; Sun, L.; Liu, J. Dating the origin and dispersal of global hepatitis B virus genotype C in humans. Drug Discov. Ther. 2022, 16, 85–92. [Google Scholar] [CrossRef]
- O’neil, K.; Hamilton, K.; Papademetriou, D. Migration in the Americas. 2005 Sep. Available online: https://www.iom.int/sites/g/files/tmzbdl486/files/jahia/webdav/site/myjahiasite/shared/shared/mainsite/policy_and_research/gcim/rs/RS1.pdf (accessed on 1 May 2025).
- OECD. International Migration in the Americas. 2015. Available online: https://www.oecd.org/en/publications/international-migration-in-the-americas_e5d3aa63-en.html (accessed on 11 August 2025).
- Jose-Abrego, A.; Roman, S.; Laguna-Meraz, S.; Rebello-Pinho, J.R.; Justo Arevalo, S.; Panduro, A. Tracing the evolutionary history of hepatitis B virus genotype H endemic to Mexico. Front. Microbiol. 2023, 14, 1180931. [Google Scholar] [CrossRef]
- Holzmayer, V.; Hance, R.; Defechereux, P.; Grant, R.; Kuhns, M.C.; Cloherty, G.; Rodgers, M.A. Identification of hepatitis B virus genotype I in Thailand. J. Med. Virol. 2019, 91, 717–721. [Google Scholar] [CrossRef]
- Shen, T.; Yan, X.M.; Liu, H.; Zhang, B.X.; Li, L.; Zhang, J.P.; Wang, J.L.; Xiao, C.J. Genotype I of hepatitis B virus was found in east Xishuangbanna, China and molecular dynamics of HBV/I. J. Viral Hepat. 2015, 22, 37–45. [Google Scholar] [CrossRef]
- Osiowy, C.; Kaita, K.; Solar, K.; Mendoza, K. Molecular characterization of hepatitis B virus and a 9-year clinical profile in a patient infected with genotype I. J. Med. Virol. 2010, 82, 942–948. [Google Scholar] [CrossRef]
- Andernach, I.E.; Hunewald, O.E.; Muller, C.P. Bayesian inference of the evolution of HBV/E. PLoS ONE 2013, 8, e81690. [Google Scholar] [CrossRef]
- Mokaya, J.; Vasylyeva, T.I.; Barnes, E.; Ansari, M.A.; Pybus, O.G.; Matthews, P.C. Global prevalence and phylogeny of hepatitis B virus (HBV) drug and vaccine resistance mutations. J. Viral Hepat. 2021, 28, 1110–1120. [Google Scholar] [CrossRef]
- Nguyen, T.K.; Van Le, D. Resistant mutations within the hepatitis B virus reverse transcriptase sequence in treatment failure patients with chronic HBV infection in Vietnam. J. Glob. Antimicrob. Resist. 2023, 33, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Jose-Abrego, A.; Roman, S.; Rebello Pinho, J.R.; Gomes-Gouvêa, M.S.; Panduro, A. High Frequency of Antiviral Resistance Mutations in HBV Genotypes A2 and H: Multidrug Resistance Strains in Mexico. J. Clin. Transl. Hepatol. 2023, 11, 1023–1034. [Google Scholar] [CrossRef] [PubMed]
- Bassit, L.; Ono, S.K.; Schinazi, R.F. Moving Fast Toward Hepatitis B Virus Elimination. Adv. Exp. Med. Biol. 2021, 1322, 115–138. [Google Scholar] [PubMed]
- Liu, T.; Sun, Q.; Gu, J.; Cen, S.; Zhang, Q. Characterization of the tenofovir resistance-associated mutations in the hepatitis B virus isolates across genotypes A to D. Antivir. Res. 2022, 203, 105348. [Google Scholar] [CrossRef]
- Lada, O.; Gervais, A.; Branger, M.; Peytavin, G.; Roquebert, B.; Collin, G.; Fraqueiro, G.; Moucari, R.; Leclerc, L.; Martinot-Peignoux, M.; et al. Quasispecies analysis and in vitro susceptibility of HBV strains isolated from HIV-HBV-coinfected patients with delayed response to tenofovir. Antivir. Ther. 2012, 17, 61–70. [Google Scholar] [CrossRef]
- Sánchez, L.V.; Tanaka, Y.; Maldonado, M.; Mizokami, M.; Panduro, A. Difference of hepatitis B virus genotype distribution in two groups of mexican patients with different risk factors: High prevalence of genotype H and G. Intervirology 2007, 50, 9–15. [Google Scholar] [CrossRef]
- Osiowy, C.; Gordon, D.; Borlang, J.; Giles, E.; Villeneuve, J.P. Hepatitis B virus genotype G epidemiology and co-infection with genotype A in Canada. J. Gen. Virol. 2008, 89 Pt 12, 3009–3015. [Google Scholar] [CrossRef]
- Cornelissen, M.; Zorgdrager, F.; Bruisten, S.M.; Bakker, M.; Berkhout, B.; van der Kuyl, A.C. Widespread hepatitis B virus genotype G (HBV-G) infection during the early years of the HIV epidemic in the Netherlands among men who have sex with men. BMC Infect. Dis. 2016, 16, 268. [Google Scholar] [CrossRef]
- Araujo, N.M.; Araujo, O.C.; Silva, E.M.; Villela-Nogueira, C.A.; Nabuco, L.C.; Parana, R.; Bessone, F.; Gomes, S.A.; Trepo, C.; Kay, A. Identification of novel recombinants of hepatitis B virus genotypes F and G in human immunodeficiency virus-positive patients from Argentina and Brazil. J. Gen. Virol. 2013, 94 Pt 1, 150–158. [Google Scholar] [CrossRef]
- Jose-Abrego, A.; Roman, S.; Rebello Pinho, J.R.; de Castro, V.F.D.; Panduro, A. Hepatitis B Virus (HBV) Genotype Mixtures, Viral Load, and Liver Damage in HBV Patients Co-infected With Human Immunodeficiency Virus. Front. Microbiol. 2021, 12, 640889. [Google Scholar] [CrossRef] [PubMed]
- World Bank Open Data. Available online: https://data.worldbank.org (accessed on 11 August 2025).
- Pierra Rouviere, C.; Dousson, C.B.; Tavis, J.E. HBV replication inhibitors. Antivir. Res. 2020, 179, 104815. [Google Scholar] [CrossRef] [PubMed]
- McNaughton, A.L.; Revill, P.A.; Littlejohn, M.; Matthews, P.C.; Azim Ansari, M. Analysis of genomic-length HBV sequences to determine genotype and subgenotype reference sequences. J. Gen. Virol. 2020, 101, 271–283. [Google Scholar] [CrossRef]
- Mojsiejczuk, L.; Torres, C.; Flichman, D.; Campos, R.H. Long-term evolution of hepatitis B virus genotype F: Strong association between viral diversification and the prehistoric settlement of Central and South America. J. Viral. Hepat. 2020, 27, 620–630. [Google Scholar] [CrossRef]
- Yin, Y.; He, K.; Wu, B.; Xu, M.; Du, L.; Liu, W.; Liao, P.; Liu, Y.; He, M. A systematic genotype and subgenotype re-ranking of hepatitis B virus under a novel classification standard. Heliyon 2019, 5, e02556. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Harrison, T.J.; He, X.; Chen, Q.Y.; Li, G.J.; Liu, M.H.; Li, H.; Yang, J.Y.; Fang, Z.L. The prevalence of mutations in the major hydrophilic region of the surface antigen of hepatitis B virus varies with subgenotype. Epidemiol. Infect. 2015, 143, 3572–3582. [Google Scholar] [CrossRef]
- Chen, L.; Shi, Y.; Yang, W.; Zhang, Y.; Xie, Q.; Li, Y.; Li, X.; Li, J.; Zhang, Z. Differences in Cpg Island Distribution Between Subgenotypes of the Hepatitis B Virus Genotype. Med. Sci. Monit. 2018, 24, 6781–6794. [Google Scholar] [CrossRef]
- Thijssen, M.; Trovão, N.S.; Mina, T.; Maes, P.; Pourkarim, M.R. Novel hepatitis B virus subgenotype A8 and quasi-subgenotype D12 in African–Belgian chronic carriers. Int. J. Infect. Dis. 2020, 93, 98–101. [Google Scholar] [CrossRef]
- Gionda, P.O.; Gomes-Gouvea, M.; Malta Fde, M.; Sebe, P.; Salles, A.P.M.; Francisco Rdos, S.; José-Abrego, A.; Roman, S.; Panduro, A.; Pinho, J.R.R. Analysis of the complete genome of HBV genotypes F and H found in Brazil and Mexico using the next generation sequencing method. Ann. Hepatol. 2022, 27 (Suppl. 1), 100569. [Google Scholar] [CrossRef]
- Zhu, H.L.; Wang, C.T.; Xia, J.B.; Li, X.; Zhang, Z.H. Establishment of reference sequences of hepatitis B virus genotype C subgenotypes. Genet. Mol. Res. 2015, 14, 16521–16534. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.H.; Chen, Q.Y.; Jiang, Z.H.; Wang, X.Y.; Zhang, W.J.; He, X.; Harrison, T.J.; Jackson, J.B.; Fang, Z.-L. A novel subgenotype I3 of hepatitis B virus in Guangxi, China: A 15-year follow-up study. Virus Genes 2023, 59, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Zhu, H.; Zhang, Y.; Li, X.; Zhang, Z. Hepatitis B virus genotype A: Design of reference sequences for sub-genotypes. Virus Genes 2016, 52, 325–333. [Google Scholar] [CrossRef]
- McNaughton, A.L.; D’Arienzo, V.; Ansari, M.A.; Lumley, S.F.; Littlejohn, M.; Revill, P.; McKeating, J.A.; Matthews, P.C. Insights from Deep Sequencing of the HBV Genome-Unique, Tiny, and Misunderstood. Gastroenterology 2019, 156, 384–399. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. NCBI. 2004 Genotyping. Available online: https://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi (accessed on 22 March 2024).
- Bell, T.G.; Yousif, M.; Kramvis, A. Bioinformatic curation and alignment of genotyped hepatitis B virus (HBV) sequence data from the GenBank public database. Springerplus 2016, 5, 1896. [Google Scholar] [CrossRef] [PubMed]




| Subtype | S106C | H126Y | D134E | V173L | A181T/V | A194T | S202C/G/I | M204V/I | N236T | L269I | L269I + H126Y | L180M + M204V/I | L180M + M204V/I + V173L |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | 0.25 (0.09–0.67) | 3.55 (3.11–4.04) | NS | NS | NS | NS | NS | 0.36 (0.19–0.68) | - | 0.18 (0.14–0.22) | 4.64 (4.05–5.31) | 0.36 (0.19–0.68) | 0.26 (0.13–0.51) |
| A2 | - | 12.22 (10.78–13.85) | NS | 5.97 (2.89–12.32) | - | - | NS | NS | NS | 0.02 (0.01–0.03) | 4.35 (3.84–4.93) | 4.77 (3.94–5.77) | 15.80 (11.20–22.28) |
| A5 | - | NS | - | - | - | - | - | - | - | - | 117.24 (15.82–868.9) | - | - |
| B2 | - | - | - | - | NS | NS | - | NS | NS | 6.37 (5.20–7.79) | - | 0.21 (0.09–0.47) | - |
| B3 | - | - | - | - | - | - | - | - | - | 8.77 (3.5–22.0) | - | - | - |
| B4 | - | 0.01 (0.002–0.09) | NS | - | 18.12 (8.34–39.39) | - | NS | 7.67 (5.21–11.31) | - | NS | - | 0.26 (0.11–0.64) | - |
| B5 | 71.34 (34.6–147.07) | - | - | - | - | 87.24 (16.94–449.13) | - | NS | - | 6.38 (3.03–13.42) | - | - | - |
| B6 | - | - | - | - | NS | - | - | - | - | 14. 36 (7.91–26.08) | - | - | - |
| B10 | - | - | - | - | - | - | - | - | - | - | - | 16.49 (1.03–264.04) | - |
| C1 | 2.99 (1.74–5.15) | 0.01 (0.004–0.06) | 5.18 (2.86–9.37) | NS | NS | NS | - | 2.03 (1.26–3.27) | - | 3.37 (2.82–4.04) | - | 0.28 (0.014–0.54) | - |
| C2 | NS | 0.05 (0.02–0.09) | 2.15 (1.05–4.39) | NS | - | - | - | NS | - | 0.27 (0.21–0.36) | 0.01 (0.004–0.06) | NS | 0.06 (0.008–0.41) |
| C5 | - | NS | 15.74 (3.57–69.42) | - | - | - | - | NS | - | NS | - | - | - |
| D1 | - | - | NS | - | 4.92 (1.15–20.99) | - | - | NS | 30.60 (2.76–339.6) | 1.58 (1.11–2.27) | - | NS | - |
| D2 | - | - | - | NS | - | - | NS | NS | - | 4.90 (3.89–6.16) | - | 2.81 (2.02–3.91) | NS |
| D3/D6 | NS | 0.02 (0.006–0.06) | NS | NS | - | - | 3.89 (1.28–11.85) | NS | - | 4.34 (3.64–5.18) | 0.02 (0.004–0.07) | 0.34 (0.19–0.61) | 0.13 (0.03–0.53) |
| D4 | - | 0.02 (0.002–0.11) | - | - | - | NS | - | - | - | 3.28 (2.56–4.2) | - | - | - |
| D9 | - | - | - | NS | NS | - | - | NS | - | NS | - | NS | 4.41 (1.73–11.24) |
| D12 | - | - | - | - | - | - | 14.89 (1.92–115.28) | - | - | 0.18 (0.04–0.74) | - | 3.69 (1.52–8.99) | - |
| E | - | 2.86 (2.14–3.82) | - | - | - | - | - | NS | - | 0.01 (0.002–0.1) | 6.97 (5.22–9.31) | 0.25 (0.08–0.79) | - |
| F1b | 7.03 (4.43–11.14) | - | - | - | NS | - | NS | - | - | NS | 0.01 (0.002–0.08) | NS | NS |
| F1c | 3.74 (1.79–7.82) | - | - | - | - | - | - | - | - | 0.19 (0.11–0.33) | - | - | - |
| F2a | NS | 0.03 (0.004–0.18) | - | - | - | - | - | - | - | 3.66 (2.66–5.05) | - | NS | - |
| F2b | - | - | - | - | - | - | - | - | - | 8.26 (1.67–40.98) | - | 5.51 (1.11–27.35) | - |
| F3 | 7.23 (3.67–14.25) | - | - | NS | - | - | - | NS | - | 5.38 (3.77–7.69) | - | NS | - |
| F4 | 4.39 (2.37–8.14) | - | - | - | - | - | NS | - | - | 1.90 (1.48–2.44) | - | 0.19 (0.06–0.58) | NS |
| F5 | - | - | - | - | - | - | - | - | - | 5.50 (1.007–30.07) | - | - | - |
| F6 | - | - | - | - | - | - | 23.87 (3.03–187.99) | - | - | NS | - | NS | - |
| G | - | 1.81 (1.27–2.59) | - | - | 9.85 (3.36–28.85) | - | - | 9.56 (5.91–15.49) | - | - | 11.94 (8.36–17.07) | NS | 2.33 (1.13–4.81) |
| H | NS | - | - | 6.76 (1.59–28.8) | NS | 13.87 (1.69–114.03) | 12.27 (2.78–54.24) | NS | - | 6.27 (3.94–9.99) | 0.06 (0.008–0.43) | NS | NS |
| I | - | NS | - | - | - | - | - | - | - | - | 50.48 (6.46–394.69) | - | - |
| Subtype | Adefovir | Entecavir | Lamivudine | Telbivudine | Tenofovir |
|---|---|---|---|---|---|
| A1 | NS | 0.30 (0.17–0.56) | 0.46 (0.36–0.60) | 0.48 (0.37–0.62) | NS |
| A2 | 0.13 (0.02–0.98) | 12.04 (8.82–16.43) | 5.45 (4.70–6.32) | 5.12 (4.41–5.95) | - |
| B2 | NS | - | 0.18 (0.10–0.35) | 0.19 (0.10–0.36) | NS |
| B4 | 16.10 (7.54–34.40) | 0.12 (0.02–0.82) | NS | 1.40 (1–1.96) | - |
| B5 | - | - | NS | NS | 87.24 (16.94–449.14) |
| C1 | NS | - | 0.50 (0.34–0.74) | 0.52 (0.36–0.76) | NS |
| C2 | - | 0.05 (0.007–0.38) | 0.52 (0.37–0.73) | 0.54 (0.38–0.75) | - |
| D1 | 4.55 (1.07–19.35) | - | NS | NS | - |
| D2 | - | NS | 1.86 (1.38–2.51) | 1.93 (1.43–2.59) | - |
| D3/D6 | - | 0.38 (0.17–0.86) | 0.35 (0.23–0.54) | 0.37 (0.24–0.56) | - |
| D9 | NS | 4.14 (1.63–10.52) | NS | NS | - |
| E | - | - | 0.18 (0.07–0.48) | 0.18 (0.07–0.50) | - |
| F1b | NS | NS | 0.55 (0.37–0.83) | 0.57 (0.38–0.86) | - |
| F2a | - | - | 0.34 (0.15–0.78) | 0.36 (0.16–0.81) | - |
| F3 | - | - | 0.40 (0.18–0.90) | 0.41 (0.18–0.93) | - |
| F4 | - | NS | 0.13 (0.05–0.35) | 0.14 (0.05–0.36) | - |
| G | 9.06 (3.11–26.37) | 2.18 (1.05–4.50) | 3.56 (2.46–5.14) | 3.68 (2.54–5.32) | - |
| H | NS | NS | NS | NS | 13.87 (1.69–114.03) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruvalcaba, I.A.; Diaz-Palomera, C.D.; Silva-Ríos, A.A.; Muñoz-Valle, J.F.; Viera-Segura, O. The Molecular Epidemiology of Hepatitis B Virus and Its Resistance-Associated Mutations in the Polymerase Gene in the Americas. Microorganisms 2025, 13, 1913. https://doi.org/10.3390/microorganisms13081913
Ruvalcaba IA, Diaz-Palomera CD, Silva-Ríos AA, Muñoz-Valle JF, Viera-Segura O. The Molecular Epidemiology of Hepatitis B Virus and Its Resistance-Associated Mutations in the Polymerase Gene in the Americas. Microorganisms. 2025; 13(8):1913. https://doi.org/10.3390/microorganisms13081913
Chicago/Turabian StyleRuvalcaba, Itzel A., Carlos Daniel Diaz-Palomera, Adrián Alejandro Silva-Ríos, José Francisco Muñoz-Valle, and Oliver Viera-Segura. 2025. "The Molecular Epidemiology of Hepatitis B Virus and Its Resistance-Associated Mutations in the Polymerase Gene in the Americas" Microorganisms 13, no. 8: 1913. https://doi.org/10.3390/microorganisms13081913
APA StyleRuvalcaba, I. A., Diaz-Palomera, C. D., Silva-Ríos, A. A., Muñoz-Valle, J. F., & Viera-Segura, O. (2025). The Molecular Epidemiology of Hepatitis B Virus and Its Resistance-Associated Mutations in the Polymerase Gene in the Americas. Microorganisms, 13(8), 1913. https://doi.org/10.3390/microorganisms13081913







