Hepatitis E Virus Detection in Swine Livers and Feces in Heilongjiang, Northeastern China
Abstract
1. Introduction
2. Material and Methods
2.1. Sample Collection and Ethical Considerations
2.2. Viral RNA Extraction
2.3. Reverse Transcription Polymerase Chain Reaction (RT-PCR)/Nested PCR
2.4. Sequencing and Phylogenetic Analysis
2.5. Nucleotide Sequence Accession Numbers
3. Results
3.1. Prevalence of HEV in Pigs on Farms
3.2. Genotypes of HEV in Pigs on Farms in Heilongjiang Province
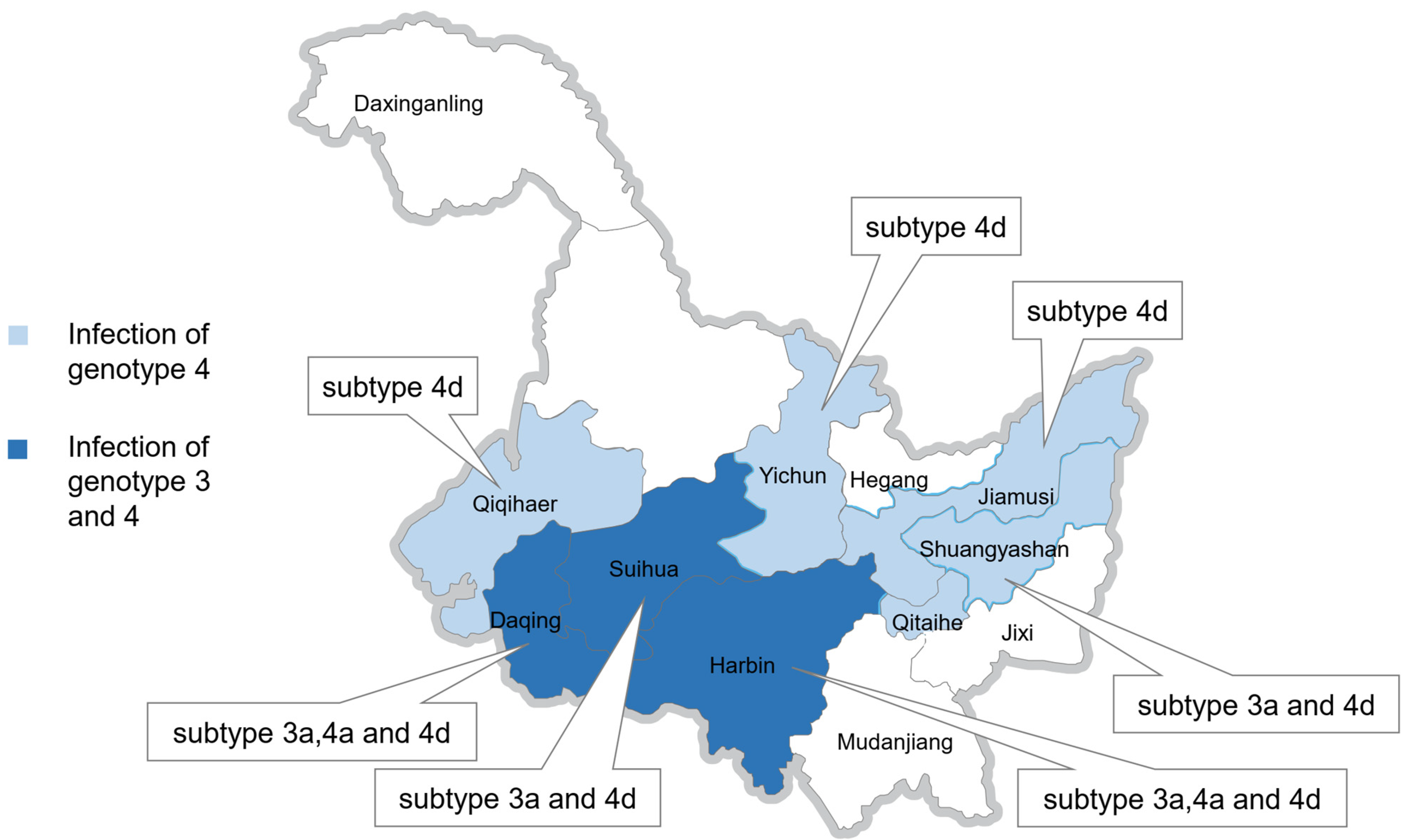
3.3. Subtypes of HEV Genotypes 3 and 4 on Farms in Heilongjiang Province
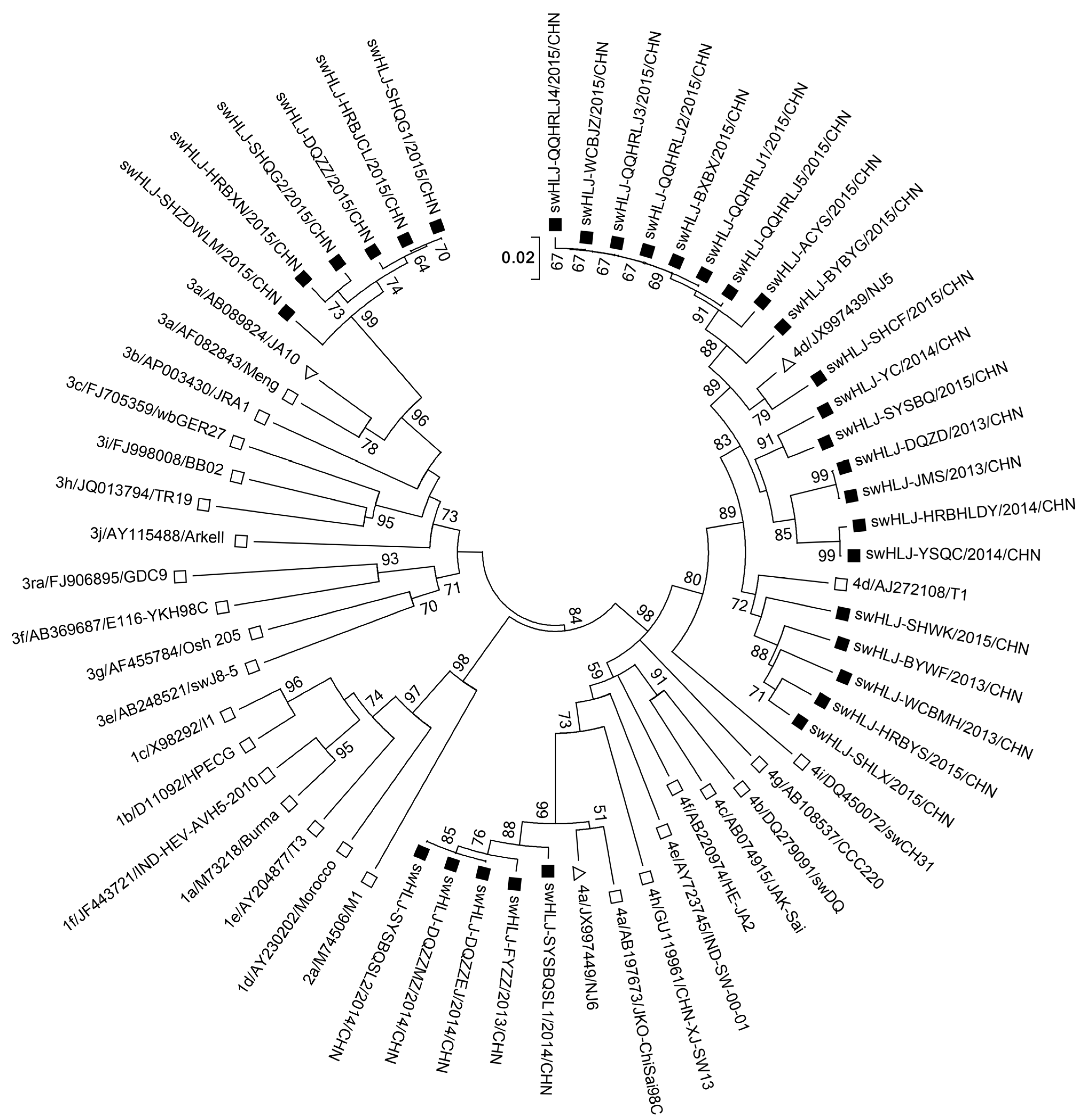
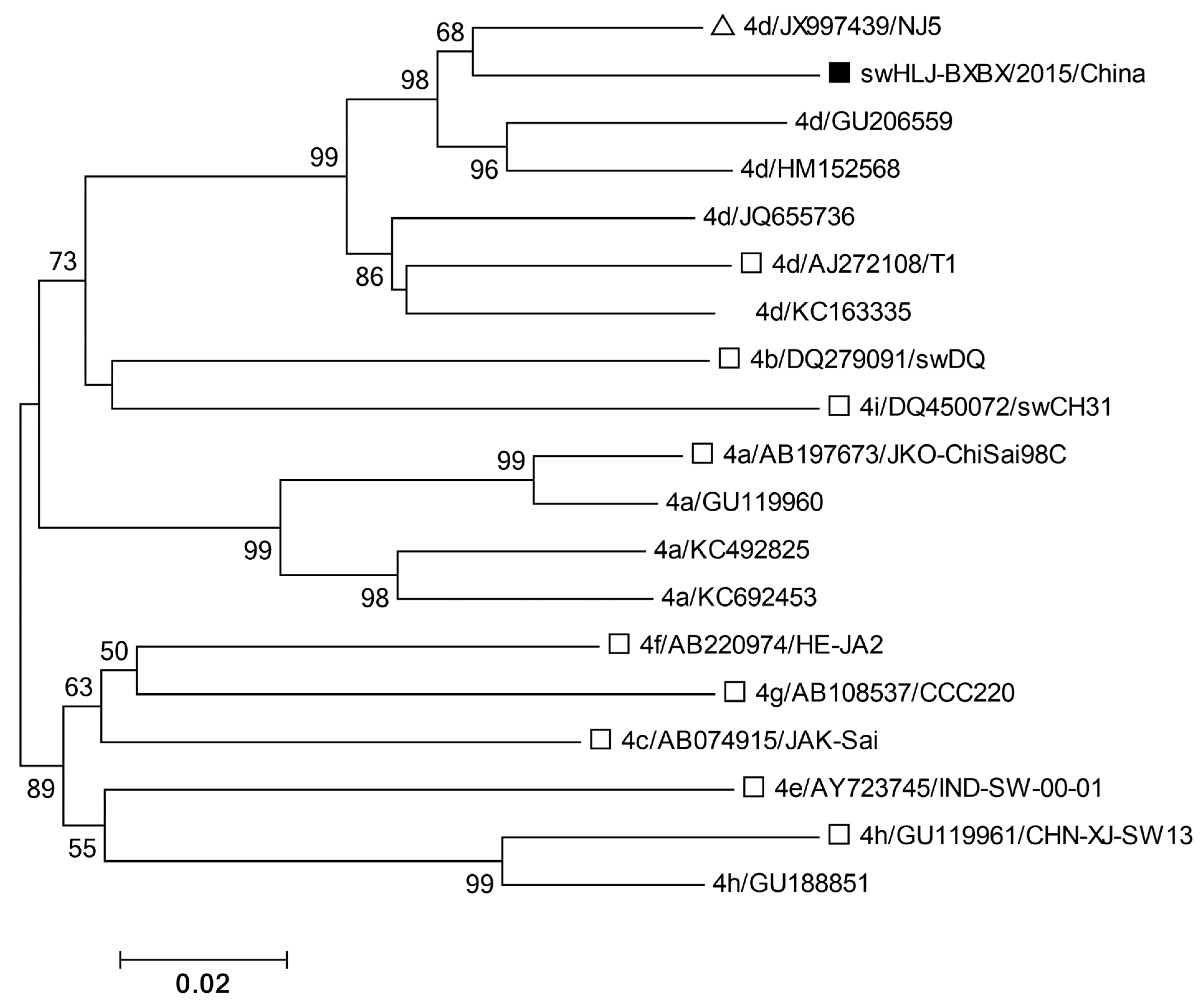
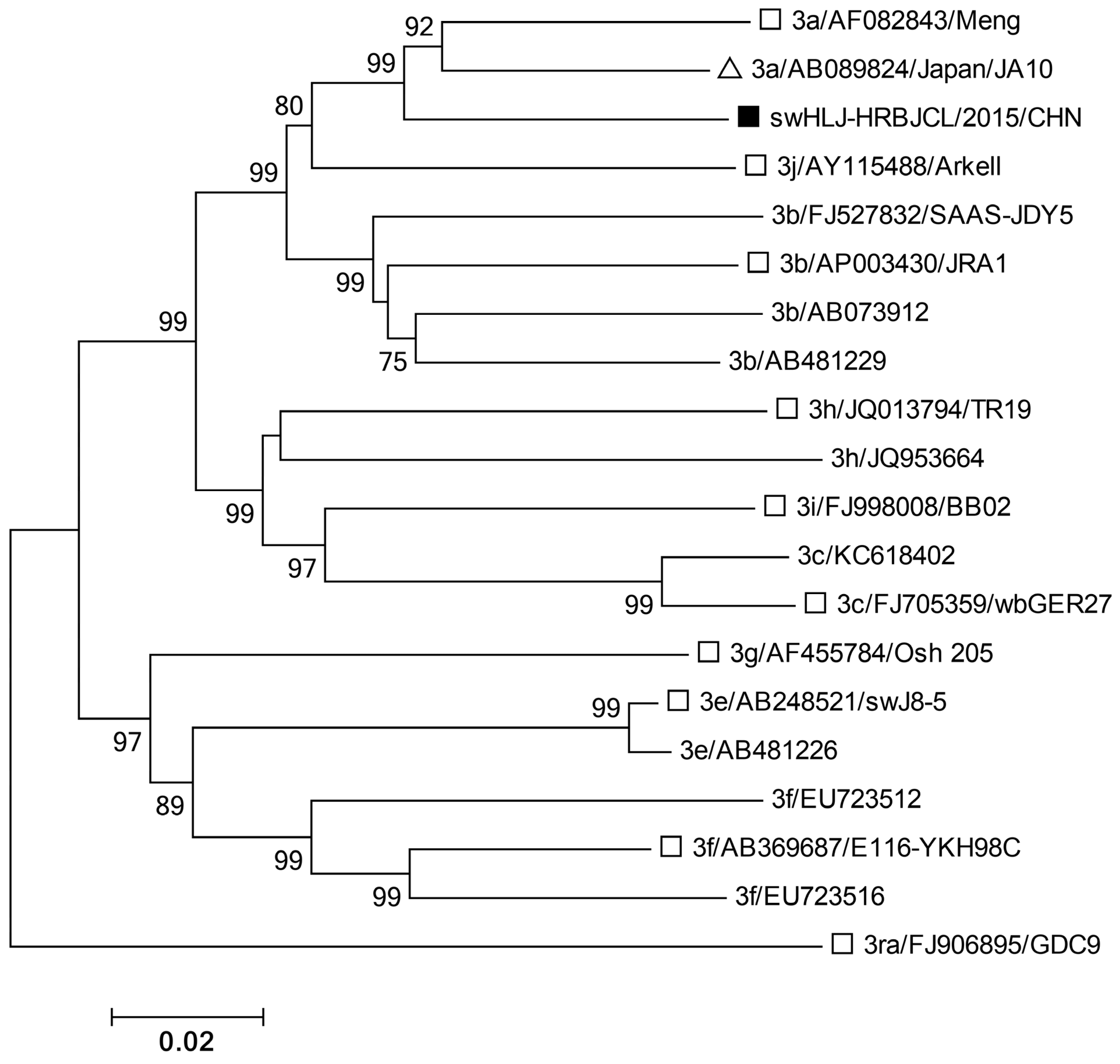
3.4. Phylogenetic Analysis of HEV ORF2 Sequence
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Heo, N.Y. Hepatitis E Virus: Epidemiology, Diagnosis, and Management. Korean J. Gastroenterol. 2019, 74, 130–136. [Google Scholar] [CrossRef][Green Version]
- Ma, Z.; de Man, R.A.; Kamar, N.; Pan, Q. Chronic hepatitis E: Advancing research and patient care. J. Hepatol. 2022, 77, 1109–1123. [Google Scholar] [CrossRef]
- Cancela, F.; Noceti, O.; Arbiza, J.; Mirazo, S. Structural aspects of hepatitis E virus. Arch. Virol. 2022, 167, 2457–2481. [Google Scholar] [CrossRef] [PubMed]
- Larrue, H.; Abravanel, F.; Péron, J.M.; Hepatitis, E. what’s the real issue? Liver Int. Off. J. Int. Assoc. Study Liver 2020, 40 (Suppl. 1), 43–47. [Google Scholar]
- Paronetto, O.; Allioux, C.; Diméglio, C.; Lobjois, L.; Jeanne, N.; Ranger, N.; Boineau, J.; Pucelle, M.; Demmou, S.; Abravanel, F.; et al. Characterization of virus–host recombinant variants of the hepatitis E virus. J. Virol. 2024, 98, e0029524. [Google Scholar] [CrossRef]
- Alexandrova, R.; Tsachev, I.; Kirov, P.; Abudalleh, A.; Hristov, H.; Zhivkova, T.; Dyakova, L.; Baymakova, M. Hepatitis E Virus (HEV) Infection Among Immunocompromised Individuals: A Brief Narrative Review. Infect. Drug Resist. 2024, 17, 1021–1040. [Google Scholar] [CrossRef]
- Mushahwar, I.K. Hepatitis E virus: Molecular virology, clinical features, diagnosis, transmission, epidemiology, and prevention. J. Med. Virol. 2008, 80, 646–658. [Google Scholar] [CrossRef]
- Kamar, N.; Bendall, R.; Legrand-Abravanel, F.; Xia, N.S.; Ijaz, S.; Izopet, J.; Dalton, H.R. Hepatitis E. Lancet 2012, 379, 2477–2488. [Google Scholar] [CrossRef] [PubMed]
- Teshale, E.H.; Hu, D.J. Hepatitis E: Epidemiology and prevention. World J. Hepatol. 2011, 3, 285–291. [Google Scholar] [CrossRef]
- Aslan, A.T.; Balaban, H.Y. Hepatitis E virus: Epidemiology, diagnosis, clinical manifestations, and treatment. World J. Gastroenterol. 2020, 26, 5543–5560. [Google Scholar] [CrossRef]
- Khuroo, M.S.; Khuroo, M.S.; Khuroo, N.S. Hepatitis E: Discovery, global impact, control and cure. World J. Gastroenterol. 2016, 22, 7030–7045. [Google Scholar] [CrossRef]
- Pavio, N.; Meng, X.-J.; Doceul, V. Zoonotic origin of hepatitis E. Curr. Opin. Virol. 2015, 10, 34–41. [Google Scholar] [CrossRef]
- Smith, D.B.; Simmonds, P.; Jameel, S.; Emerson, S.U.; Harrison, T.J.; Meng, X.J.; Okamoto, H.; Van der Poel, W.H.M.; Purdy, M.A.; Members of The International Committee on The Taxonomy of Viruses Study Group. Consensus proposals for classification of the family Hepeviridae. J. Gen. Virol. 2014, 95, 2223–2232. [Google Scholar] [CrossRef]
- Meng, X.J.; Purcell, R.H.; Halbur, P.G.; Lehman, J.R.; Webb, D.M.; Tsareva, T.S.; Haynes, J.S.; Thacker, B.J.; Emerson, S.U. A novel virus in swine is closely related to the human hepatitis E virus. Proc. Natl. Acad. Sci. USA 1997, 94, 9860–9865. [Google Scholar] [CrossRef]
- Wang, B.; Subramaniam, S.; Tian, D.; Mahsoub, H.M.; Heffron, C.L.; Meng, X.-J.; Patton, J.T. Phosphorylation of Ser711 residue in the hypervariable region of zoonotic genotype 3 hepatitis E virus is important for virus replication. mBio 2024, 15, e0263524. [Google Scholar] [CrossRef] [PubMed]
- Tam, A.W.; Smith, M.M.; Guerra, M.E.; Huang, C.-C.; Bradley, D.W.; Fry, K.E.; Reyes, G.R. Hepatitis E virus (HEV): Molecular cloning and sequencing of the full-length viral genome. Virology 1991, 185, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Parvez, M.K. The hepatitis E virus nonstructural polyprotein. Future Microbiol. 2017, 12, 915–924. [Google Scholar] [CrossRef]
- Sayed, I.M.; Ramadan, H.K.; Hafez, M.H.R.; Elkhawaga, A.A.; El-Mokhtar, M.A. Hepatitis E virus (HEV) open reading frame 2: Role in pathogenesis and diagnosis in HEV infections. Rev. Med. Virol. 2022, 32, e2401. [Google Scholar] [CrossRef]
- LeDesma, R.; Heller, B.; Biswas, A.; Maya, S.; Gili, S.; Higgins, J.; Ploss, A. Structural features stabilized by divalent cation coordination within hepatitis E virus ORF1 are critical for viral replication. eLife 2023, 12, e80529. [Google Scholar] [CrossRef]
- LeDesma, R.; Nimgaonkar, I.; Ploss, A. Hepatitis E Virus Replication. Viruses 2019, 11, 719. [Google Scholar] [CrossRef] [PubMed]
- Kalia, M.; Chandra, V.; Rahman, S.A.; Sehgal, D.; Jameel, S. Heparan sulfate proteoglycans are required for cellular binding of the hepatitis E virus ORF2 capsid protein and for viral infection. J. Virol. 2009, 83, 12714–12724. [Google Scholar] [CrossRef]
- Xing, L.; Wang, J.C.; Li, T.-C.; Yasutomi, Y.; Lara, J.; Khudyakov, Y.; Schofield, D.; Emerson, S.U.; Purcell, R.H.; Takeda, N.; et al. Spatial configuration of hepatitis E virus antigenic domain. J. Virol. 2011, 85, 1117–1124. [Google Scholar] [CrossRef]
- Yamada, K.; Takahashi, M.; Hoshino, Y.; Takahashi, H.; Ichiyama, K.; Nagashima, S.; Tanaka, T.; Okamoto, H. ORF3 protein of hepatitis E virus is essential for virion release from infected cells. J. Gen. Virol. 2009, 90, 1880–1891. [Google Scholar] [CrossRef]
- Kenney, S.P. The Current Host Range of Hepatitis E Viruses. Viruses 2019, 11, 452. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, H.; Xia, N.; Peng, G.; Lan, H.; Zhuang, H.; Zhu, Y.; Li, S.; Tian, K.; Gu, W.; et al. Prevalence, isolation, and partial sequence analysis of hepatitis E virus from domestic animals in China. J. Med. Virol. 2002, 67, 516–521. [Google Scholar] [CrossRef]
- Lu, Y.-H.; Qian, H.-Z.; Qin, X.; Jiang, Q.-W.; Zheng, Y.-J. Subtypes of genotype 3 hepatitis E virus in pigs. Vet. J. 2013, 197, 509–511. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Li, L.; Wang, L.; Bu, Q.; Fu, H.; Han, J.; Zhu, Y.; Lu, F.; Zhuang, H. Phylogenetic analysis of 626 hepatitis E virus (HEV) isolates from humans and animals in China (1986–2011) showing genotype diversity and zoonotic transmission. Infect. Genet. Evol. 2012, 12, 428–434. [Google Scholar] [CrossRef]
- Si, F.-S.; Zhu, Y.-M.; Dong, S.-J.; Yu, S.-S.; Yu, R.-S.; Shen, S.-Y.; Yang, Q.; Li, Z. Full genomic sequence analysis of swine genotype 3 hepatitis E virus isolated from Shanghai. Virus Res. 2009, 144, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-J.; Zhao, Q.; Jiang, F.-L.; Liu, B.-Y.; Zhao, J.-N.; Dang, L.; Sun, Y.-N.; Mu, Y.; Xiao, S.-Q.; Wang, C.-B.; et al. Genetic characterization and serological prevalence of swine hepatitis E virus in Shandong Province, China. Vet. Microbiol. 2014, 172, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Lu, L.; Li, C.; Hagedorn, C.H. Phylogenetic analysis of global hepatitis E virus sequences: Genetic diversity, subtypes and zoonosis. Rev. Med. Virol. 2006, 16, 5–36. [Google Scholar] [CrossRef]
- Smith, D.B.; Izopet, J.; Nicot, F.; Simmonds, P.; Jameel, S.; Meng, X.-J.; Norder, H.; Okamoto, H.; van der Poel, W.H.; Reuter, G.; et al. Update: Proposed reference sequences for subtypes of hepatitis E virus (species Orthohepevirus A). J. Gen. Virol. 2020, 101, 692–698. [Google Scholar] [CrossRef]
- Dai, X.; Dong, C.; Zhou, Z.; Liang, J.; Dong, M.; Yang, Y.; Fu, J.; Tian, H.; Wang, S.; Fan, J.; et al. Hepatitis E virus genotype 4, Nanjing, China, 2001-2011. Emerg. Infect. Dis. 2013, 19, 1528–1530. [Google Scholar] [CrossRef] [PubMed]
- Tokita, H.; Harada, H.; Gotanda, Y.; Takahashi, M.; Nishizawa, T.; Okamoto, H. Molecular and serological characterization of sporadic acute hepatitis E in a Japanese patient infected with a genotype III hepatitis E virus in 1993. J. Gen. Virol. 2003, 84, 421–427. [Google Scholar] [CrossRef]
- Purcell, R.H.; Emerson, S.U. Hepatitis E: An emerging awareness of an old disease. J. Hepatol. 2008, 48, 494–503. [Google Scholar] [CrossRef] [PubMed]
- Abraham, P. Viral hepatitis in India. Clin. Lab. Med. 2012, 32, 159–174. [Google Scholar] [CrossRef]
- Wielick, C.; Ludwig-Begall, L.; Faes, C.; Ribbens, S.; Saegerman, C.; Thiry, E. A Randomized Large-Scale Cross-Sectional Serological Survey of Hepatitis E Virus Infection in Belgian Pig Farms. Microorganisms 2023, 11, 129. [Google Scholar] [CrossRef]
- Monini, M.; Di Bartolo, I.; Ianiro, G.; Angeloni, G.; Magistrali, C.F.; Ostanello, F.; Ruggeri, F.M. Detection and molecular characterization of zoonotic viruses in swine fecal samples in Italian pig herds. Arch. Virol. 2015, 160, 2547–2556. [Google Scholar] [CrossRef] [PubMed]
- Izopet, J.; Tremeaux, P.; Marion, O.; Migueres, M.; Capelli, N.; Chapuy-Regaud, S.; Mansuy, J.M.; Abravanel, F.; Kamar, N.; Lhomme, S. Hepatitis E virus infections in Europe. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2019, 120, 20–26. [Google Scholar] [CrossRef]
- Raji, Y.E.; Toung, O.P.; Taib, N.M.; Bin Sekawi, Z. A systematic review of the epidemiology of Hepatitis E virus infection in South-Eastern Asia. Virulence 2021, 12, 114–129. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.B.; Simmonds, P.; Izopet, J.; Oliveira-Filho, E.F.; Ulrich, R.G.; Johne, R.; Koenig, M.; Jameel, S.; Harrison, T.J.; Meng, X.-J.; et al. Proposed reference sequences for hepatitis E virus subtypes. J. Gen. Virol. 2016, 97, 537–542. [Google Scholar] [CrossRef]
- Ma, H.; Geng, Y.; Li, Z.; Harrison, T.J.; Huang, W.; Zhao, C.; Wang, Y. Analysis of the complete genome sequences of one swine and two human hepatitis E virus genotype 4 strains isolated in Beijing, China. Infect. Genet. Evol. 2013, 18, 42–47. [Google Scholar] [CrossRef]
- Yu, Y.; Sun, J.; Liu, M.; Xia, L.; Zhao, C.; Harrison, T.J.; Wang, Y. Seroepidemiology and genetic characterization of hepatitis E virus in the northeast of China. Infect. Genet. Evol. 2009, 9, 554–561. [Google Scholar] [CrossRef]
- Zhang, W.; He, Y.; Wang, H.; Shen, Q.; Cui, L.; Wang, X.; Shao, S.; Hua, X. Hepatitis E virus genotype diversity in eastern China. Emerg. Infect. Dis. 2010, 16, 1630–1632. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Li, T.; Xia, Y.; Cong, C.; Chen, S.; Zhang, Y.; Gong, S.; Wang, W.; Liu, H.; Chen, D.; et al. Genotype 4 Hepatitis E virus replicates in the placenta, causes severe histopathological damage, and vertically transmits to fetuses. J. Infect. 2023, 87, 34–45. [Google Scholar] [CrossRef] [PubMed]
| Accession | Strain | Host | Genotype | Subtype |
|---|---|---|---|---|
| AB089824 | 3a/AB089824/JA10 | Homo sapiens | 3 | 3a |
| AF082843 | 3a/AF082843/Meng | Swine | 3 | 3a |
| AP003430 | 3b/AP003430/JRA1 | Homo sapiens | 3 | 3b |
| FJ705359 | 3c/FJ705359/wbGER27 | Wild boar | 3 | 3c |
| FJ998008 | 3i/FJ998008/BB02 | Wild boar | 3 | 3i |
| JQ013794 | 3h/JQ013794/TR19 | Homo sapiens | 3 | 3h |
| AY115488 | 3j/AY115488/ArKell | Swine | 3 | 3j |
| FJ906895 | 3ra/FJ906895/GDC9 | Rabbit | 3 | 3ra |
| AB369687 | 3f/AB369687/E116-YKH98C | Homo sapiens | 3 | 3f |
| AF455784 | 3g/AF455784/Osh 205 | Swine | 3 | 3g |
| AB248521 | 3e/AB248521/swJ8-5 | Swine | 3 | 3e |
| X98292 | 1c/X98292/I1 | Homo sapiens | 1 | 1c |
| D11092 | 1b/D11092/HPECG | Homo sapiens | 1 | 1b |
| JF443721 | 1f/JF443721/IND-HEV-AVH5-2010 | Homo sapiens | 1 | 1f |
| M73218 | 1a/M73218/Burma | Homo sapiens | 1 | 1a |
| AY204877 | 1e/AY204877/T3 | Homo sapiens | 1 | 1e |
| AY230202 | 1d/AY230202/Morocco | Homo sapiens | 1 | 1d |
| M74506 | 2a/M74506/M1 | Homo sapiens | 2 | 2a |
| JX997449 | 4a/JX997449/NJ6 | Homo sapiens | 4 | 4a |
| AB197673 | 4a/AB197673/JKO-ChiSai98C | Homo sapiens | 4 | 4a |
| GU119961 | 4h/GU119961/CHN-XJ-SW13 | Swine | 4 | 4a |
| AY723745 | 4e/AY723745/IND-SW-00-01 | Swine | 4 | 4e |
| AB220974 | 4f/AB220974/HE-JA2 | Homo sapiens | 4 | 4f |
| AB074915 | 4c/AB074915/JAK-Sai | Homo sapiens | 4 | 4c |
| DQ279091 | 4b/DQ279091/swDQ | Swine | 4 | 4b |
| AB108537 | 4g/AB108537/CCC220 | Homo sapiens | 4 | 4g |
| DQ450072 | 4i/DQ450072/swCH31 | Swine | 4 | 4i |
| AJ272108 | 4d/AJ272108/T1 | Homo sapiens | 4 | 4d |
| JX997439 | 4d/JX997439/NJ5 | Homo sapiens | 4 | 4d |
| Accession | Strain | Isolation Source | Genotype: | Subtype |
|---|---|---|---|---|
| KU130364 | swHLJ-FYZZ/2013/CHN | Liver | 4 | 4a |
| KU130365 | swHLJ-DQZZEJ/2014/CHN | Liver | 4 | 4a |
| KU130366 | swHLJ-DQZZMZ/2014/CHN | Liver | 4 | 4a |
| KU130367 | swHLJ-SYSBQSL1/2014/CHN | Liver | 4 | 4a |
| KU130368 | swHLJ-SYSBQSL2/2014/CHN | Liver | 4 | 4a |
| KU130369 | swHLJ-BYWF/2013/CHN | Liver | 4 | 4d |
| KU130370 | swHLJ-DQZD/2013/CHN | Liver | 4 | 4d |
| KU130371 | swHLJ-JMS/2013/CHN | Liver | 4 | 4d |
| KU130372 | swHLJ-WCBMH/2013/CHN | Liver | 4 | 4d |
| KU130373 | swHLJ-HRBHLDY/2014/CHN | Liver | 4 | 4d |
| KU130374 | swHLJ-YC/2014/CHN | Liver | 4 | 4d |
| KU130375 | swHLJ-YSQC/2014/CHN | Liver | 4 | 4d |
| KU130376 | swHLJ-ACYS/2015/CHN | Liver | 4 | 4d |
| KU130377 | swHLJ-BXBX/2015/CHN | Liver | 4 | 4d |
| KU130378 | swHLJ-BYBYG/2015/CHN | Liver | 4 | 4d |
| KU130379 | swHLJ-HRBYS/2015/CHN | Liver | 4 | 4d |
| KU130380 | swHLJ-QQHRLJ1/2015/CHN | Fecal sample | 4 | 4d |
| KU130381 | swHLJ-QQHRLJ2/2015/CHN | Fecal sample | 4 | 4d |
| KU130382 | swHLJ-QQHRLJ3/2015/CHN | Fecal sample | 4 | 4d |
| KU130383 | swHLJ-QQHRLJ4/2015/CHN | Fecal sample | 4 | 4d |
| KU130384 | swHLJ-QQHRLJ5/2015/CHN | Fecal sample | 4 | 4d |
| KU130385 | swHLJ-SHCF/2015/CHN | Liver | 4 | 4d |
| KU130386 | swHLJ-SHLX/2015/CHN | Liver | 4 | 4d |
| KU130387 | swHLJ-SHWK/2015/CHN | Liver | 4 | 4d |
| KU130388 | swHLJ-SYSBQ/2015/CHN | Liver | 4 | 4d |
| KU130389 | swHLJ-WCBJZ/2015/CHN | Liver | 4 | 4d |
| KU130390 | swHLJ-DQZZ/2015/CHN | Liver | 3 | 3a |
| KU130391 | swHLJ-HRBJCL/2015/CHN | Liver | 3 | 3a |
| KU130392 | swHLJ-HRBXN/2015/CHN | Liver | 3 | 3a |
| KU130393 | swHLJ-SHQG1/2015/CHN | Liver | 3 | 3a |
| KU130394 | swHLJ-SHQG2/2015/CHN | Liver | 3 | 3a |
| KU130395 | swHLJ-SHZDWLM/2015/CHN | Liver | 3 | 3a |
| Number of Samples (n = 82) | Number of Pig Farms (n = 63) | No. Positive/No. Tested (%) | HEV Subtypes | Location (city) | Source |
|---|---|---|---|---|---|
| 27/82(32.9) | |||||
| 43 | 30 | 12/43(27.9) | 3a, 4a, 4d | Harbin | Diseased/deceased pigs, Liver samples |
| 16 | 10 | 6/16(37.5) | 3a, 4d | Suihua | Diseased/deceased pigs, Liver samples |
| 7 | 7 | 4/7(57.1) | 3a, 4a, 4d | Daqing | Diseased/deceased pigs, Liver samples |
| 7 | 7 | 3/7(42.8) | 4a, 4d | Shuangyashan | Diseased/deceased pigs, Liver samples |
| 1 | 1 | 1/1(100) | 4d | Jiamusi | Diseased/deceased pigs, Liver samples |
| 5 | 5 | 1/5(20) | 4d | Yichun | Diseased/deceased pigs, Liver samples |
| 3 | 3 | 0/3(0) | -- | Mudanjiang | Diseased/deceased pigs, Liver samples |
| Number of Samples (n = 86) | Name of Pig Farms (n = 3) | No. Positive/No. Tested (%) | HEV Subtypes | Location (city) | Source |
|---|---|---|---|---|---|
| 5/86(5.8) | |||||
| 28 | Farm 1 | 1/28(3.5) | 4d | Qiqihar | Healthy pigs, Fecal samples |
| 28 | Farm 2 | 2/28(7.1) | 4d | Qiqihar | Healthy pigs, Fecal samples |
| 30 | Farm 3 | 2/30(6.7) | 4d | Qiqihar | Healthy pigs, Fecal samples |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, H.; Li, H.; Yan, L.; Wang, G.; Liu, Y.; An, T.; Tu, Y.; Wang, S.; Cai, X. Hepatitis E Virus Detection in Swine Livers and Feces in Heilongjiang, Northeastern China. Microorganisms 2025, 13, 1899. https://doi.org/10.3390/microorganisms13081899
He H, Li H, Yan L, Wang G, Liu Y, An T, Tu Y, Wang S, Cai X. Hepatitis E Virus Detection in Swine Livers and Feces in Heilongjiang, Northeastern China. Microorganisms. 2025; 13(8):1899. https://doi.org/10.3390/microorganisms13081899
Chicago/Turabian StyleHe, Haijuan, Hai Li, Lei Yan, Gang Wang, Yonggang Liu, Tongqing An, Yabin Tu, Shujie Wang, and Xuehui Cai. 2025. "Hepatitis E Virus Detection in Swine Livers and Feces in Heilongjiang, Northeastern China" Microorganisms 13, no. 8: 1899. https://doi.org/10.3390/microorganisms13081899
APA StyleHe, H., Li, H., Yan, L., Wang, G., Liu, Y., An, T., Tu, Y., Wang, S., & Cai, X. (2025). Hepatitis E Virus Detection in Swine Livers and Feces in Heilongjiang, Northeastern China. Microorganisms, 13(8), 1899. https://doi.org/10.3390/microorganisms13081899






