The Role of Coffee Microbiomes in Pathogen Resistance Across Varieties and Ecological Niches
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Plant Materials
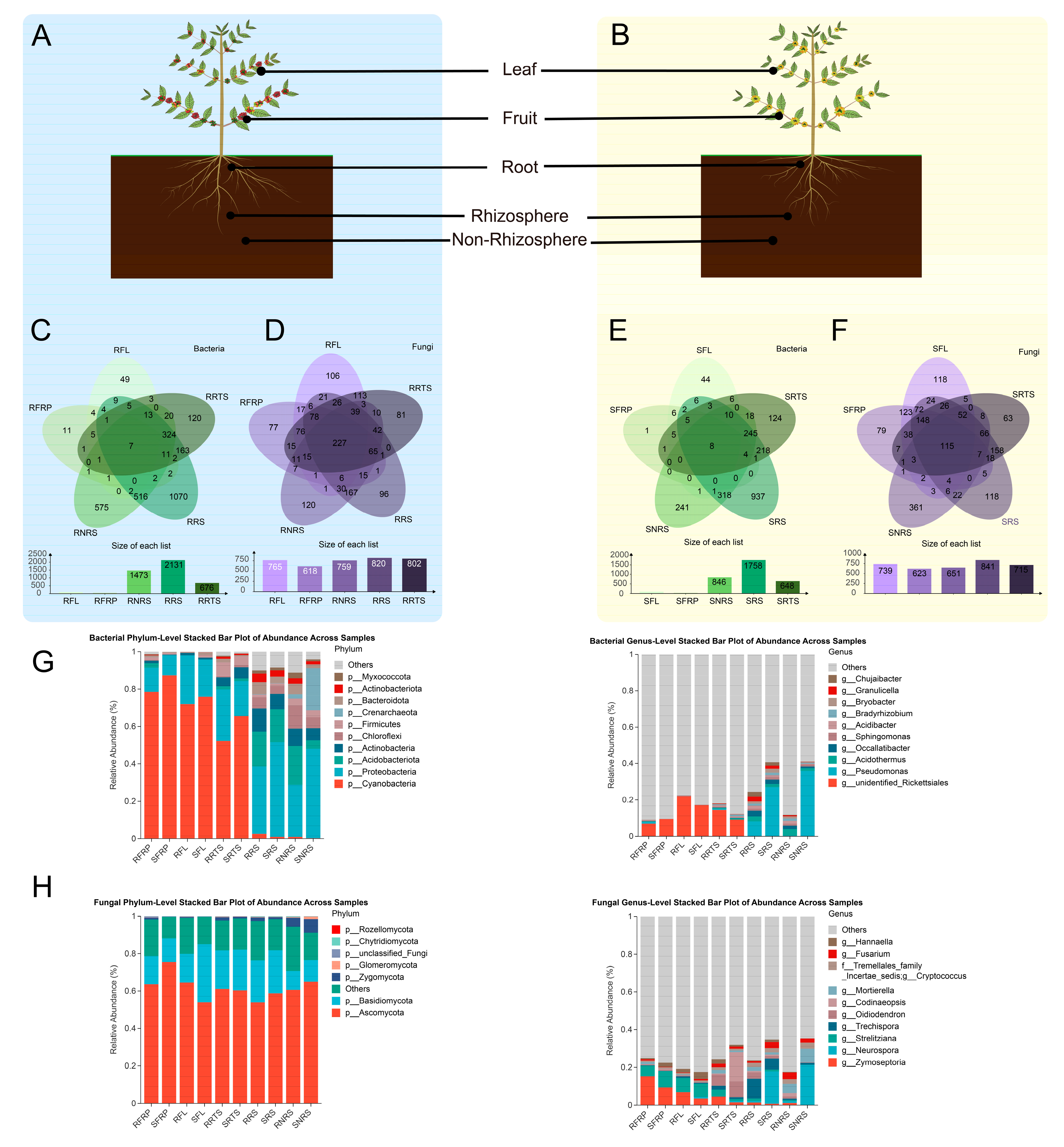
2.2. Sampling Collection
2.3. DNA Extraction and High-Throughput Sequencing
2.4. DNA Extraction and High-Throughput Sequencing Protocols
2.5. Alpha and Beta Diversity Analyses of Microbial Communities
2.6. Identification of Significantly Different Microbial Communities
2.7. Microbial Co-Occurrence Network Analysis
2.8. Data Statistics
3. Results
3.1. Microbial Sequencing and De Novo Assembly of Resistance Varieties of Coffea arabica Across Diverse Ecological Niches
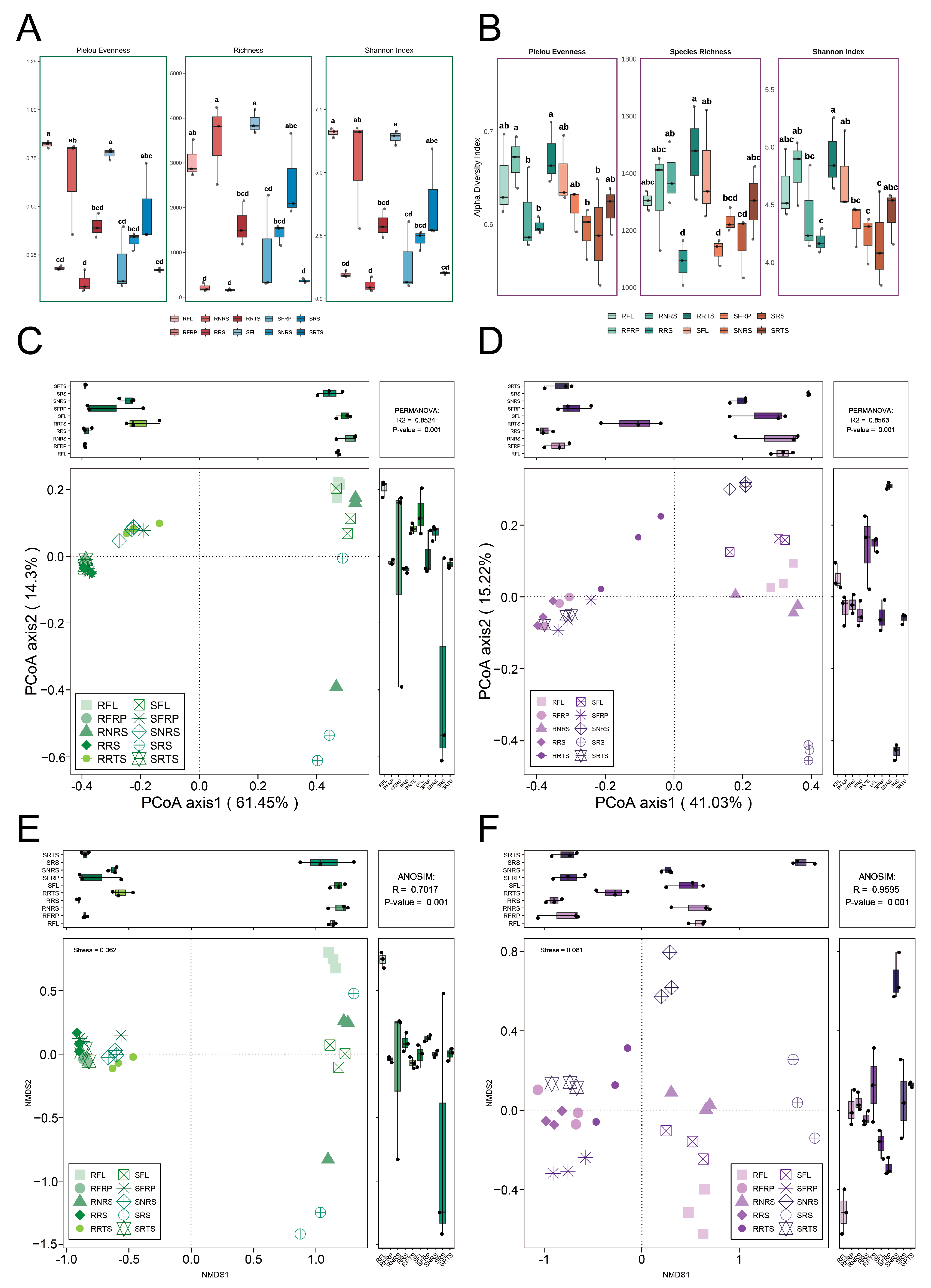
3.2. Alpha Diversity Analysis Reveals the Joint Influence of Ecological Niches and Species on Microbial Community Diversity
3.3. Beta Diversity Analysis Reveals the Co-Shaping of Microbial Community Structure by Niche and Species
3.4. Characteristics of Microbial Community Composition
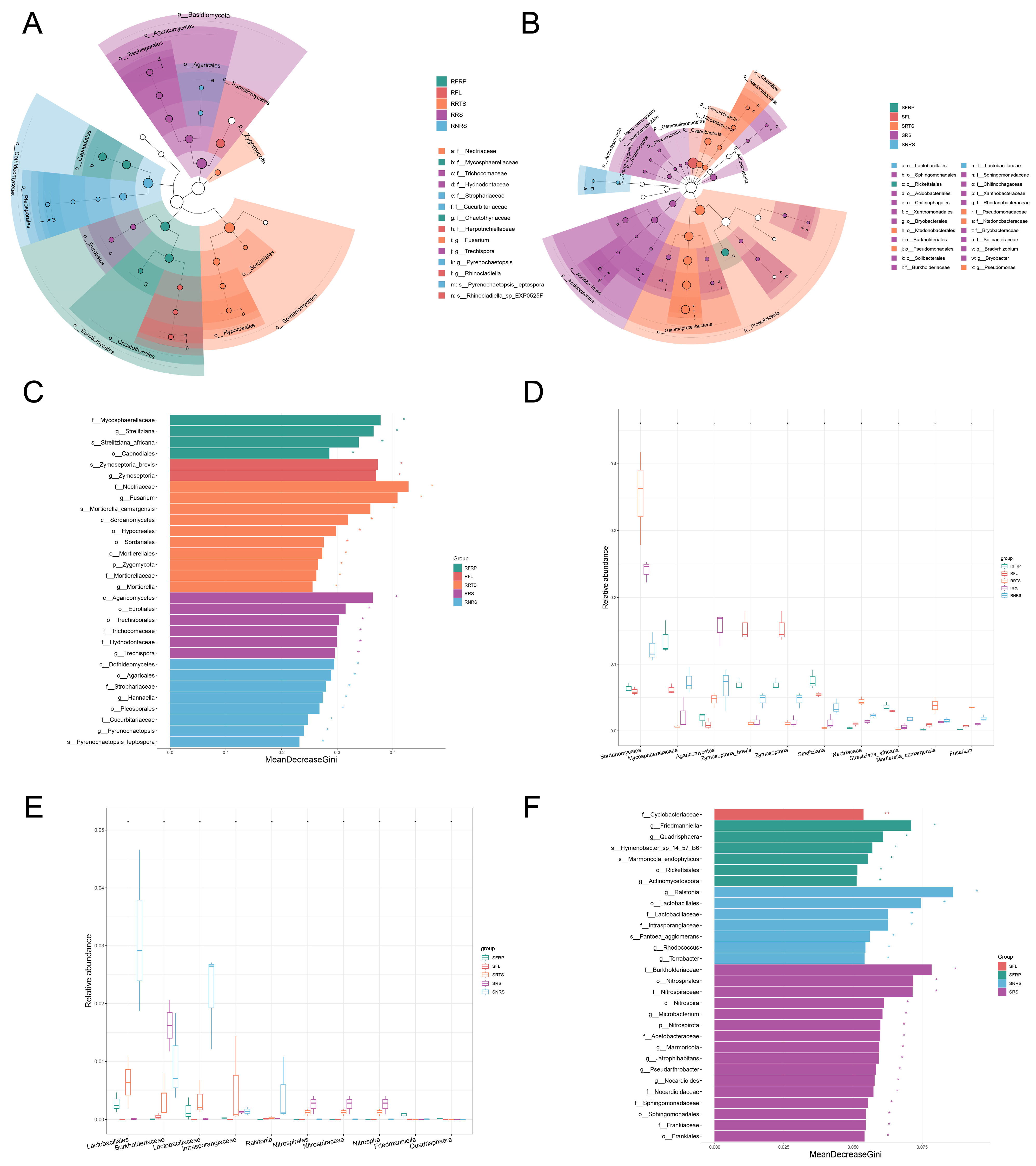
3.5. Changes in Dominant Species Markers of Microbial Communities and Taxonomic Level Analysis
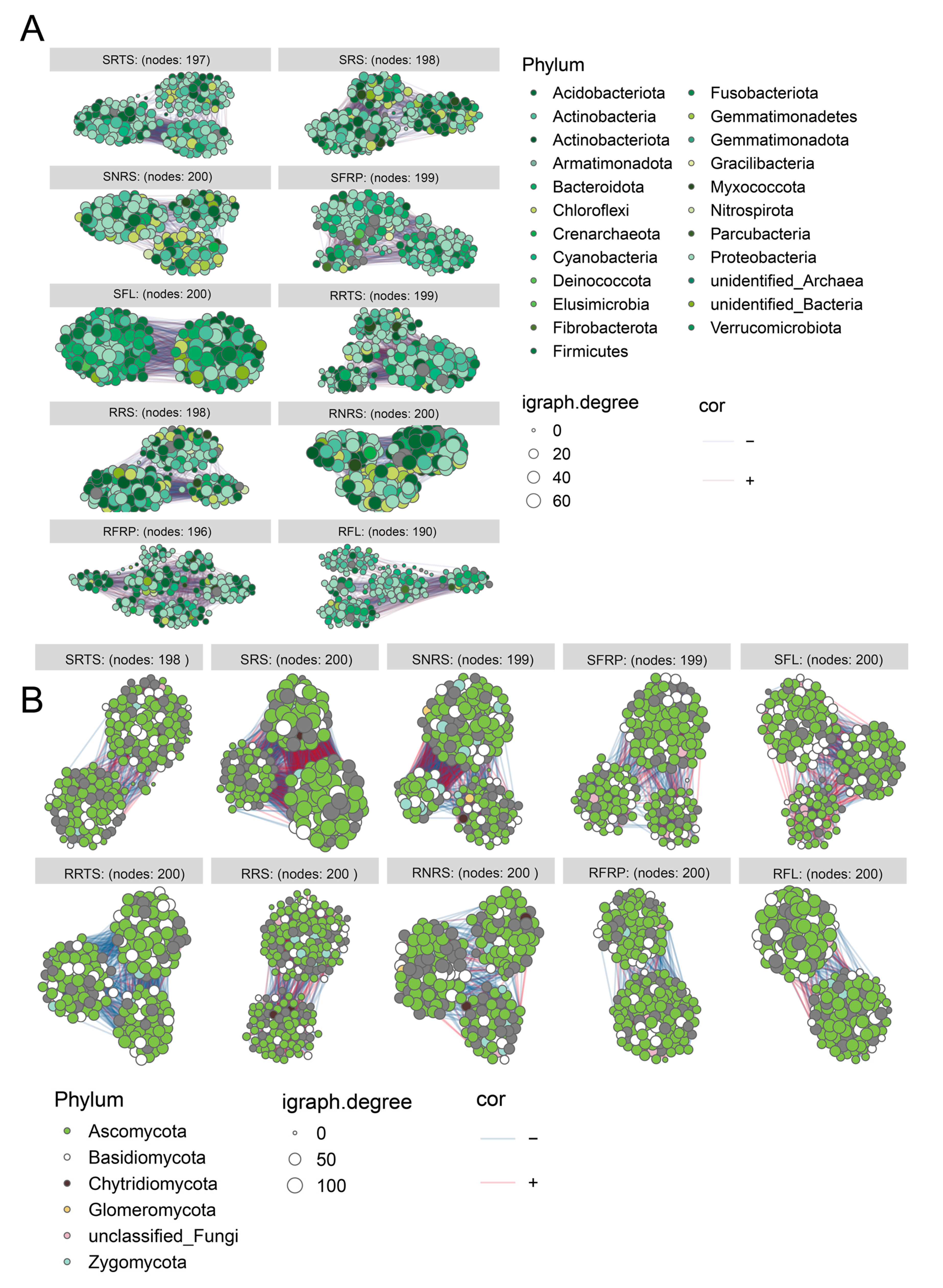
3.6. Network Analysis of Microbial Co-Occurrence Patterns in Coffee-Associated Ecological Niches
3.7. Functional Prediction of Bacterial and Fungal Communities Exhibiting Varied Resistance Traits
4. Discussion
4.1. Dual Driving Forces of Ecological Niche and Host Genotype
4.2. Microbiome Characteristics of Disease-Resistant Varieties: A Stable, Diverse, and Functionally Redundant “Defence Alliance”
4.3. Microbiome Characteristics of Disease-Susceptible Varieties: Ecological Imbalance, Pathogen Enrichment, and Fragile Networks
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Salojärvi, J.; Rambani, A.; Yu, Z.; Guyot, R.; Strickler, S.; Lepelley, M.; Wang, C.; Rajaraman, S.; Rastas, P.; Zheng, C. The genome and population genomics of allopolyploid Coffea arabica reveal the diversification history of modern coffee cultivars. Nat. Genet. 2024, 56, 721–731. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, X.; Li, R.; Wang, X.; Cheng, J.; Yang, Q.; Kong, H. AHP-GIS and MaxEnt for delineation of potential distribution of Arabica coffee plantation under future climate in Yunnan, China. Ecol. Indic. 2021, 132, 108339. [Google Scholar] [CrossRef]
- Rhiney, K.; Guido, Z.; Knudson, C.; Avelino, J.; Bacon, C.M.; Leclerc, G.; Aime, M.C.; Bebber, D.P. Epidemics and the future of coffee production. Proc. Natl. Acad. Sci. USA 2021, 118, e2023212118. [Google Scholar] [CrossRef]
- Torres Castillo, N.E.; Melchor-Martínez, E.M.; Ochoa Sierra, J.S.; Ramirez-Mendoza, R.A.; Parra-Saldívar, R.; Iqbal, H.M.N. Impact of climate change and early development of coffee rust—An overview of control strategies to preserve organic cultivars in Mexico. Sci. Total Environ. 2020, 738, 140225. [Google Scholar] [CrossRef]
- DaMatta, F.M.; Avila, R.T.; Cardoso, A.A.; Martins, S.C.V.; Ramalho, J.C. Physiological and Agronomic Performance of the Coffee Crop in the Context of Climate Change and Global Warming: A Review. J. Agric. Food Chem. 2018, 66, 5264–5274. [Google Scholar] [CrossRef]
- Silva, D.N.; Várzea, V.; Paulo, O.S.; Batista, D. Population genomic footprints of host adaptation, introgression and recombination in coffee leaf rust. Mol. Plant Pathol. 2018, 19, 1742–1753. [Google Scholar] [CrossRef]
- Ogata-Gutiérrez, K.; Chumpitaz-Segovia, C.; Lirio-Paredes, J.; Zúñiga-Dávila, D. Antifungal Activity of Phyllospheric Bacteria Isolated from Coffea arabica against Hemileia vastatrix. Microorganisms 2024, 12, 582. [Google Scholar] [CrossRef]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Man, L.; Palni, L. The rhizosphere effect in trees of the Indian Central Himalaya with special reference to altitude. Appl. Ecol. Environ. Res. 2007, 5, 93–102. [Google Scholar] [CrossRef]
- Wang, C.; Kuzyakov, Y. Soil organic matter priming: The pH effects. Glob. Chang. Biol. 2024, 30, e17349. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Dinesen, C.; Pioppi, A.; Kovács, Á.T.; Lozano-Andrade, C.N. Composing a microbial symphony: Synthetic communities for promoting plant growth. Trends Microbiol. 2025, 33, 738–751. [Google Scholar] [CrossRef]
- Wang, X.; Liang, C.; Dini-Andreote, F.; Zhou, S.; Jiang, Y. Impacts of trophic interactions on carbon accrual in soils. Trends Microbiol. 2025, 33, 277–284. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef]
- Kivlin, S.N.; Hawkes, C.V.; Papeş, M.; Treseder, K.K.; Averill, C. The future of microbial ecological niche theory and modeling. New Phytol. 2021, 231, 508–511. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Fathy, W.A.; Techen, N.; Elsayed, K.N.; Essawy, E.A.; Tawfik, E.; Alwutayd, K.M.; Abdelhameed, M.S.; Hammouda, O.; Ross, S.A. Applying an internal transcribed spacer as a single molecular marker to differentiate between Tetraselmis and Chlorella species. Front. Microbiol. 2023, 14, 1228869. [Google Scholar] [CrossRef]
- Aldrete-Tapia, A.; Escobar-Ramírez, M.C.; Tamplin, M.L.; Hernández-Iturriaga, M. High-throughput sequencing of microbial communities in Poro cheese, an artisanal Mexican cheese. Food Microbiol. 2014, 44, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Aminu, S.; Ascandari, A.; Laamarti, M.; Safdi, N.E.H.; El Allali, A.; Daoud, R. Exploring microbial worlds: A review of whole genome sequencing and its application in characterizing the microbial communities. Crit. Rev. Microbiol. 2024, 50, 805–829. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Martin, I.; Doolittle-Hall, J.; Teles, R.P.; Patel, M.; Belibasakis, G.N.; Hämmerle, C.H.; Jung, R.E.; Teles, F.R. Exploring the microbiome of healthy and diseased peri-implant sites using Illumina sequencing. J. Clin. Periodontol. 2017, 44, 1274–1284. [Google Scholar] [CrossRef]
- Degnan, P.H.; Ochman, H. Illumina-based analysis of microbial community diversity. ISME J. 2012, 6, 183–194. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Sun, T.; Zou, W.; Luo, R.; Li, C.; Zhang, C.; Yu, H. Compositional and functional diversities of core microbial communities in wild and artificial Ophiocordyceps sinensis. Int. Microbiol. 2023, 26, 791–806. [Google Scholar] [CrossRef]
- Xu, X.; Chen, Y.; Li, B.; Zhang, Z.; Qin, G.; Chen, T.; Tian, S. Molecular mechanisms underlying multi-level defense responses of horticultural crops to fungal pathogens. Hortic. Res. 2022, 9, uhac066. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621, Erratum in Nat Rev Microbiol.2021, 19, 72.. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Ji, N.; Liang, D.; Clark, L.V.; Sacks, E.J.; Kent, A.D. Host genetic variation drives the differentiation in the ecological role of the native Miscanthus root-associated microbiome. Microbiome 2023, 11, 216. [Google Scholar] [CrossRef]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- Nadarajah, K.; Abdul Rahman, N.S.N. Plant–microbe interaction: Aboveground to belowground, from the good to the bad. Int. J. Mol. Sci. 2021, 22, 10388. [Google Scholar] [CrossRef]
- Mercado-Blanco, J.; Abrantes, I.; Barra Caracciolo, A.; Bevivino, A.; Ciancio, A.; Grenni, P.; Hrynkiewicz, K.; Kredics, L.; Proença, D.N. Belowground microbiota and the health of tree crops. Front. Microbiol. 2018, 9, 1006. [Google Scholar] [CrossRef]
- Dos Santos Gomes, W.; Pereira, L.L.; Rodrigues da Luz, J.M.; Soares da Silva, M.d.C.; Reis Veloso, T.G.; Partelli, F.L. Exploring the microbiome of coffee plants: Implications for coffee quality and production. Food Res. Int. 2024, 179, 113972. [Google Scholar] [CrossRef]
- Pino, A.F.S.; Espinosa, Z.Y.D.; Cabrera, E.V.R. Characterization of the Rhizosphere Bacterial Microbiome and Coffee Bean Fermentation in the Castillo-Tambo and Bourbon Varieties in the Popayan-Colombia Plateau. BMC Plant Biol. 2023, 23, 13. [Google Scholar] [CrossRef]
- Sera, G.H.; de Carvalho, C.H.S.; de Rezende Abrahão, J.C.; Pozza, E.A.; Matiello, J.B.; de Almeida, S.R.; Bartelega, L.; dos Santos Botelho, D.M. Coffee leaf rust in Brazil: Historical events, current situation, and control measures. Agronomy 2022, 12, 496. [Google Scholar] [CrossRef]
- Yang, J.; Li, X.N.; Dong, Y.P.; Lyu, Z.J.; Wang, P.B.; He, A.L.; Hou, J.Z.; Luo, Z.Y. Agronomic and Yield Traits of 7 Arabica Coffee Hybrids F1 in Different Regions of Pu’er, Yunnan, China. Chin. J. Trop. Crops 2025, 46, 310–321. [Google Scholar] [CrossRef]
- Bie, W.; Qi, Z.Y.; Zeng, L.B.; Cui, F.Y.; Li, B.B.; Qiao, L.N.; Zhang, Z.H. Quality Characteristics and Standardization Research of Pu’er Coffee in Yunnan. China Port Sci. Technol. 2023, 5, 80–88. [Google Scholar]
- Huang, J.X.; Li, W.R.; Xia, B.; Hu, F.G. Strategies on How to Promote the Development of Coffee Industry with High Quality in Yunnan Province. Trop. Agric. Sci. Technol. 2022, 45, 21–29. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Deng, Y.; Wang, P.; Li, C.; Shi, D.; Wang, S. Assessing riverine fish community diversity and stability by eDNA metabarcoding. Ecol. Indic. 2023, 157, 111222. [Google Scholar] [CrossRef]
- Liao, H.; Liu, C.; Zhou, S.; Liu, C.; Eldridge, D.J.; Ai, C.; Wilhelm, S.W.; Singh, B.K.; Liang, X.; Radosevich, M.; et al. Prophage-encoded antibiotic resistance genes are enriched in human-impacted environments. Nat. Commun. 2024, 15, 8315. [Google Scholar] [CrossRef]
- Wen, T.; Liu, Y.X.; Liu, L.; Niu, G.; Ding, Z.; Teng, X.; Ma, J.; Liu, Y.; Yang, S.; Xie, P.; et al. ggClusterNet 2: An R package for microbial co-occurrence networks and associated indicator correlation patterns. Imeta 2025, 4, e70041. [Google Scholar] [CrossRef]
- Chaloner, T.M.; Gurr, S.J.; Bebber, D.P. Geometry and evolution of the ecological niche in plant-associated microbes. Nat. Commun. 2020, 11, 2955. [Google Scholar] [CrossRef] [PubMed]
- Coleman-Derr, D.; Desgarennes, D.; Fonseca-Garcia, C.; Gross, S.; Clingenpeel, S.; Woyke, T.; North, G.; Visel, A.; Partida-Martinez, L.P.; Tringe, S.G. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol. 2016, 209, 798–811. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, W.; Bu, J.; Lin, Y.; Bai, Y. Host genetics regulate the plant microbiome. Curr. Opin. Microbiol. 2023, 72, 102268. [Google Scholar] [CrossRef] [PubMed]
- Mendes, L.W.; Raaijmakers, J.M.; De Hollander, M.; Sepo, E.; Gómez Expósito, R.; Chiorato, A.F.; Mendes, R.; Tsai, S.M.; Carrión, V.J. Impact of the fungal pathogen Fusarium oxysporum on the taxonomic and functional diversity of the common bean root microbiome. Environ. Microbiome 2023, 18, 68. [Google Scholar] [CrossRef]
- Gao, M.; Xiong, C.; Gao, C.; Tsui, C.K.; Wang, M.-M.; Zhou, X.; Zhang, A.-M.; Cai, L. Disease-induced changes in plant microbiome assembly and functional adaptation. Microbiome 2021, 9, 187. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.L.; Zhu, Y.; Li, X.N.; Qin, Y.C.; Li, G.F.; Ye, Y.F.; He, Y.; Huang, J.Y.; Yang, S.D. Comparison of Soil Microbial Composition in Rhizospheres Between Wilt Disease-Resistant and Susceptible Melon Varieties. Microorganisms 2025, 13, 15. [Google Scholar] [CrossRef]
- Araujo, A.S.F.; Pereira, A.P.A.; de Medeiros, E.V.; Mendes, L.W. Restoring unbalanced rhizosphere: Microbiome transplants combatting leaf diseases. Trends Plant Sci. 2025, 30, 451–453. [Google Scholar] [CrossRef]
- Wu, C.F.; Liu, H.W.; Lai, L.Y.; Mei, Z.C.; Cai, P.; Zhang, H.Q.; Yang, J.; Chen, J.P.; Ge, T.D. Host genotype-specific plant microbiome correlates with wheat disease resistance. Biol. Fertil. Soils 2025, 61, 277–291. [Google Scholar] [CrossRef]
- Oliver, T.H.; Heard, M.S.; Isaac, N.J.B.; Roy, D.B.; Procter, D.; Eigenbrod, F.; Freckleton, R.; Hector, A.; Orme, C.D.L.; Petchey, O.L.; et al. Biodiversity and Resilience of Ecosystem Functions. Trends Ecol. Evol. 2015, 30, 673–684. [Google Scholar] [CrossRef]
- Dinesh, R.; Srinivasan, V.; Sheeja, T.E.; Anandaraj, M.; Srambikkal, H. Endophytic actinobacteria: Diversity, secondary metabolism and mechanisms to unsilence biosynthetic gene clusters. Crit. Rev. Microbiol. 2017, 43, 546–566. [Google Scholar] [CrossRef]
- Pascale, A.; Proietti, S.; Pantelides, I.S.; Stringlis, I.A. Modulation of the Root Microbiome by Plant Molecules: The Basis for Targeted Disease Suppression and Plant Growth Promotion. Front. Plant Sci. 2020, 10, 23. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, Y.; Dai, Z.; Liu, T.; Qin, Y.; Sun, D.; Liu, C.; Li, S. Ecological niche and environmental adaptability of denitrifiers in the yellow river estuary. Mar. Pollut. Bull. 2025, 213, 117693. [Google Scholar] [CrossRef]
- Ma, B.; Wang, Y.; Ye, S.; Liu, S.; Stirling, E.; Gilbert, J.A.; Faust, K.; Knight, R.; Jansson, J.K.; Cardona, C.; et al. Earth microbial co-occurrence network reveals interconnection pattern across microbiomes. Microbiome 2020, 8, 82. [Google Scholar] [CrossRef]
- Liu, C.; Li, C.; Jiang, Y.; Zeng, R.J.; Yao, M.; Li, X. A guide for comparing microbial co-occurrence networks. Imeta 2023, 2, e71. [Google Scholar] [CrossRef]
- Gao, C.; Xu, L.; Montoya, L.; Madera, M.; Hollingsworth, J.; Chen, L.; Purdom, E.; Singan, V.; Vogel, J.; Hutmacher, R.B.; et al. Co-occurrence networks reveal more complexity than community composition in resistance and resilience of microbial communities. Nat. Commun. 2022, 13, 3867. [Google Scholar] [CrossRef] [PubMed]
- Mendes, L.W.; Raaijmakers, J.M.; de Hollander, M.; Mendes, R.; Tsai, S.M. Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. ISME J. 2017, 12, 212–224. [Google Scholar] [CrossRef] [PubMed]
- De Sousa, L.P.; Guerreiro, O.; Mondego, J.M.C. The Rhizosphere Microbiomes of Five Species of Coffee Trees. Microbiol. Spectr. 2022, 10, 12. [Google Scholar] [CrossRef]
- Viegas, C.; Pacífico, C.; Faria, T.; de Oliveira, A.C.; Caetano, L.A.; Carolino, E.; Gomes, A.Q.; Viegas, S. Fungal contamination in green coffee beans samples: A public health concern. J. Toxicol. Env. Health Part A 2017, 80, 719–728. [Google Scholar] [CrossRef]
- Costa, G.X.R.; Silva, L.C.F.; de Oliveira, L.M.; Santos, L.D. Microbiota of arabica coffee: Insights from soil to fruit. World J. Microbiol. Biotechnol. 2024, 40, 18. [Google Scholar] [CrossRef]
- Zhao, S.G.; Zhang, A.; Zhao, Q.Y.; Zhang, Y.Y.; Wang, D.; Su, L.X.; Lin, X.J.; Sun, Y.; Yan, L.; Wang, X.W.; et al. Effects of coffee pericarp and litter mulsching on soil microbiomes diversity and functions in a tropical coffee plantation, South China. Front. Microbiol. 2024, 14, 13. [Google Scholar] [CrossRef]
- Das, P.; Effmert, U.; Baermann, G.; Quella, M.; Piechulla, B. Impact of bacterial volatiles on phytopathogenic fungi: An in vitro study on microbial competition and interaction. J. Exp. Bot. 2022, 73, 596–614. [Google Scholar] [CrossRef] [PubMed]
- Preston, G.M. Plant perceptions of plant growth-promoting Pseudomonas. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004, 359, 907–918. [Google Scholar] [CrossRef]
- Mesny, F.; Hacquard, S.; Thomma, B.P. Co-evolution within the plant holobiont drives host performance. EMBO Rep. 2023, 24, e57455. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Cañizares, C.; Jorrín, B.; Poole, P.S.; Tkacz, A. Understanding the holobiont: The interdependence of plants and their microbiome. Curr. Opin. Microbiol. 2017, 38, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Flörl, L.; Lin, J.; Griggs, R.G.; Massonnet, M.; Cochetel, N.; Figueroa-Balderas, R.; Cantù, D.; Bokulich, N.A. The genetic basis of microbiome recruitment in grapevine and its association with fermentative and pathogenic taxa. New Phytol. 2025. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Zhao, X.; Wang, Z.; Li, X.; Zhang, X.; Xie, C.; Du, H.; Jiang, K.; Qu, P.; Zhang, C. The Role of Coffee Microbiomes in Pathogen Resistance Across Varieties and Ecological Niches. Microorganisms 2025, 13, 1909. https://doi.org/10.3390/microorganisms13081909
Wu Y, Zhao X, Wang Z, Li X, Zhang X, Xie C, Du H, Jiang K, Qu P, Zhang C. The Role of Coffee Microbiomes in Pathogen Resistance Across Varieties and Ecological Niches. Microorganisms. 2025; 13(8):1909. https://doi.org/10.3390/microorganisms13081909
Chicago/Turabian StyleWu, Yihong, Xiu Zhao, Zuquan Wang, Xuejun Li, Xuesong Zhang, Chun Xie, Huabo Du, Kuaile Jiang, Peng Qu, and Chuanli Zhang. 2025. "The Role of Coffee Microbiomes in Pathogen Resistance Across Varieties and Ecological Niches" Microorganisms 13, no. 8: 1909. https://doi.org/10.3390/microorganisms13081909
APA StyleWu, Y., Zhao, X., Wang, Z., Li, X., Zhang, X., Xie, C., Du, H., Jiang, K., Qu, P., & Zhang, C. (2025). The Role of Coffee Microbiomes in Pathogen Resistance Across Varieties and Ecological Niches. Microorganisms, 13(8), 1909. https://doi.org/10.3390/microorganisms13081909




