Dynamics and Assembly Mechanisms of Bacterial Communities During Larval Development of Macrobrachium rosenbergii: A High-Frequency Sampling Study Based on 16S rRNA Absolute Quantification Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Sample Collection
2.2. DNA Extraction and Amplicon Sequencing
2.3. Illumina Read Data Processing and Analysis
2.4. Data Analyses
3. Results
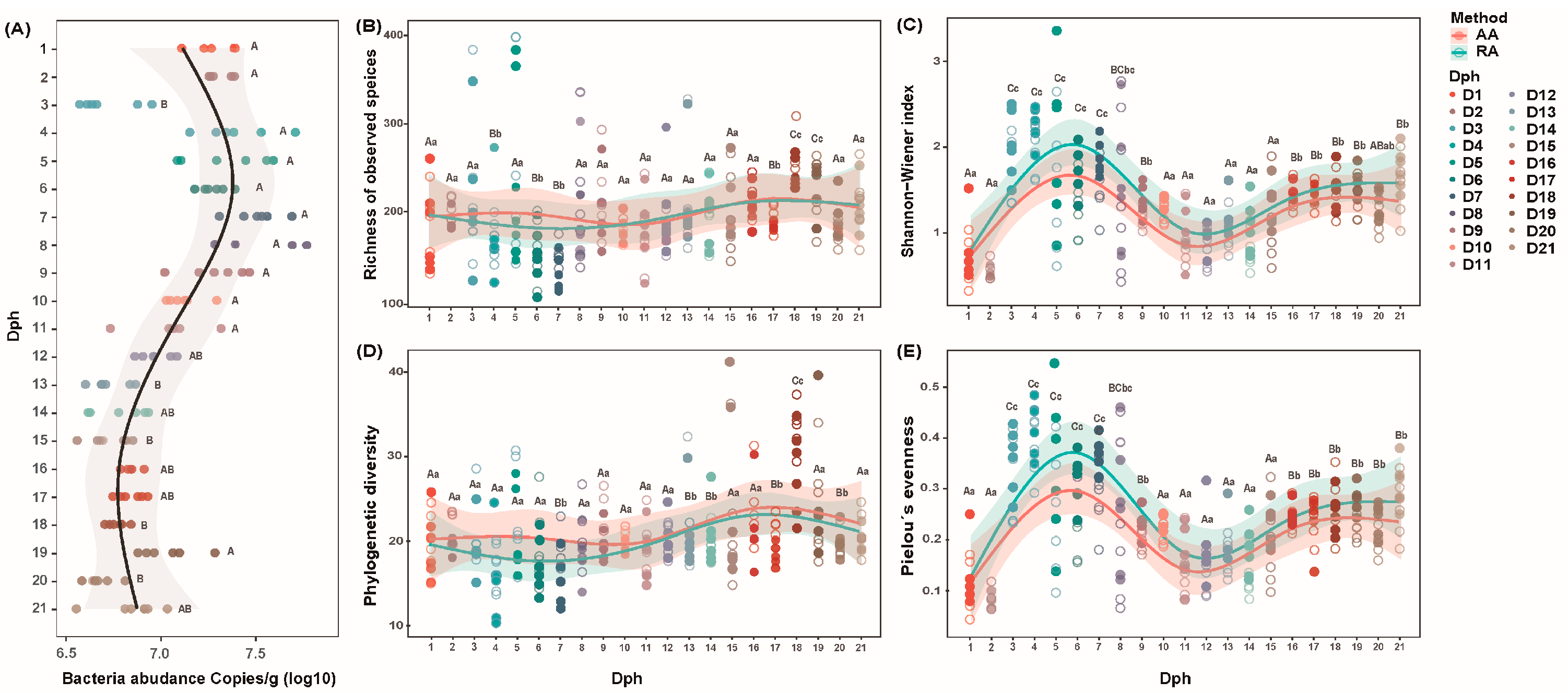
3.1. Abundance and Alpha-Diversity of Bacterial Community
3.2. Dynamics of Bacterial Composition of GFP Larvae
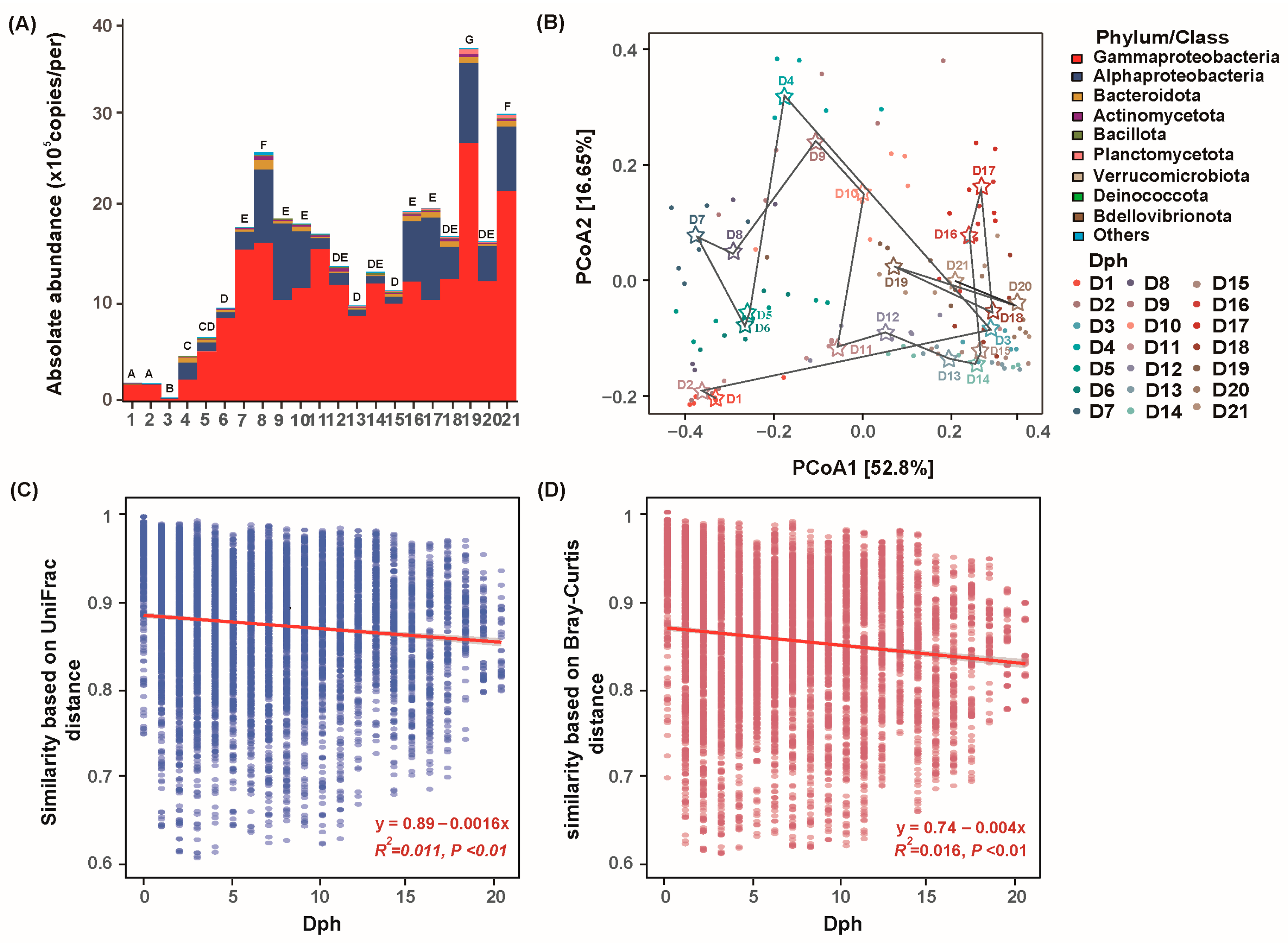
3.3. Taxonomic and Phylogenetic Turnover of Bacterial Community with GFP Larval Development
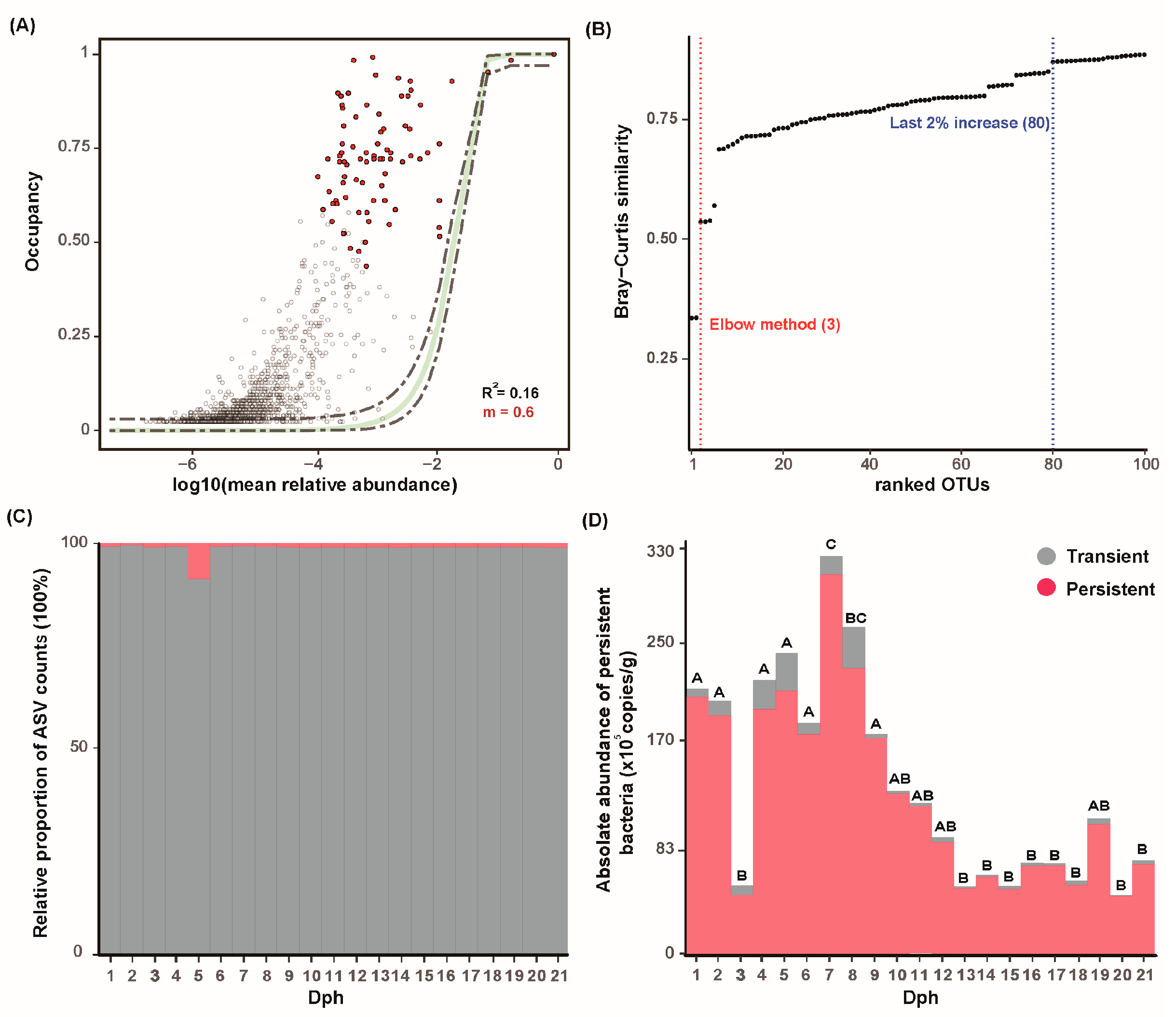
3.4. Persistent Microbiome Is Detected Across GFP Larval Development
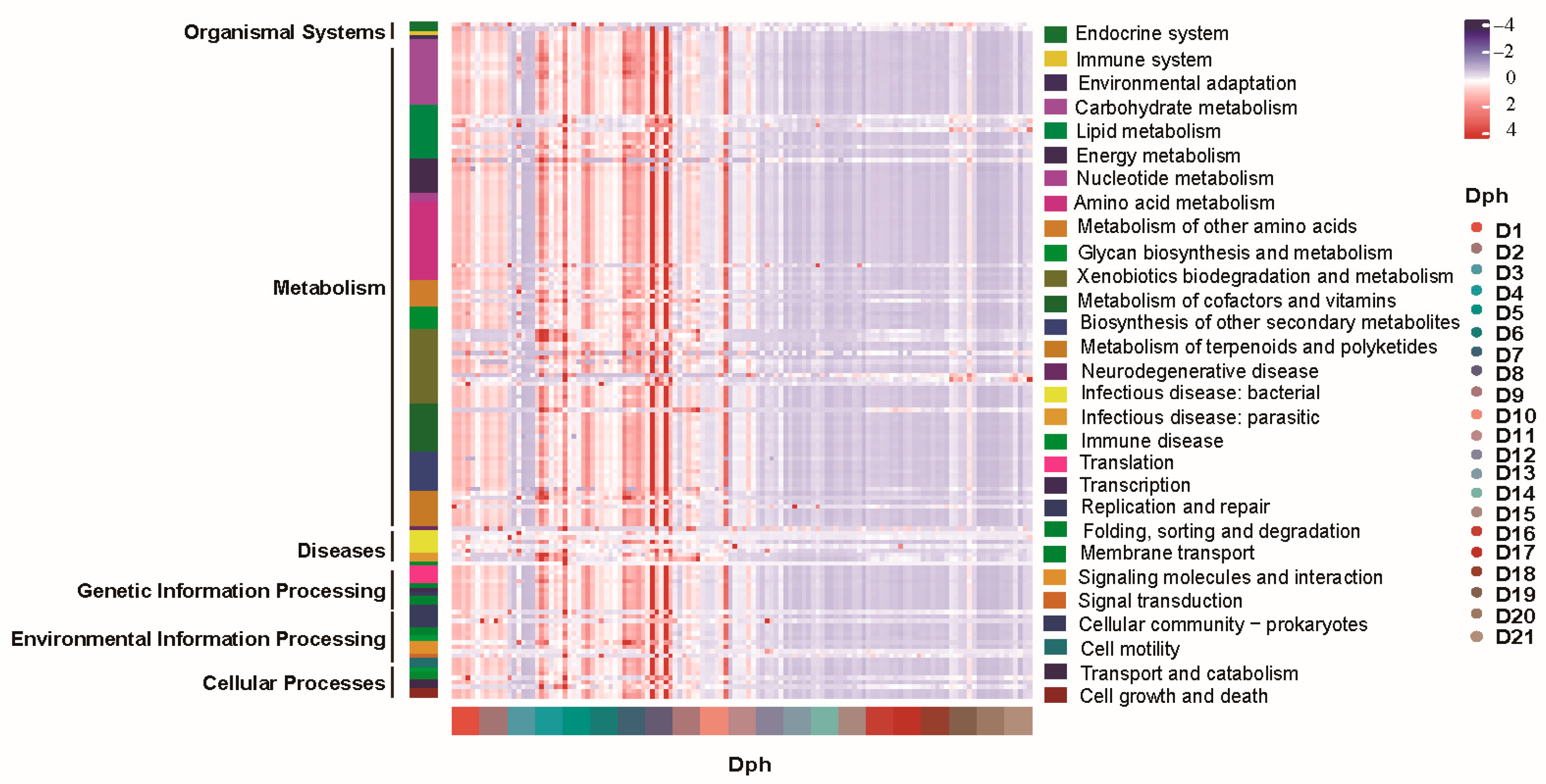
3.5. Functional Potential of GFP Larval Bacterial Community and Change Patterns
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pillai, B.R.; Ponzoni, R.W.; Das Mahapatra, K.; Panda, D. Genetic improvement of giant freshwater prawn 638 Macrobrachium rosenbergii: A review of global status. Rev. Aquac. 2022, 14, 1285–1299. [Google Scholar] [CrossRef]
- Zhao, C.; Zhou, X.; Yuan, X.; Xi, Q. Morphological characteristics of zoaea larva of Macrobrachium rosenbergii. J. Guangdong Ocean Univ. 2010, 4, 30. [Google Scholar]
- Vadstein, O.; Attramadal, K.J.K.; Bakke, I.; Olsen, Y. K-selection as microbial community management strategy: A method for improved viability of larvae in aquaculture. Front. Microbiol. 2018, 9, 2730. [Google Scholar] [CrossRef]
- Vestrum, R.I.; Attramadal, K.J.K.; PER, W.; Li, K.; Olsen, Y.; Bones, A.M.; Vasdtein, O.; Bakke, I. Rearing water treatment induces microbial selection influencing the microbiota and pathogen associated transcripts of cod (Gadus morhua) larvae. Front. Microbiol. 2018, 9, 851. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Wang, K.; Wu, J.; Qiuqian, L.; Yang, K.; Qian, Y.; Zhang, D. Changes in intestinal bacterial communities are closely associated with shrimp disease severity. Appl. Microbiol. Biotechnol. 2015, 99, 6911–6919. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Zhu, J.; Dai, W.; Dong, C.; Qiu, Q.; Li, C. Integrating gut microbiota immaturity and disease-discriminatory taxa to diagnose the initiation and severity of shrimp disease. Environ. Microbiol. 2017, 19, 1490–1501. [Google Scholar] [CrossRef]
- Xiong, J.; Li, X.; Yan, M.; Lu, J.; Qiu, Q.; Chen, J. Comparable ecological processes govern the temporal succession of gut bacteria and microeukaryotes as shrimp aged. Microb. Ecol. 2020, 80, 935–945. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, K.; Huang, L.; Dong, P.; Wang, S.; Chen, H.; Zhang, D. Fine-scale succession patterns and assembly mechanisms of bacterial community of Litopenaeus vannamei larvae across the developmental cycle. Microbiome 2020, 8, 106. [Google Scholar] [CrossRef]
- Xu, Y.; Cheng, H.; Gu, Z.; Li, X.; Shen, P.; Gao, Q. Study on microflora structure of Macrobrachium rosenbergii larvae and environmental water at different developmental stages. J. Freshw. Ecol. 2022, 37, 363–372. [Google Scholar] [CrossRef]
- Ma, R.; Wang, Y.; Zhao, S.; Yin, M.; Fang, W. The composition of the microbial community associated with Macrobrachium rosenbergii zoeae varies throughout larval development. J. Fish. Dis. 2020, 43, 413–421. [Google Scholar] [CrossRef]
- Shirsalimian, M.S.; Akhavan, S.A.; Dabbagh, A.M.A.R. Isolation of two radiation resistant and desiccation tolerant bacteria, Modestobacter sp. A2 and Maritalea sp. B9, from gandom beryan hill in the lut desert of Iran. Microbiology 2018, 87, 363–371. [Google Scholar] [CrossRef]
- Gao, X.; Zhou, Y.; Zhu, X.; Tang, H.; Li, X.; Jiang, Q.; Wei, W.; Zhang, X. Enterobacter cloacae: A probable etiological agent associated with slow growth in the giant freshwater prawn Macrobrachium rosenbergii. Aquaculture 2021, 530, 735826. [Google Scholar] [CrossRef]
- Li, C.; Jin, L.; Zhang, C.; Li, S.; Zhou, T.; Hua, Z.; Wang, L.; Ji, S.; Wang, Y.; Gan, Y.; et al. Destabilized microbial networks with distinct performances of abundant and rare biospheres in maintaining networks under increasing salinity stress. iMeta 2023, 2, e79. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xu, S.; Zhang, Z.; Zhang, X.; Liu, X. Transcriptomic profiling and characterization of microRNAs in Macrobrachium rosenbergii potentially involved in immune response to Enterobacter cloacae infection. Microb. Pathog. 2023, 183, 106291. [Google Scholar] [CrossRef]
- Rao, C.; Coyte, K.Z.; Bainter, W.; Geha, R.S.; Martin, C.R.; Rakoff-Nahoum, S. Multi-kingdom ecological drivers of microbiota assembly in preterm infants. Nature 2021, 591, 633–638.3. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.; Zhang, N.; Hu, B.; Jin, T.; Xu, H.; Qin, Y.; Yan, P.; Zhang, X.; Guo, X.; et al. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 2019, 37, 676–684. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, M.; Liu, Y.; Yang, X.; Wei, F.; Jin, X.; Liu, D.; Guo, Y.; Hu, Y. Quantitative microbiome profiling reveals the developmental trajectory of the chicken gut microbiota and its connection to host metabolism. iMeta 2023, 2, e105. [Google Scholar] [CrossRef]
- Qiao, Y.; Wang, Z.; Sun, H.; Guo, H.; Song, Y.; Zhang, H.; Ruan, Y.; Xu, Q.; Huang, Q.; Shen, Q.; et al. Synthetic community derived from grafted watermelon rhizosphere provides protection for ungrafted watermelon against Fusarium oxysporum via microbial synergistic effects. Microbiome 2024, 12, 101. [Google Scholar] [CrossRef]
- Li, S.; Delgado-Baquerizo, M.; Ding, J.; Hu, H.; Huang, W.; Sun, Y.; Ni, H.; Kuang, Y.; Yuan, M.M.; Zhou, J.; et al. Intrinsic microbial temperature sensitivity and soil organic carbon decomposition in response to climate change. Glob. Change Biol. 2024, 30, 7395. [Google Scholar] [CrossRef]
- Yajima, D.; Fujita, H.; Hayashi, I.; Shima, G.; Suzuki, K.; Toju, H. Core species and interactions prominent in fish-associated microbiome dynamics. Microbiome 2023, 11, 53. [Google Scholar] [CrossRef]
- Stopnisek, N.; Shade, A. Persistent microbiome members in the common bean rhizosphere: An integrated analysis of space, time, and plant genotype. ISME J. 2021, 9, 2708–2722. [Google Scholar] [CrossRef]
- Shade, A.; Stopnisek, N. Abundance-occupancy distributions to prioritize plant core microbiome membership. Curr. Opin. Microbiol. 2019, 49, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, D.; Qi, J.; Peng, Z.; Chen, W.; Wei, G.; Jiao, S. Stochastic processes shape the biogeographic variations in core bacterial communities between aerial and belowground compartments of common bean. Environ. Microbiol. 2021, 23, 949–964. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Ren, Z.; Lin, W.; Shi, C.; Mu, C.; Wang, C.; Wu, Q.; Ye, Y. Succession, sources, and assembly of bacterial community in the developing crab larval microbiome. Aquaculture 2022, 548, 737600. [Google Scholar] [CrossRef]
- DeSantis, T.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Edgar, R. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package, R package version 2.5-3; The R Project for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Burns, A.R.; Stephens, W.Z.; Stagaman, K.; Wong, S.; Rawls, J.F.; Guillemin, K.; Bohannan, B.J.M. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. 2016, 10, 655–664. [Google Scholar] [CrossRef]
- Xiong, J.; Dai, W.; Qiu, Q.; Zhu, J.; Yang, W.; Li, C. Response of host-bacterial colonization in shrimp to developmental stage, environment and disease. Microb. Ecol. 2018, 27, 3686–3699. [Google Scholar] [CrossRef]
- Gundersen, M.S.; Vadstein, O.; De Schryver, P.; Attramadal, K.J.K. Aquaculture rearing systems induce no legacy effects in Atlantic cod larvae or their rearing water bacterial communities. Sci. Rep. 2022, 12, 19812. [Google Scholar] [CrossRef]
- Tang, Y.; Wang, R.; Tan, L.; Guo, L.; Duan, Y.; Yang, L.; Jiang, S.; Zhou, F.; Jiang, S.; Huang, J. Effects of live microalgae and algae powder on microbial community, survival, metamorphosis and digestive enzyme activity of Penaeus monodon larvae at different growth stages. Aquaculture 2020, 526, 735344. [Google Scholar] [CrossRef]
- Estefanía, G.V.; Marcel, M.P.; Kadiya, C.; Francisco, V.A.; Teresa, G.G.; Luis, M.C. Taxonomic and functional changes in the microbiota of the white shrimp (Litopenaeus vannamei) associated with postlarval ontogenetic development. Aquaculture 2020, 518, 734842. [Google Scholar] [CrossRef]
- Kumar, T.S.; Vidya, R.; Kumar, S.; Alavandi, S.V.; Vijayan, K.K. Zoea-2 syndrome of Penaeus vannamei in shrimp hatcheries. Aquaculture 2017, 479, 759–767. [Google Scholar] [CrossRef]
- Liu, B.; Song, C.; Gao, Q.; Liu, B.; Zhou, Q.; Sun, C.; Zhang, H.; Liu, M.; Tadese, W.A. Maternal and environmental microbes dominate offspring microbial colonization in the giant freshwater prawn Macrobrachium rosenbergii. Sci. Total Environ. 2021, 790, 148062. [Google Scholar] [CrossRef]
- Mente, E.; Gannon, A.T.; Nikouli, E.; Hammer, H.; Kormas, K.A. Gut microbial communities associated with the molting stages of the giant freshwater prawn Macrobrachium rosenbergii. Aquaculture 2016, 463, 181–188. [Google Scholar] [CrossRef]
- Xiong, J.; Xuan, L.; Yu, W.; Zhu, J.; Qiu, Q.; Chen, J. Spatiotemporal successions of shrimp gut microbial colonization: High consistency despite distinct species pool. Environ. Microbiol. 2019, 21, 1383–1394. [Google Scholar] [CrossRef] [PubMed]
- Lan, X.; Peng, X.; Du, T.; Xia, Z.; Gao, Q.; Tang, Q.; Yi, S.; Yang, G. Alterations of the gut microbiota and metabolomics associated with the different growth performances of Macrobrachium rosenbergii families. Animals 2023, 13, 1539. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; van der Gast, C.J.; Yu, Y. Bacterial community assembly and turnover within the intestines of developing zebrafish. PLoS ONE 2012, 7, e30603. [Google Scholar] [CrossRef]
- Li, E.; Xu, C.; Wang, X.; Wang, S.; Zhao, Q.; Zhang, M.; Qin, J.; Chen, L. Gut microbiota and its modulation for healthy farming of Pacific white shrimp Litopenaeus vannamei. Rev. Fish. Sci. Aquac. 2018, 26, 381–399. [Google Scholar] [CrossRef]
- Ricotta, C.; Podani, J.; Schmera, D.; Bacaro, G.; Maccherini, S.; Pavoine, S. The ternary diagram of functional diversity. Methods Ecol. Evol. 2023, 14, 1168–1174. [Google Scholar] [CrossRef]
- Davies, T.J.; Urban, M.C.; Rayfield, B.; Cadotte, M.W.; Peres-Neto, P.R. Deconstructing the relationships between phylogenetic diversity and ecology: A case study on ecosystem functioning. Ecology 2016, 97, 2212–2222. [Google Scholar] [CrossRef] [PubMed]
- Tzeng, T.D.; Pao, Y.; Chen, P.; Weng, F.; Jean, W.; Wang, D. Effects of host phylogeny and habitats on gut microbiomes of oriental river prawn (Macrobrachium nipponense). PLoS ONE 2015, 10, e0132860. [Google Scholar] [CrossRef]
- Zheng, Y.; Yu, M.; Liu, J.; Qiao, Y.; Wang, L.; Li, Z.; Zhang, X.; Yu, M. Bacterial community associated with healthy and diseased Pacific white shrimp (Litopenaeus vannamei) larvae and rearing water across different growth stages. Front. Microbiol. 2017, 8, 1362. [Google Scholar] [CrossRef]
- Zhou, J.; Ning, D. Stochastic community assembly: Does it matter in microbial ecology? Microbiol. Mol. Biol. Rev. 2017, 81, e00002-17. [Google Scholar] [CrossRef]
- Dini-Andreote, F.; Stegen, J.; Elsas, J.; Salles, J. Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc. Natl. Acad. Sci. USA 2015, 112, 1326–1332. [Google Scholar] [CrossRef]
- Fernandes, C.; Rainey, F.A.; Fernanda, N.M.; Pinhal, I.; Folhas, F.; Costa, M.S. Herminiimonas fonticola gen. nov., sp. nov., a Betaproteobacterium isolated from a source of bottled mineral water. Syst. Appl. Microbiol. 2005, 28, 596–603. [Google Scholar] [CrossRef]
- Dopson, M.; Johnson, D.B. Biodiversity, metabolism and applications of acidophilic sulfur-metabolizing microorganism. Environ. Microbiol. 2012, 14, 2620–2631. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Z.; Hao, J.; Meng, J.; Liu, C.; Ye, T.; Yan, J.; Li, G.; Zheng, Y.; Xu, P.; Gu, Z. Dynamics and Assembly Mechanisms of Bacterial Communities During Larval Development of Macrobrachium rosenbergii: A High-Frequency Sampling Study Based on 16S rRNA Absolute Quantification Sequencing. Microorganisms 2025, 13, 1881. https://doi.org/10.3390/microorganisms13081881
Lu Z, Hao J, Meng J, Liu C, Ye T, Yan J, Li G, Zheng Y, Xu P, Gu Z. Dynamics and Assembly Mechanisms of Bacterial Communities During Larval Development of Macrobrachium rosenbergii: A High-Frequency Sampling Study Based on 16S rRNA Absolute Quantification Sequencing. Microorganisms. 2025; 13(8):1881. https://doi.org/10.3390/microorganisms13081881
Chicago/Turabian StyleLu, Zhibin, Jingwen Hao, Jilun Meng, Cui Liu, Tiantian Ye, Junjun Yan, Guo Li, Yutong Zheng, Pao Xu, and Zhimin Gu. 2025. "Dynamics and Assembly Mechanisms of Bacterial Communities During Larval Development of Macrobrachium rosenbergii: A High-Frequency Sampling Study Based on 16S rRNA Absolute Quantification Sequencing" Microorganisms 13, no. 8: 1881. https://doi.org/10.3390/microorganisms13081881
APA StyleLu, Z., Hao, J., Meng, J., Liu, C., Ye, T., Yan, J., Li, G., Zheng, Y., Xu, P., & Gu, Z. (2025). Dynamics and Assembly Mechanisms of Bacterial Communities During Larval Development of Macrobrachium rosenbergii: A High-Frequency Sampling Study Based on 16S rRNA Absolute Quantification Sequencing. Microorganisms, 13(8), 1881. https://doi.org/10.3390/microorganisms13081881




