“Encyclopaedia Cloacae”—Mapping Wastewaters from Pathogen A to Z
Abstract
1. Introduction
2. Materials and Methods
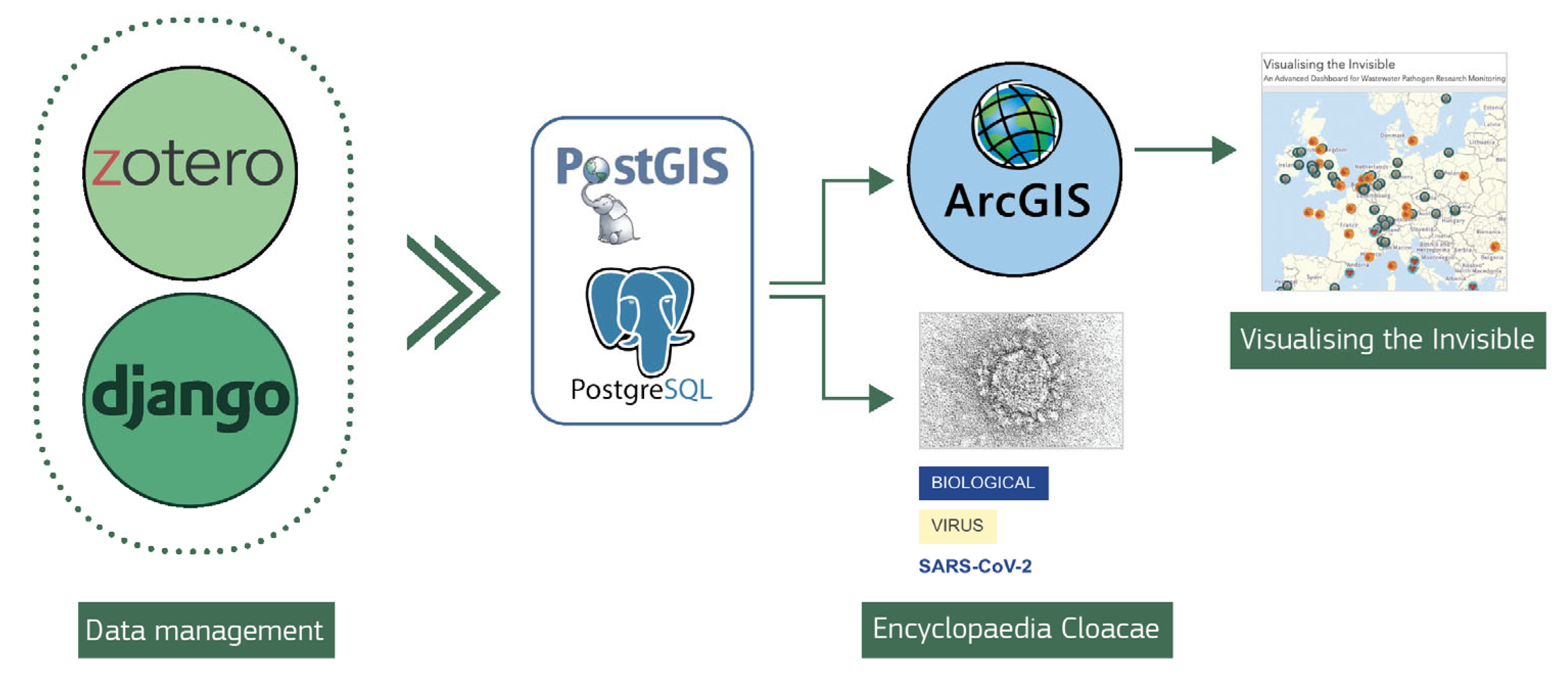
2.1. General Website Architecture
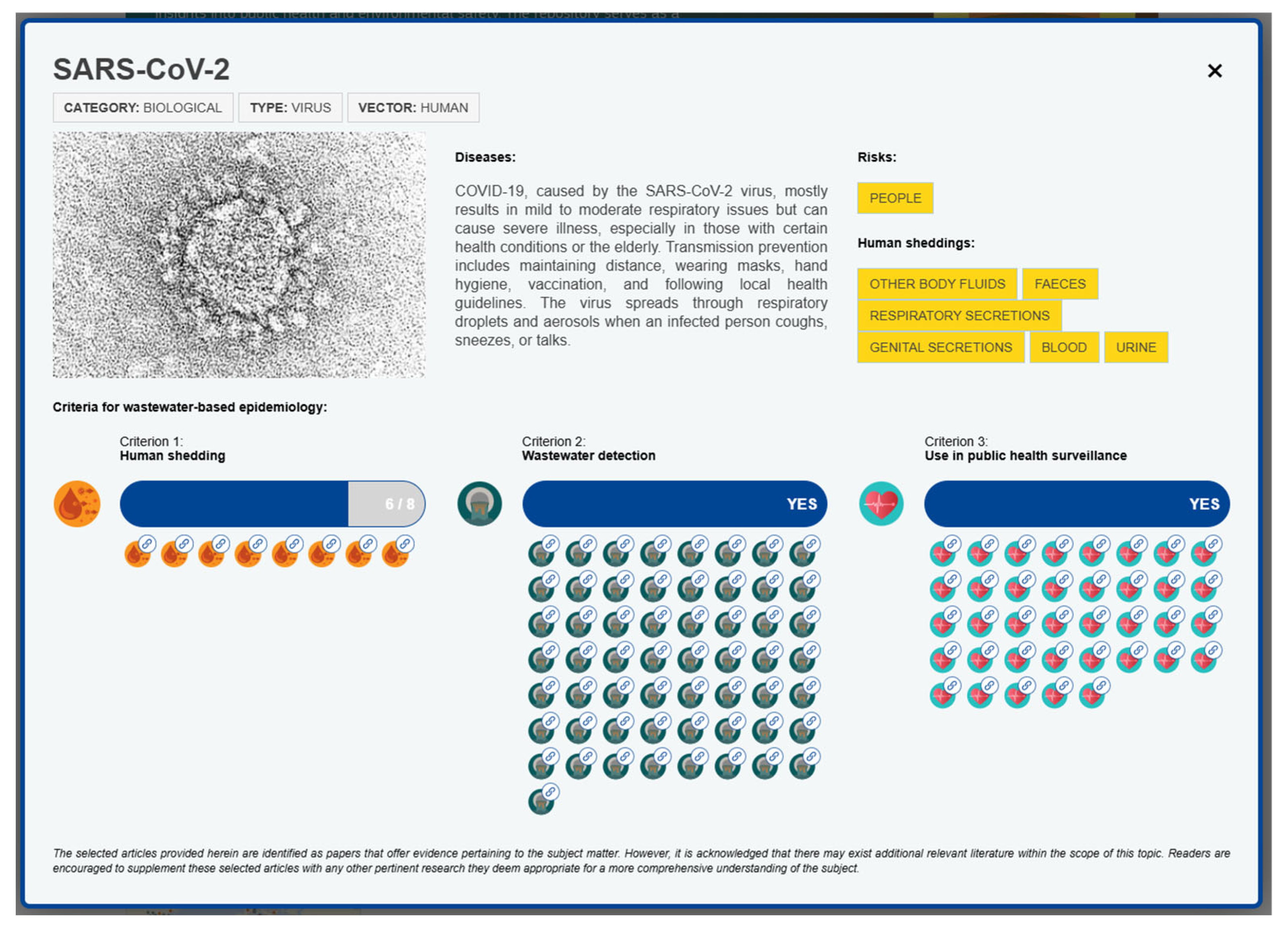
2.2. Criteria Description
- Criterion 1: Detection in human excreta focuses on the detection of pathogens in various forms of human excreta, providing insights into transmission dynamics and potential disease spread.
- Criterion 2: Detection in wastewater emphasises the importance of detecting pathogens in wastewater and establishing reliable detection methods, crucial for effective surveillance and public health interventions.
- Criterion 3: Public health surveillance highlights the integration of pathogen detection into public health surveillance systems, supporting early detection and continuous monitoring of public health threats.
2.2.1. Criterion 1: Detection in Human Excreta
2.2.2. Criterion 2: Detection in Wastewater
2.2.3. Criterion 3: Public Health Surveillance
2.3. Workflow: Population of Dataset and Maintenance
2.3.1. Addition of Pathogens and Related Information
- The name of the pathogen to be added.
- The type of pathogen (such as bacterium, virus, parasite, fungus, etc.).
- The vector, defined as the source of pathogen diffusion (such as animal, animal/human, human, environment, or none).
- A brief and easy-to-understand description (up to 500 characters), avoiding the use of acronyms.
- An optional thumbnail image with copyright clearance. If no image is available, a temporary generic clipart may be used.
- Information on the pathogen’s level of risk to humans, ecosystems, and/or farmed animals.
2.3.2. Publication Entry
2.3.3. Geo-Referencing Publications
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
Abbreviations
| ADMIN | Administration |
| AI | Artificial intelligence |
| AMR | Antimicrobial resistance |
| DOI | Digital Object Identifier |
| EU4S | EU Wastewater Observatory for Public Health |
| ISBN | International Standard Book Number |
| NGS | Next-generation sequencing |
| qPCR | Quantitative polymerase chain reaction |
| RSV | Respiratory syncytial virus |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| WBE | Wastewater-based epidemiology |
Appendix A
- Giuseppina La Rosa—Department of Environment and Health, Istituto Superiore di Sanità, Rome, Italy.
- Elisabetta Suffredini—Department of Food Safety, Nutrition and Veterinary Public Health, Istituto Superiore di Sanità, Rome, Italy.
- Timo Greiner—Robert Koch Institute, Berlin, Germany.
- Sophia Beyer—Department of Infectious Diseases, Robert Koch Institute, Berlin, Germany.
- Timo Greiner—Department of Infectious Disease Epidemiology, Robert Koch Institute, Berlin, Germany.
- Sophie Schneitler—Institute of Pneumology, University of Cologne, Köln, Germany.
- Sören L. Becker—Institute of Medical Microbiology and Hygiene, Saarland University, Homburg, Germany.
- Dr. Hagen Reichert—Gemeinschaftspraxis für Kinder- und Jugendmedizin, Homburg, Germany.
- Ulrike Braun—Umweltbundesamt, Berlin, Germany.
- Bilge Alpaslan Kocamemi—Turkish Water Institute (SUEN); Faculty of Engineering, Environmental Engineering Department, Marmara University, Istanbul, Turkey.
- Kata Farkas—School of Environmental and Natural Sciences, Bangor University, Bangor, United Kingdom.
References
- Levy, J.I.; Andersen, K.G.; Knight, R.; Karthikeyan, S. Wastewater Surveillance for Public Health. Science 2023, 379, 26–27. [Google Scholar] [CrossRef]
- The Potential of Wastewater-Based Epidemiology. Nat. Water 2023, 1, 399. [CrossRef]
- Medema, G.; Heijnen, L.; Elsinga, G.; Italiaander, R.; Brouwer, A. Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020, 7, 511–516. [Google Scholar] [CrossRef]
- La Rosa, G.; Iaconelli, M.; Mancini, P.; Bonanno Ferraro, G.; Veneri, C.; Bonadonna, L.; Lucentini, L.; Suffredini, E. First Detection of SARS-CoV-2 in Untreated Wastewaters in Italy. Sci. Total Environ. 2020, 736, 139652. [Google Scholar] [CrossRef]
- Castiglioni, S.; Schiarea, S.; Pellegrinelli, L.; Primache, V.; Galli, C.; Bubba, L.; Mancinelli, F.; Marinelli, M.; Cereda, D.; Ammoni, E.; et al. SARS-CoV-2 RNA in Urban Wastewater Samples to Monitor the COVID-19 Pandemic in Lombardy, Italy (March–June 2020). Sci. Total Environ. 2022, 806, 150816. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Ahmed, A.I.; Almansoori, S.; Alameri, S.; Adlan, A.; Odivilas, G.; Chattaway, M.A.; Salem, S.B.; Brudecki, G.; Elamin, W. A Narrative Review of Wastewater Surveillance: Pathogens of Concern, Applications, Detection Methods, and Challenges. Front. Public Health 2024, 12, 1445961. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Moshi, M.A.; Quinones, O.; Trenholm, R.A.; Chang, C.-L.; Cordes, D.; Vanderford, B.J.; Vo, V.; Gerrity, D.; Oh, E.C. Drug Use Patterns in Wastewater and Socioeconomic and Demographic Indicators. JAMA Netw. Open 2024, 7, e2432682. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Wang, D.; Li, P.; Deng, H.; Deng, Z. Advances in Wastewater-Based Epidemiology for Pandemic Surveillance: Methodological Frameworks and Future Perspectives. Microorganisms 2025, 13, 1169. [Google Scholar] [CrossRef] [PubMed]
- Sharara, N.; Endo, N.; Duvallet, C.; Ghaeli, N.; Matus, M.; Heussner, J.; Olesen, S.W.; Alm, E.J.; Chai, P.R.; Erickson, T.B. Wastewater Network Infrastructure in Public Health: Applications and Learnings from the COVID-19 Pandemic. PLoS Glob. Public Health 2021, 1, e0000061. [Google Scholar] [CrossRef]
- Bivins, A.; North, D.; Ahmad, A.; Ahmed, W.; Alm, E.; Been, F.; Bhattacharya, P.; Bijlsma, L.; Boehm, A.B.; Brown, J.; et al. Wastewater-Based Epidemiology: Global Collaborative to Maximize Contributions in the Fight against COVID-19. Environ. Sci. Technol. 2020, 54, 7754–7757. [Google Scholar] [CrossRef]
- Keenum, I.; Lin, N.J.; Logan-Jackson, A.; Gushgari, A.J.; D’Souza, N.; Steele, J.A.; Kaya, D.; Gushgari, L.R. Optimizing Wastewater Surveillance: The Necessity of Standardized Reporting and Proficiency for Public Health. Am. J. Public Health 2024, 114, 859–863. [Google Scholar] [CrossRef]
- Robins, K.; Leonard, A.F.C.; Farkas, K.; Graham, D.W.; Jones, D.L.; Kasprzyk-Hordern, B.; Bunce, J.T.; Grimsley, J.M.S.; Wade, M.J.; Zealand, A.M.; et al. Research Needs for Optimising Wastewater-Based Epidemiology Monitoring for Public Health Protection. J. Water Health 2022, 20, 1284–1313. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, M.; Ahmed, W.; Bibby, K.; Carducci, A.; Gerba, C.P.; Hamilton, K.A.; Haramoto, E.; Rose, J.B. SARS-CoV-2 in Wastewater: State of the Knowledge and Research Needs. Sci. Total Environ. 2020, 739, 139076. [Google Scholar] [CrossRef] [PubMed]
- Singer, A.C.; Thompson, J.R.; Filho, C.R.M.; Street, R.; Li, X.; Castiglioni, S.; Thomas, K.V. A World of Wastewater-Based Epidemiology. Nat. Water 2023, 1, 408–415. [Google Scholar] [CrossRef]
- Gawlik, B.M.; Comero, S.; Maffettone, R.; Glowacka, N.; Pierannunzi, F.; Sion, S.; Casado Poblador, T.; Bausa Lopez, L.; Philipp, W.; Tessarolo, A.; et al. The International Conference “Towards a Global Wastewater Surveillance System for Public Health”; Publications Office of the European Union: Luxembourg, 2024. [Google Scholar]
- Launching GLOWACON: A Global Initiative for Wastewater Surveillance for Public Health-European Commission. Available online: https://health.ec.europa.eu/latest-updates/launching-glowacon-global-initiative-wastewater-surveillance-public-health-2024-03-21_en (accessed on 18 June 2025).
- Murakami, M.; Kitajima, M.; Endo, N.; Ahmed, W.; Gawlik, B.M. The Growing Need to Establish a Global Wastewater Surveillance Consortium for Future Pandemic Preparedness. J. Travel Med. 2023, 30, taad035. [Google Scholar] [CrossRef]
- Gordeev, V.; Hölzer, M.; Desirò, D.; Goraichuk, I.V.; Knyazev, S.; Solo-Gabriele, H.; Skums, P.; Karthikeyan, S.; Evans, A.; Agrawal, S.; et al. Leveraging Wastewater Sequencing to Strengthen Global Public Health Surveillance. BMC Glob. Public Health 2025, 3, 23. [Google Scholar] [CrossRef]
- Servetas, S.L.; Parratt, K.H.; Brinkman, N.E.; Shanks, O.C.; Smith, T.; Mattson, P.J.; Lin, N.J. Standards to Support an Enduring Capability in Wastewater Surveillance for Public Health: Where Are We? Case Stud. Chem. Environ. Eng. 2022, 6, 100247. [Google Scholar] [CrossRef]
- Thakur, P.; Jadon, P. Django: Developing Web Using Python. In Proceedings of the 2023 3rd International Conference on Advance Computing and Innovative Technologies in Engineering (ICACITE), Greater Noida, India, 12–13 May 2023; pp. 303–306. [Google Scholar]
- Ahmed, K.K.M.; Al Dhubaib, B.E. Zotero: A Bibliographic Assistant to Researcher. J. Pharmacol. Pharmacother. 2011, 2, 303–305. [Google Scholar] [CrossRef]
- ESRI ArcGIS Server. ESRI’s Complete Server GIS White Paper; ESRI: Redlands, CA, USA, 2006. [Google Scholar]
- Carmo dos Santos, M.; Cerqueira Silva, A.C.; dos Reis Teixeira, C.; Pinheiro Macedo Prazeres, F.; Fernandes dos Santos, R.; de Araújo Rolo, C.; de Souza Santos, E.; Santos da Fonseca, M.; Oliveira Valente, C.; Saraiva Hodel, K.V.; et al. Wastewater Surveillance for Viral Pathogens: A Tool for Public Health. Heliyon 2024, 10, e33873. [Google Scholar] [CrossRef]
- Tiwari, A.; Radu, E.; Kreuzinger, N.; Ahmed, W.; Pitkänen, T. Key Considerations for Pathogen Surveillance in Wastewater. Sci. Total Environ. 2024, 945, 173862. [Google Scholar] [CrossRef] [PubMed]
- Grassly, N.C.; Shaw, A.G.; Owusu, M. Global Wastewater Surveillance for Pathogens with Pandemic Potential: Opportunities and Challenges. Lancet Microbe 2025, 6, 100939. [Google Scholar] [CrossRef] [PubMed]
- Diamond, M.B.; Keshaviah, A.; Bento, A.I.; Conroy-Ben, O.; Driver, E.M.; Ensor, K.B.; Halden, R.U.; Hopkins, L.P.; Kuhn, K.G.; Moe, C.L.; et al. Wastewater Surveillance of Pathogens Can Inform Public Health Responses. Nat. Med. 2022, 28, 1992–1995. [Google Scholar] [CrossRef] [PubMed]
- National Academies of Sciences, E. Wastewater-Based Disease Surveillance for Public Health Action; The National Academies Press: Washington, DC, USA, 2023; ISBN 978-0-309-69551-0. [Google Scholar]
- Endo, N.; Hisahara, A.; Kameda, Y.; Mochizuki, K.; Kitajima, M.; Yasojima, M.; Daigo, F.; Takemori, H.; Nakamura, M.; Matsuda, R.; et al. Enabling Quantitative Comparison of Wastewater Surveillance Data across Methods through Data Standardization without Method Standardization. Sci. Total Environ. 2024, 953, 176073. [Google Scholar] [CrossRef] [PubMed]
- Bailey, O.; Williams, V.; Shuttleworth, J.; Khan, L.; Morcillo de Amuedo, P.; Earl, E.; Walsh, P. Wastewater for Health-A Guide for Implementing Wastewater-Based Epidemiology to Improve Public Health in Low-Resource Settings; ARUP: Sydney, Australia, 2021. [Google Scholar]





| Proposed Harmonised Categories | Existing Categories |
|---|---|
| Faeces | Rectal bleeding |
| Faeces | |
| Stool | |
| Rectal swabs | |
| Urine | Urine |
| Vomit | Vomit |
| Respiratory secretions | Mucus |
| Nasal throat | |
| Nasal secretions | |
| Respiratory fluids | |
| Respiratory | |
| Respiratory droplets | |
| Respiratory specimens | |
| Saliva | |
| Throat secretions | |
| Sputum | |
| Spittle | |
| Blood | Blood |
| Serum | |
| Plasma | |
| Genital secretions | Semen |
| Vaginal fluids | |
| Tissues | Tissues |
| Skin | |
| Other body fluids | Cerebrospinal fluid |
| Sweat | |
| Breast milk | |
| Tears | |
| Fluids or excretions from abscesses |
| Name | Description |
|---|---|
| New | New publications not yet reviewed. |
| Checked | Publications reviewed for content and linked to hazard(s) and criteria. |
| Published | Folder automatically populated with publications registered on the website. |
| Errors | Publications rejected from automatic registration on the website. |
| Info | Contains reference materials such as the guidance manual and lists of pathogens and excreta. |
| For Later | Stores publications for future developments of the Encyclopaedia Cloacae. |
| Name | Description |
|---|---|
| pathogen | The name of the pathogen(s), separated by commas and registered according to standard nomenclature. |
| criteria | The Encyclopaedia Cloacae criterion or criteria the publication refers to, separated by commas. |
| excreta | Required only for criterion 1. The excreta where the selected pathogen has been found, separated by commas and registered according to the harmonised excreta categories reported in Table 1. |
| Microbial Class | Number of Pathogens (%) | Number of Publications (%) | |||
|---|---|---|---|---|---|
| Criterion 1: Detection in Human Excreta | Criterion 2: Detection in Wastewater | Criterion 3: Public Health Surveillance | Total | ||
| Viruses | 55 (51) | 101 (68) | 210 (76) | 122 (89) | 433 (77) |
| Bacteria | 34 (32) | 34 (23) | 48 (17) | 11 (8) | 93 (16) |
| Parasites | 17 (16) | 14 (9) | 18 (6) | 4 (3) | 36 (6) |
| Fungi | 1 (1) | 0 | 2 (1) | 0 | 2 (<1) |
| Total | 107 | 149 (26) | 278 (49) | 137 (24) | 564 |
| Microbial Class | Number of Publications (%) | ||||
|---|---|---|---|---|---|
| Human | Animal/Human | Animal | Environment | Total | |
| Viruses | 36 (34) | 13 (12) | 4 (4) | 2 (2) | 55 (51) |
| Bacteria | 13 (12) | 14 (13) | 4 (4) | 3 (3) | 34 (32) |
| Parasites | 0 | 10 (9) | 3 (3) | 4 (4) | 17 (16) |
| Fungi | 0 | 1 (1) | 0 | 0 | 1 (1) |
| Total | 49 (46) | 38 (36) | 11 (10) | 9 (8) | 107 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hirvonen, A.; Comero, S.; Tavazzi, S.; Mariani, G.; Cacciatori, C.; Maffettone, R.; Pierannunzi, F.; Panzarella, G.; Bausa-Lopez, L.; Sion, S.; et al. “Encyclopaedia Cloacae”—Mapping Wastewaters from Pathogen A to Z. Microorganisms 2025, 13, 1900. https://doi.org/10.3390/microorganisms13081900
Hirvonen A, Comero S, Tavazzi S, Mariani G, Cacciatori C, Maffettone R, Pierannunzi F, Panzarella G, Bausa-Lopez L, Sion S, et al. “Encyclopaedia Cloacae”—Mapping Wastewaters from Pathogen A to Z. Microorganisms. 2025; 13(8):1900. https://doi.org/10.3390/microorganisms13081900
Chicago/Turabian StyleHirvonen, Aurora, Sara Comero, Simona Tavazzi, Giulio Mariani, Caterina Cacciatori, Roberta Maffettone, Francesco Pierannunzi, Giulia Panzarella, Luis Bausa-Lopez, Sorin Sion, and et al. 2025. "“Encyclopaedia Cloacae”—Mapping Wastewaters from Pathogen A to Z" Microorganisms 13, no. 8: 1900. https://doi.org/10.3390/microorganisms13081900
APA StyleHirvonen, A., Comero, S., Tavazzi, S., Mariani, G., Cacciatori, C., Maffettone, R., Pierannunzi, F., Panzarella, G., Bausa-Lopez, L., Sion, S., Casado Poblador, T., Głowacka, N., Jones, D. L., Petrillo, M., Marchini, A., Querci, M., Gawlik, B. M., & on behalf of the Encyclopaedia Cloacae Collaborators. (2025). “Encyclopaedia Cloacae”—Mapping Wastewaters from Pathogen A to Z. Microorganisms, 13(8), 1900. https://doi.org/10.3390/microorganisms13081900






