Abstract
Gram-negative Burkholderia bacteria are known for causing diseases in humans, animals, and plants, and high intrinsic resistance to antibiotics. Phage therapy is a promising alternative to control multidrug-resistant bacterial pathogens. Here, we present an overview of Burkholderia phage characteristics, host specificity, genomic classification, and therapeutic potentials across medical, veterinary, and agricultural systems. We evaluate the efficacy and limitations of current phage candidates, the biological and environmental barriers of phage applications, and the phage cocktail strategy. We highlight the innovations on the development of targeted phage delivery systems and the transition from the exploration of clinical phage therapy to plant disease management, advocating integrated disease control strategies.
1. Introduction
Burkholderia sensu lato are Gram-negative bacteria within the family Burkholderiaceae, order Burkholderiales, and class Betaproteobacteria. Burkholderia sensu lato has been divided into Burkholderia sensu stricto and other six genera named Paraburkholderia, Caballeronia, Robbsia, Mycetohabitans, Trinickia, and Pararobbsia [1]. The genus Burkholderia sensu stricto currently comprises 36 validly published species (https://lpsn.dsmz.de/genus/burkholderia) (accessed on 10 May 2025). Burkholderia sensu stricto has wide metabolic versatility and adaptation to versatile lifestyles as free-living bacteria in soil or water and as commensals of plants, animals, or fungi [1]. The genus Burkholderia sensu stricto is phylogenetically divided into three major species complexes: Burkholderia cepacia complex (Bcc), Burkholderia pseudomallei complex (Bpc), and Burkholderia glumae complex (Bgc). Bcc includes B. cepacia, B. cenocepacia, B. ambifaria, B. contaminans, B. multivorans, B. stabilis, and B. vietnamiensis; Bpc includes B. pseudomallei, B. mallei, B. thailandensis, B. oklahomensis, and B. singularis; and Bgc includes B. glumae, B. gladioli, and B. plantarii [2].
Although some members of the genus Burkholderia sensu stricto show biotechnological potentials of plant growth promotion, biocontrol, antibiotic production, biodegradation, and bioremediation, major members are pathogens to human, animals, and plants. Bcc members, such as B. cenocepacia and B. multivorans, are well-known pathogens that cause chronic pulmonary infections in cystic fibrosis (CF) [3,4]. Bpc members, such as B. pseudomallei, are etiological agents of melioidosis, a potentially fatal disease endemic to tropical regions [5]. Bgc members B. glumae and B. gladioli cause bacterial panicle blight (BPB) in rice, while B. plantarii causes rice seedling blight and grain rot [2]. Of particular concern to Bgc is their potential to be opportunistic human pathogens to various immunocompromised populations [6,7,8,9].
The genus Burkholderia sensu stricto is characterized by large and complex genomes, typically comprising multiple replicons (chromosomes and plasmids) that encode extensive repertoires of genes for environmental adaptation and metabolic plasticity [10]. The large and complex genomes facilitate environmental adaptation, allowing for the colonization of diverse niches, including soil ecosystems, aquatic environments, plant rhizospheres, and even intracellular compartments of eukaryotic hosts [11]. This genomic architecture also facilitates high levels of antibiotic resistance, posing serious challenges for both clinical treatment and agricultural disease control [11,12]. Of particular concern is the increasing prevalence of multidrug-resistant Burkholderia strains in clinical settings where therapeutic options are severely constrained [13]. In agriculture, overreliance on chemical pesticides has further driven resistance and raised environmental hazards. These challenges have spurred interest in alternative approaches, notably phage therapy [14].
Phages, also known as bacteriophages, are viruses that specifically infect and lyse bacteria, offering a targeted biocontrol strategy against Burkholderia infections. Phage action involves specific recognition of bacterial surface receptors, the injection of viral genetic material, and hijacking of the host’s cellular machinery for replication. This process culminates in cell lysis, releasing new phages to continue the cycle [15]. Due to the high specificity, phages offer an alternative to conventional antibiotics, particularly for managing multidrug-resistant strains. In clinical settings, phages have shown efficacy against multidrug-resistant strains where antibiotics fail [15,16,17,18,19]. For example, phage C34 targeting B. pseudomallei significantly reduced bacterial load and improved survival rates in infected mice [17]. In agriculture, phage NBP4-7 and jumbo phage S13 reduced BPB severity in rice by targeting key virulence factors like flagella [20,21].
Phage therapy presents a promising cross-disciplinary solution for managing Burkholderia-induced diseases in humans, animals, and plants. The advance of phage therapy in human and veterinary medicine provides an adaptation strategy for plant disease management. In clinical settings, phages are administered through intravenous, oral, and topical routes with formulations optimized for stability and therapeutic efficacy [22]. In agriculture, phages are typically applied via foliar sprays, seed treatments, or soil drenches depending on the crops and pathogens [23,24]. However, phage application in cropland is vulnerable to environmental inactivation and degradation by high temperature, UV radiation, drought, agrochemicals, and soil absorption. Recent development in delivery technologies, particularly phage encapsulation in nanocarriers, enhances phage viability and site-specific release [25]. These delivery technologies can be translated to protect phages targeting Bgc members in cropland [26].
Here, we review the advances in research on Burkholderia phages, particularly on Burkholderia phage therapeutic potential across host systems. We highlight the delivery innovations and cross-application strategies that may enhance the integration of phage therapy into sustainable disease management programs.
2. Pathogenic Burkholderia Species
2.1. Human and Animal Pathogens
The genus Burkholderia sensu stricto includes pathogenic species that pose significant threats to human and animal health (Table S1). Among these, members of Bcc such as B. cepacia, B. multivorans, and B. cenocepacia are well-known opportunistic pathogens. They are most frequently isolated from individuals with CF and chronic granulomatous disease, where they are associated with severe respiratory infections, including necrotizing pneumonia and septicemia [27,28]. B. dolosa and B. anthina have been linked to accelerated pulmonary decline and chronic obstructive pulmonary diseases, respectively [29,30]. Beyond respiratory infections, Bcc contributes to bloodstream infections, wound contaminations, and sepsis. B. stabilis and B. contaminans have been associated with nosocomial infections and bacteremia, posing challenges in hospital settings [31,32,33,34]. B. pseudomultivorans was first isolated from clinical CF sputum and rhizosphere soil [35]. Recently, B. pseudomultivorans was identified as the cause of sepsis in cats, suggesting zoonotic potential [36].
Within Bpc, B. pseudomallei causes melioidosis, a severe zoonotic disease endemic to Southeast Asia and northern Australia. It infects a broad-host range including humans, domestic animals, wildlife, and pets. Clinical presentations include pneumonia, sepsis, abscesses, and chronic infections [37,38]. B. mallei causes glanders, a zoonotic disease primarily affecting horses, donkeys, and mules. B. mallei was weaponized during World War I due to its high infectivity [39,40,41]. B. thailandensis is less virulent and typically causes opportunistic infections in immunocompromised individuals [42,43,44].
Members of the Bcc possess multiple virulence factors (Table 1) including biofilm formation, motility, quorum sensing, pili, LPS variation, secretion systems, and extracellular enzymes that enable colonization and immune evasion. They exhibit intrinsic resistance mechanisms such as efflux pumps, β-lactamases, low membrane permeability, modified LPS, and polymyxin resistance in some species [45,46,47,48,49,50,51]. Similarly, Bpc species like B. pseudomallei display virulence traits including biofilm formation, motility, intracellular survival, capsular polysaccharide, quorum sensing, diverse secretion systems (Type III, V, VI), and adhesins [14,52,53,54]. Both groups share resistance features, including multidrug efflux pumps and β-lactam resistance. Effective management typically requires carbapenems or β-lactam/β-lactamase inhibitor combinations. However, persistent infections and relapse are common due to biofilm formation and adaptive resistance [14].
2.2. Plant Pathogens
Several Bgc members (B. glumae, B. gladioli, and B. plantarii) and Bcc members (B. cepacia, B. orbicola, B. semiarida and B. sola) are recognized as plant pathogens, causing substantial agricultural losses (Table S1). B. glumae is a major pathogen responsible for BPB in rice, causing symptoms like aborted seeds, empty grains, and seedling rot, significantly reducing rice yield [55,56]. B. glumae also infects other crops such as pepper, eggplant, tomato, sesame, and perilla [50]. Notably, B. glumae has also been isolated from human clinical cases, indicating its potential for cross-kingdom pathogenicity [6,57]. Likewise, B. gladioli infects rice and a variety of other crops, causing grain rot and seedling blight. B. gladioli also acts as an opportunistic human pathogen, causing bacteremia, pneumonia, and lung infections in CF patients [58,59,60,61]. B. plantarii primarily infects rice, leading to seedling blight, grain rot, chlorosis, and stunting [62,63]. B. cepacia causes bulb rot in onions [64]. B. orbicola reduces bean seed germination and impairs insect survival [59]. B. semiarida and B. sola are associated with onion sour skin disease [65,66]. These pathogens exhibit biofilm formation, motility (flagella), quorum sensing systems, and secretion systems (including type III), and produce toxins like toxoflavin and extracellular enzymes that facilitate host tissue colonization and damage [67,68,69,70,71,72]. They possess resistance to conventional control measures, making them agriculturally significant threats.

Table 1.
Comparative summary of major pathogenic Burkholderia.
Table 1.
Comparative summary of major pathogenic Burkholderia.
| Species | Host Range | Key Virulence Factor | Resistance Trait | Zoonotic Risk | Reference |
|---|---|---|---|---|---|
| B. cepacia | Humans, occasionally animals, plants | Biofilm formation, motility, pili, lipopolysaccharide variation, quorum sensing (QS), extracellular enzymes | Efflux pumps, β-lactamases, low permeability, modified lipopolysaccharide | Opportunistic zoonotic risk | [45,46] |
| B. multivorans | Humans (CF) | Biofilm formation, motility, cable pili, QS-controlled virulence | Aminoglycoside, β-lactam resistance, efflux pumps, polymyxin resistance | No known zoonotic transmission | [14,46] |
| B. cenocepacia | Humans (CF, immunocompromised) | Biofilm formation, motility, QS-regulated proteases, cable pili, secretion systems, siderophore production | Efflux pumps, β-lactamases, polymyxin resistance | Potential zoonotic pathogen | [14,46,47,48] |
| B. dolosa | Humans (CF) | Biofilm and capsule formation, motility, adhesins and proteases, secretion systems | Extensive multidrug resistance, multiple efflux pumps, β-lactamases | No known zoonotic transmission | [49,50] |
| B. contaminans | Humans (nosocomial) | Biofilm formation, motility, hemolysins, antifungal activity, secretion systems | β-lactams, disinfectants, efflux pumps | Potential zoonotic pathogen | [33,46,51] |
| B. pseudomallei | Humans and animals | Biofilm formation, motility, intracellular survival, polysaccharides, QS, secretion systems, immune evasion | Aminoglycosides, macrolides, β-lactamases, efflux pumps, polymyxin resistance | Confirmed zoonotic agent | [14,52,53,54] |
| B. mallei | Equids, zoonotic to humans | Biofilm formation, motility, secretion systems, immune evasion, novel virulence proteins, modulation of ubiquitination, actin-cytoskeleton rearrangement | Aminoglycosides, β-lactams, efflux pumps | Confirmed zoonotic agent | [52,73,74,75] |
| B. thailandensis | Environment, immunocompromised hosts | Biofilm formation, motility, attenuated virulence, secretion systems, QS, siderophore (malleobactin) production | Limited resistance, efflux pumps, β-lactamases | Opportunistic zoonotic risk | [76,77,78] |
| B. glumae | Plants, rare human cases | Biofilm formation, motility, toxoflavin, lipase, QS, flagella, extracellular polysaccharides, lipase, secretion systems | Multidrug resistance, efflux pumps, β-lactamases | No known zoonotic transmission | [55,67,68,69] |
| B. gladioli | Plants, humans (CF, immunocompromised) | Biofilm formation, protein secretion systems (T2SS, T3SS), motility, proteases, toxoflavin, QS | β-lactams, aminoglycosides, multidrug efflux | Potential zoonotic pathogen | [70,71,72] |
3. Characterization of Burkholderia Phages
3.1. Isolation
Burkholderia phages have been isolated from a wide range of environmental samples, including soil, water, plant tissues, compost, and clinical settings. Common isolation methods involve enrichment using selective media and plaque assays, where samples are mixed with host bacterial strains and plated onto agar to identify lytic and lysogenic phages through plaque formation [79,80]. For instance, Jungkhun et al. isolated 61 phages using direct plating and plaque assays, selecting NBP1-1, NBP4-7, and NBP4-8 as effective lytic agents against B. glumae [20]. Adachi et al. used filtration and ultracentrifugation to isolate phages BGPP-Ar, BGPP-Sa, and BGPP-Ya from water and puddles, demonstrating their potential for controlling bacterial seedling blight in rice [81]. Kanaizuka et al. obtained jumbo phages FLC8, FLC9, and FLC10 from fallen leaf compost, highlighting the natural abundance of Burkholderia phages in decaying plant material [82]. Jumbo phages Chiangavirus FLC6 and FLC8 infecting B. glumae were isolated from rice fields and compost samples [82,83]. Lessievirus BcepIL02 and Aptresvirus vB_BceM_AP3 infecting B. cenocepacia were obtained from soil sample planted with corns and irrigated fields [84,85]. These diverse isolations highlight the natural abundance and ecological adaptability of Burkholderia phages.
3.2. Morphology
Most Burkholderia phages possess icosahedral heads and exhibit either contractile or non-contractile (long or short) tails, morphologically classified into the families Myoviridae, Podoviridae, and Siphoviridae. For examples, jumbo Burkholderia phage FLC6 and non-jumbo Burkholderia phages NBP1-1, NBP4-7, and NBP4-8 infecting B. glumae possess icosahedral heads and contractile tails typical of the family Myoviridae [20,83]; Burkholderia phage Bp-AMP1 infecting B. pseudomallei has an icosahedral capsid and a short non-contractile tail typical of the family Podoviridae; Burkholderia phages phiE125 and phi1026b targeting B. mallei are characterized by icosahedral heads and long non-contractile tails typical of the family Siphoviridae [86,87,88]. Morphology-based phage classification depends on transmission electron microscopy to visualize phage particles and determine phage particle size, shape, and structural features [89].
3.3. Life Cycle
Burkholderia phages possess lytic or lysogenic life cycles (Table 2), impacting their use in phage therapy and biocontrol. Lytic Burkholderia phages hijack the host cellular machinery to replicate and lyse bacterial cells and are effective against pathogenic Burkholderia. For example, the jumbo phage Chiangavirus FLC6 shows strong lytic activity against B. glumae, B. plantarii, and even Ralstonia pseudosolanacearum, indicating broad-host range and cross-genus infectivity [83]. While promising, this broad-host range requires further validation through in vivo studies and testing against diverse environmental isolates, as current evidence are mainly derived from in vitro assays. Lysogenic Burkholderia phages integrate their genome into the host genome as prophages [90,91]. This lysogenic conversion drives horizontal gene transfer and phage–host co-evolution, where integrated phage genes may enhance bacterial virulence, stress tolerance, or antibiotic resistance. Most characterized Burkholderia phages within the family Peduoviridae, such as Kisquattuordecimvirus KS14, Kisquinquevirus KS5, and Tigrvirus phiE202, are lysogenic (Table 2). Interestingly, Ampunavirus phage Bp-AMP1 has a temperature-dependent life cycle, remaining lysogenic at 25 °C but switching to a lytic cycle at 37 °C [92,93]. This thermally controlled behavior suggests its potential in temperature-regulated phage therapies. Although temperate phages have limited direct therapeutic use, synthetic biology allows for conversion into obligate lytic forms by disrupting lysogeny-related genes (e.g., integrases, repressors), expanding their clinical and agricultural applications [94]. Overall, lytic Burkholderia phages with broad-host ranges are promising for phage therapy. Temperate Burkholderia phages may require genetic modification or lytic derivative selection to ensure therapeutic safety and efficacy.
3.4. Host Range and Specificity
Burkholderia phages exhibit diverse host specificities, ranging from narrow-host to broad-host. Narrow-host phages infect very limited strains within one species, such as Kayeltresvirus KL3 infecting only B. ambifaria LMG 17828, and temperate phages KS4 and KS9 infecting only two out of 24 tested Bcc strains. In contrast, broad-host phages can infect multiple bacterial species even genera, such as the jumbo phage FLC6, which can lyse multiple strains of B. glumae, B. plantarii, and Ralstonia pseudosolanacearum [83,90,95]. This specificity is primarily governed by tail fiber proteins (TFPs), which mediate phage–host interactions by recognizing bacterial surface receptors such as lipopolysaccharides (LPSs) and outer membrane proteins [96]. Variations in TFPs, including C-terminal extensions and single amino acid mutations, significantly impact host range [97]. Structural and genetic modifications in TFPs play a critical role in host adaptation. For example, Burkholderia phage AP3 possesses a unique 365-amino-acid C-terminal extension in its TFP that enhances its specificity for B. cenocepacia IIIA LPS variants, contributing to its narrow-host range [85]. Furthermore, engineered chimeric phages, such as Pseudomonas aeruginosa phage PaP1-rec1, acquire expanded host ranges through tail fiber gene swaps, demonstrating the potential of genetic modifications in customizing phage infectivity [98]. Recent advances, such as targeted point mutations (e.g., G→C in Acinetobacter phage Abp4-M) [99] and domain swapping (e.g., STyj5-1 with BD13 tail fibers) [100], have further expanded host ranges while maintaining adsorption efficiency. A rational therapeutic approach could involve phage cocktails, combining highly specific phages with engineered broad-range variants to balance efficacy and safety in treating multidrug-resistant infections.
3.5. Genomic Taxonomy
Bacteriophage taxonomy has evolved from a discipline based mainly on morphology to genome [101]. The morphology-based families Myoviridae, Podoviridae, and Siphoviridae were abolished and the order Caudovirales was replaced by the class Caudoviricetes to group all tailed bacterial and archaeal viruses with icosahedral capsids and a double-stranded DNA genome [102]. The advances of next-generation sequencing techniques promote the genome-based classification to generate a more accurate evolutionary framework and better reflection of the diversity and phylogeny of the abundant and diverse viruses, and establishment of new genome-based taxa recognized by the International Committee on Taxonomy of Viruses (ICTV) (Figure S1). Nowadays, ICTV uses a holistic approach to classify prokaryote viruses by considering morphotype, host, lifestyle, genome characteristics (such as size, mol% G + C), % protein homologs, overall DNA and protein similarity, and phylogeny based on core genes [101]. Prokaryote viruses belonging to the same taxonomy rank form a cohesive and monophyletic group. Two phages are assigned to the same species if their genomes are more than 95% identical at the nucleotide level over their full genome length, while 70% of nucleotide identity of the full genome length is the cut-off for genera [101]. Members of a viral family share a significant number of orthologous genes, forming a cohesive and monophyletic group based on common proteomes. The sequencing and analyzing of phage genomes revealed a much higher genomic diversity than had previously been considered, leaving a significant fraction of sequenced phages unclassified at the family level [101].
Almost all Burkholderia phages whose whole-genome sequences are deposited in the GenBank database of the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/) (accessed on 25 February 2025) belong to the class Caudoviricetes, except for Alphatectivirus BCE1, which belongs to the class Tectiliviricetes (Table 2). Another distinguished feature of Alphatectivirus BCE1 is the smallest genome size of 14,800 bp. Based on the genome size, Burkholderia phages belonging to the class Caudoviricetes are divided into two groups: jumbo Burkholderia phages and non-jumbo Burkholderia phages (Table 2, Figure 1). The genome size of the jumbo Burkholderia phages ranges from 225,545 bp to 321,833 bp. The genome size of the non-jumbo Burkholderia phages ranges from 32,090 bp to 72,415 bp.

Table 2.
Information of Burkholderia phages.
Table 2.
Information of Burkholderia phages.
| Phage Name | Morphotype | ICTV Taxonomy (Class > Order > Family > Genus) | Host | Lifestyle | GC Content (%) | Genome Length (bp) | Reference |
|---|---|---|---|---|---|---|---|
| BCE1 | / | Tectiliviricetes > Kalamavirales > Tectiviridae > Alphatectivirus | B. cepacia | / | 48.21 | 14,800 | [103] |
| Class Caudoviricetes | |||||||
| FLC6 | Myovirus | Chimalliviridae > Chiangmaivirus | B. glumae; B. plantarii; Ralstonia pseudosolanacearum | Lytic | 52.01 | 227,105 | [83] |
| FLC8 | Myovirus | Chimalliviridae > Chiangmaivirus | B. glumae; B. plantarii | Lytic | 52.05 | 225,545 | [82] |
| S13 | Myovirus | Chimalliviridae > Chiangmaivirus | B. glumae; B. gladioli; B. multivorans; B. cenocepacia; B. dolosa; | Lytic | 51.7 | 227,647 | [21] |
| FLC9 | Myovirus | Novel species 16 within a novel genus 8 * | B. glumae; B. plantarii | / | 55.97 | 321,833 | [82] |
| BcepSauron | Myovirus | Sarumanvirus | B. cenocepacia | Lytic | 58.10 | 262,653 | [104] |
| BcepSaruman | Myovirus | Sarumanvirus | B. cenocepacia | Unknown | 58.14 | 263,735 | / |
| BCSR5 | Myovirus | Novel species 4 within a novel genus 2 * | B. cepacia | / | 54.74 | 227,351 | [105] |
| KL1 | Siphovirus | Jondennisvirinae > Kilunavirus | B. cenocepacia | Lytic | 54.61 | 42,832 | [106] |
| BcepGomr | / | Novel species 7 within a novel genus 3 * | Burkholderia | Unknown | 56.29 | 52,414 | [106] |
| Bp-AMP2 | Podovirus | Autographivirales > Autonotataviridae > Ampunavirus | B. pseudomallei | / | 61.76 | 42,492 | [92] |
| Bp-AMP1 | Podovirus | Autographivirales > Autonotataviridae > Ampunavirus | B. pseudomallei; B. thaliandensis | Temperate | 61.75 | 42,409 | [92,93] |
| Bp AMP4 | Podovirus | Autographivirales > Autonotataviridae > Ampunavirus | B. pseudomallei | / | 61.79 | 42,112 | [92] |
| Bp AMP3 | Podovirus | Autographivirales > Autonotataviridae > Ampunavirus | B. pseudomallei | / | 61.77 | 41,882 | [92] |
| JG068 | Podovirus | Autographivirales > Autonotataviridae > Mguuvirus | B. multivorans; B. cenocepacia; B. stabilis; B. dolosa | Lytic | 60.69 | 41,604 | [107] |
| Paku | / | Autographivirales > Autonotataviridae > Pakuvirus | B. cenocepacia | Temperate | 61.86 | 42,727 | [107] |
| Maja | Myovirus | Lindbergviridae > Gladiolivirus | B. gladioli | Temperate | 54.50 | 68,393 | [108] |
| BcepF1 | Myovirus | Lindbergviridae > Bcepfunavirus | B. ambifaria | / | 55.89 | 72,415 | [106,109] |
| BCSR52 | Myovirus | Lindbergviridae > Irusalimvirus | B. cepacia | / | 51.45 | 70,038 | / |
| WTB | Myovirus | Bglawtbvirus | B. gladioli | Lytic | 60.04 | 68,541 | [110] |
| BCSR129 | Myovirus | Novel species 10 within a novel genus 5 * | B. cepacia | Unknown | 58.42 | 66,147 | [105] |
| BcepB1A | Myovirus | Novel species 2 within a novel genus 1 * | B. cenocepacia | Lytic | 54.45 | 47,399 | [106] |
| BcepNazgul | Siphovirus | Casjensviridae > Nazgulvirus | B. cepacia | Lytic | 60.64 | 57,455 | [111] |
| AH2 | Siphovirus | Casjensviridae > Ahduovirus | B. cenocepacia; B. gladioli | Lytic | 61.31 | 58,065 | [106,112] |
| PhiE255 | Myovirus | Bcepmuvirus | B. thailandensis | Temperate | 63.05 | 37,446 | [91] |
| BcepMu | Myovirus | Bcepmuvirus | B. cenocepacia | Temperate | 62.86 | 36,748 | [18] |
| KS10 | Myovirus | Novel species 25 within a novel genus 10 * | B. cenocepacia; B. stabilis; B. ambifaria | Temperate | 62.87 | 37,635 | [113] |
| phiX216 | Myovirus | Peduoviridae > Tigrvirus | B. pseudomallei; B. mallei | Temperate | 64.82 | 37,637 | [114] |
| phi52237 | Myovirus | Peduoviridae > Tigrvirus | B. pseudomallei | Temperate | 64.82 | 37,639 | [91] |
| BEK | Myovirus | Peduoviridae > Tigrvirus | B.pseudomallei | / | 68.82 | 37,631 | [85] |
| phiE202 | Myovirus | Peduoviridae > Tigrvirus | B. mallei; B. pseudomallei | Temperate | 65.43 | 35,741 | [91] |
| phiE094 | Myovirus | Peduoviridae > Tigrvirus | B. thailandensis; B. pseudomallei | Temperate | 64.48 | 37,727 | [115] |
| NBP1-1 | Myovirus | Peduoviridae > Tigrvirus | B. glumae | Lytic | 63.23 | 40,570 | [20] |
| NBP4-7 | Myovirus | Peduoviridae > Tigrvirus | B. glumae | Lytic | 63.23 | 40,563 | [20] |
| NBP4-8 | Myovirus | Peduoviridae > Tigrvirus | B. glumae | Lytic | 63.23 | 40,568 | [20] |
| KL3 | Myovirus | Peduoviridae > Kayeltresvirus | B. ambifaria | Temperate | 63.23 | 40,555 | [90] |
| PK23 | Myovirus | Peduoviridae > Duodecimduovirus | B. pseudomallei | Temperate | 65.12 | 35,343 | [116] |
| phiE12_2 | Myovirus | Peduoviridae > Duodecimduovirus | B. mallei | Temperate | 64.62 | 36,690 | [91] |
| FLC10 | Myovirus | Peduoviridae > Kisquattuordecimvirus | B. glumae | Lytic | 61.29 | 32,867 | [82] |
| FLC5 | Myovirus | Peduoviridae > Kisquattuordecimvirus | B. glumae; B. plantarii | Temperate | 61.79 | 32,090 | [117] |
| KS14 | Myovirus | Peduoviridae > Kisquattuordecimvirus | B. multivorans; B. cenocepacia; B. dolosa; B. ambifaria | Temperate | 62.28 | 32,317 | [90] |
| vB BceM AP3 | Myovirus | Peduoviridae > Aptresvirus | B. cenocepacia | Temperate | 64.04 | 36,499 | [85] |
| Mana | Myovirus | Peduoviridae > Aptresvirus | B. gladioli | / | 64.31 | 38,038 | [118] |
| KS5 | Myovirus | Peduoviridae > Kisquinquevirus | B. multivorans; B. cenocepacia | Temperate | 63.71 | 37,236 | [90] |
| ST79 | Myovirus | Peduoviridae > Nampongvirus | B. pseudomallei; B. mallei | Lytic | 62.50 | 35,430 | [119] |
| BcepMigl | Podovirus | Lessievirus | B. cenocepacia | / | 65.51 | 62,952 | / |
| Bcep22 | Podovirus | Lessievirus | B. cenocepacia | Temperate | 65.31 | 63,882 | [84] |
| DC1 | Podovirus | Lessievirus | B. cepacia; B. cenocepacia; B. stabilis | Temperate, unstably lysogenic | 66.21 | 61,847 | [120] |
| BcepIL02 | Podovirus | Lessievirus | B. cenocepacia | Temperate | 66.20 | 62,715 | [84] |
| Mica | Myovirus | Micavirus | B. cenocepacia | Temperate | 62.15 | 43,707 | [121] |
| Bcep781 | Myovirus | Naesvirus | B. cepacia | Lytic | 63.33 | 48,247 | [122] |
| Bcep43 | Myovirus | Naesvirus | B. cepacia | Lytic | 63.43 | 48,024 | [122] |
| BcepNY3 | / | Naesvirus | B. cenocepacia | / | 63.64 | 47,382 | / |
| Bcep1 | Myovirus | Naesvirus | B. cenocepacia | Lytic | 63.64 | 48,177 | [122] |
| phiE058 | Myovirus | Novel species 40 within a novel genus 16 * | B. mallei; B. pseudomallei; B. thailandensis | Temperate | 64.12 | 44,121 | [123] |
| PE067 | Myovirus | Novel species 39 within a novel genus 16 * | B. pseudomallei; B. thailandensis | Temperate | 64.48 | 43,649 | [123] |
| BcepC6B | Podovirus | Ryyoungvirus | B. cepacia | Temperate | 65.19 | 42,415 | [122] |
| vB BmuP KL4 | / | Kelquatrovirus | B. multivorans | / | 63.18 | 42,250 | / |
| Magia | Myovirus | Magiavirus | B. cenocepacia | Temperate | 65.06 | 44,942 | [124] |
| phiE125 | Siphovirus | Stanholtvirus | B. mallei | Temperate | 61.19 | 53,373 | [86] |
| Phi644_2 | Siphovirus | Stanholtvirus | B. mallei; B. pseudomallei | Temperate | 60.45 | 48,674 | [91] |
| PhiBP82.1 | / | Stanholtvirus | B. pseudomallei | / | 60.68 | 54,921 | / |
| Phi1026b | Siphovirus | Stanholtvirus | B. mallei; B. pseudomallei | Temperate | 60.68 | 54,865 | [87] |
| phiBt | / | Stanholtvirus | B. pseudomallei | / | 60.30 | 56,453 | / |
| Bcep176 | Siphovirus | Stanholtvirus | B. multivorans; B. cepacia | Temperate | 61.54 | 44,856 | [125] |
| KS9 | Siphovirus | Stanholtvirus | B. pyrrocinia; B. cenocepacia | Temperate | 60.68 | 39,896 | [18,126] |
* Genomic classification by VICTOR [127] in Figure 1; / data unavailable.
Sixty-one whole-genome sequences of Burkholderia phages within the class Caudoviricetes were used to generate a phylogenomic tree using the Genome BLAST Distance Phylogeny (GBDP) method implemented in VICTOR (https://ggdc.dsmz.de/victor.php) (accessed on 26 February 2025) [127], allowing genome-based classification. The 61 Burkholderia phages are classified into 54 species, 21 genera, and 3 families (Figure 1). Family 1 includes the jumbo Burkholderia phages Chiangmaivirus FLC6 and FLC8 within the ICTV family Chimalliviridae and an unclassified genus (phage FLC9), which infect B. glumae. Family 2 includes the jumbo Burkholderia phage Sarumanvirus infecting B. cenocepacia and an unclassified genus (phage BCSR5). Family 3 includes all non-jumbo Burkholderia phages belonging to 17 genomogenera, among which 8 genera were classified into 5 existing ICTV families (Jondennisvirinae, Autonotataviridae, Lindbergviridae, Casjensviridae, and Peduoviridae). In other words, one genome-based family contains all non-jumbo Burkholderia phages within the class Caudoviricetes, indicating a limited taxon range of the non-jumbo Burkholderia phages. Nonetheless, the G + C mol% of these non-jumbo Burkholderia phages ranging from 51.45% to 68.82% indicates considerable genomic diversities within the genome-based Family 3. Together, this phylogenomic overview highlights both evolutionary divergence and taxonomic coherence among Burkholderia phages.
The VICTOR phylogenomic overview (Figure 1) also shows the host ranges at three phage taxon levels. First, the host range of the Burkholderia phages within Family 3 covers the genus Burkholderia sensu stricto. Second, multiple Burkholderia phages within a virus species infects only one Burkholderia species. For example, four Burkholderia phages within Ampunavirus BpAMP1 infect B. pseudomallei; two Burkholderia phages within Stanholtvirus sv1026b infect B. pseudomallei; three Burkholderia phages within Tigrvirus phi52237 infect B. pseudomallei; and Naesvirus Bcep781 and Naesvirus Bcep43 composing a genomospecies infect B. cepacia. Third, multiple virus species within multiple genera can infect the same Burkholderia species. As just noted, Ampunavirus BpAMP1, Stanholtvirus sv1026b, and Tigrvirus phi52237 infect B. pseudomallei. Fourth, multiple virus genera can infect the same multiple species within a species complex. For example, Lessievirus and Naesvirus infect Bcc species B. cepacia and B. cenocepacia; Nazgulvirus BcepNazgul and Ahduovirus AH2 composing a genomogenus also infect B. cepacia and B. cenocepacia. Fifth, a virus genus can infect multiple Burkholderia species within multiple species complexes. For example, Bcepmuvirus infects B. thailandensis (Bpc) and B. cenocepacia (Bcc); Ampunavirus BpAMP1, Pakuvirus paku, and Mguuvirus JG068 composing a genomogenus infect B. pseudomallei (Bpc) and B. cenocepacia (Bcc); Gladiolivirus Maja, Bcepfunavirus BcepF1, Irusalimvirus BCSR52, and Bglawtbvirus WTB composing a genomogenus infect B. cepacia (Bcc), B. ambifaria (Bcc), and B. gladioli (Bgc). Tigrvirus, Kayeltresvirus, Duodecimduovirus, Kisquattuordecimvirus, Aptresvirus, Kisquinquevirus, and Nampongvirus composing a genomogenus infect B. pseudomallei (Bpc), B. thailandensis (Bpc), B. cenocepacia (Bcc), B. ambifaria (Bcc), B. glumae (Bgc), and B. gladioli (Bgc).
Together, the genetic variability of the Burkholderia phages holds significant promise for both medical and agricultural applications. The multiple virus species or genera targeting the same Burkholderia species or species complex supports the strategy of using phage cocktails to control the Burkholderia-associated human, animal, or plant diseases. The phage cocktails containing diverse Burkholderia phages may use multiple mechanisms to control Burkholderia pathogens and to avoid immune escape by the Burkholderia pathogens.
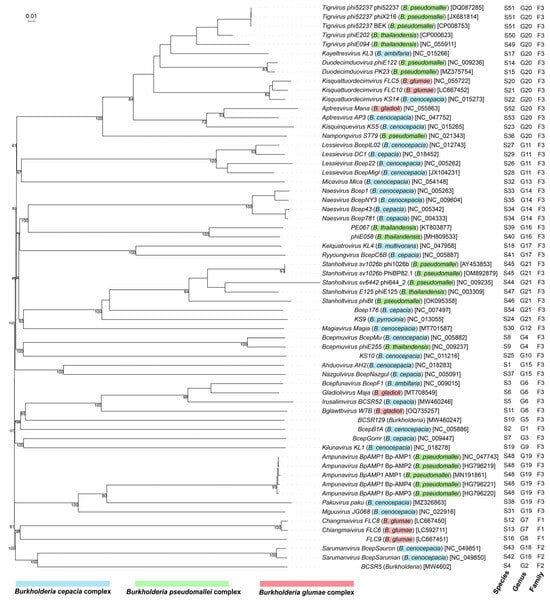
Figure 1.
Phylogenomic relationships among Burkholderia phages within the class Caudoviricetes. The balanced minimum-evolution tree inferred from intergenomic distances based on whole-genome sequence comparisons was generated using the Genome-BLAST Distance Phylogeny (GBDP) method implemented in VICTOR [127]. Branch support was inferred from 100 pseudo-bootstrap replicates via FASTME including SPR postprocessing [128]. Taxon boundaries at the species, genus, and family level were estimated with the OPTSIL program [129], the recommended clustering thresholds [127], and an F value (fraction of links required for cluster fusion) of 0.5 [130]. The branch lengths are scaled in terms of the GBDP distance formula d0. The tree was rooted at the jumbo phages and displayed using the online tool iTOL version 7 (https://itol.embl.de/) (accessed on 16 June 2025). Tree leaves were labeled with phage names (host) [nucleotide sequence accession numbers in GenBank] and genomic classification of phages into species, genus, and family. Phage host species within Burkholderia cepacia complex, Burkholderia pseudomallei complex, and Burkholderia glumae complex are highlighted in blue, green, and red, respectively.
4. Mechanism of Phage Action and Burkholderia Resistance
Phages infect Burkholderia host cells by recognizing and adsorbing to surface receptors, primarily LPS on the outer membrane. LPS consists of Lipid A (anchored in the membrane and responsible for endotoxicity), a core oligosaccharide with conserved inner and variable outer regions, and a highly variable O-antigen polysaccharide chain [21]. Additional receptors include capsular polysaccharides, flagella, and fimbriae [116]. After injection of genomes, phages hijack bacterial machinery to replicate and produce holins (membrane pore-forming proteins) and endolysins (peptidoglycan-degrading enzymes), which disrupt the cell envelope, leading to lysis and release of progeny phages [15,131].
Phage predation drives bacterial resistance primarily through modifications or loss of receptors. However, bacterial surface components are also critical to bacterial survival, motility, or virulence. As a result, receptor modifications frequently incur fitness costs including increased susceptibility to host immune factors and antibiotics [132]. For example, B. cenocepacia mutants with truncated LPS exhibit phage resistance but compromise serum resistance and increase sensitivity to colistin [133]. Similarly, infection by phage Bp-AMP1 can downregulate efflux pumps in B. thailandensis, increasing bacterial sensitivity to a broad range of antibiotics [134]. Beyond receptor modifications, Burkholderia has additional phage defense mechanisms, including excessive production of extracellular polysaccharides to physically shield receptors, activation of CRISPR-Cas systems to destroy invading phage genomes, and abortive infection systems that trigger programmed cell death to prevent phage propagation. However, these strategies also incur fitness trade-offs: overproduction of extracellular polysaccharide, reducing motility and nutrient uptake; CRISPR-Cas systems requiring metabolic resources and carrying a risk of autoimmunity; and abortive infection sacrificing the survival of individual cells [135,136]. These fitness trade-offs form the foundation of “phage steering” [133]. Strategically, phage–antibiotic synergy and phage therapy using phage cocktails can reduce bacterial resistance development and improve therapeutic outcomes. Combining phages with antibiotics such as meropenem can enhance bacterial clearance, reduce antibiotic doses, and delay resistance development [137]. Phage cocktails targeting diverse bacterial receptors and using evolving phages through directed adaptation improves treatment efficacy and mitigates Burkholderia resistance development (Figure 2).
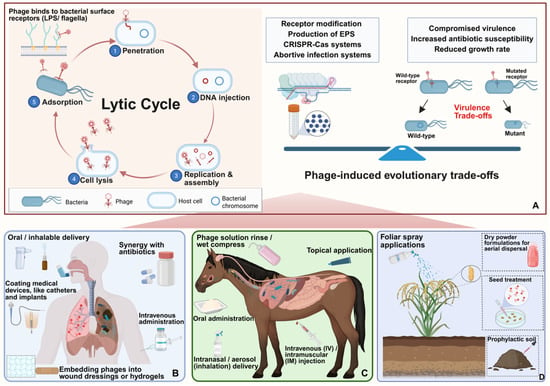
Figure 2.
Mechanisms and application of Burkholderia phages in control of human, animal, and plant diseases. (A) Mechanism and evolutionary trade-offs: The lytic cycle—from adsorption to lysis—eliminates bacteria cells. In response, bacteria evolve resistance mechanisms such as receptor modification and CRISPR-Cas systems, which incur fitness costs and can increase sensitivity to antibiotics. (B) Human therapy: Phages are administered via oral, inhalable, intravenous, and topical routes. Phages can be used with antibiotics and medical device coatings. (C) Veterinary use: Phages are delivered via oral, inhalable, intravenous, intramuscular and topical routes. (D) Agricultural application: Phages are applied via foliar sprays, soil treatments, or seed coatings to control plant diseases.
5. Biotechnological Applications of Burkholderia Phages
The success of phage therapy against Burkholderia infections relies heavily on the selection of appropriate delivery strategies that can overcome biological and environmental barriers across different systems: humans, animals, and plants.
5.1. Medical and Veterinary Applications
In human medicine, phages targeting Burkholderia species are primarily administered via inhalation or intravenous injection, tailored to infection sites. Aerosolized delivery, especially through nose-only inhalation devices, has demonstrated efficacy in murine models by significantly reducing lung bacterial loads caused by B. cenocepacia [18]. This method provides direct access to the respiratory tract, a common infection site in CF patients, and ensures phage viability post-aerosolization despite mechanical and pH stress [133]. In clinical cases, intravenous phage therapy, such as the administration of phage BdPF16phi4281, has been used compassionately to treat B. dolosa infections, resulting in temporary bacterial load reductions [138]. However, systemic administration poses risks of immune clearance and antibiotic-related toxicity, underscoring the need for improved delivery formulations.
In veterinary medicine, although Burkholderia-specific phages are yet to be tested, analogs targeting other pathogens like Salmonella have shown promising outcomes via oral and topical delivery in broilers [139]. These methods provide scalable, practical models for future adaptation to treat Burkholderia infections in livestock, especially for gastrointestinal or dermal infections.
5.2. Agricultural Applications
Phage-based biocontrol presents a sustainable alternative to chemical pesticides for managing Burkholderia-associated plant diseases. Effective deployment, however, requires consideration of environmental factors such as UV exposure, high temperature, desiccation, and phage persistence in the phyllosphere and rhizosphere [23]. Several Burkholderia-specific phages have shown potential in agricultural disease management. Phages KS12 and AH2, targeting B. gladioli, significantly reduce tissue destruction in onion and mushroom using a quantitative ex planta maceration model [112]. Phage WTB (vB_BglM_WTB), a high-efficiency lytic phage, also targets B. gladioli, offering rapid suppression of infections and potential for field deployment [110]. For B. glumae, a key pathogen of rice, the jumbo phage S13 demonstrates a unique flagella-dependent infection mechanism. By selecting for non-flagellated, less virulent mutants, S13 reduces pathogenicity while directly lysing motile bacterial populations [21]. Similarly, compost-derived jumbo phages FLC8 and FLC9 display broad-host ranges and have achieved over 77% control of rice seedling rot in greenhouse assays, while FLC10 exhibits narrower efficacy [82]. Application methods for these phages vary based on the plant–pathogen context. Foliar sprays, commonly used against epiphytic pathogens, are suitable for applying phages like KS12 and AH2 to aerial plant parts. However, foliar applications of phages are vulnerable to rapid UV inactivation; phage viability may drop below 1% within hours under sunlight [140]. To address this problem, formulations with UV-protective agents and humectants are being developed to enhance phage persistence on leaves. Soil drenching offers an effective alternative for root-associated infections by delivering phages like FLC8, FLC9, and FLC10 directly to the rhizosphere. This approach exploits phage mobility in moist soil, improving contact with root pathogens [141]. Additionally, seed coating with phages, particularly using polymer-based carriers, provides early-stage protection during germination and colonization of the rhizosphere, enhancing defense against soil-borne Burkholderia [142].
5.3. Nanotechnology-Enhanced Delivery
Nanotechnology-based delivery systems are increasingly used in phage therapy to enhance survival, targeting, and controlled release of phages [139,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157]. Alginate and chitosan nanocarriers, leveraging their pH-responsive properties, effectively protect phages during gastrointestinal transit while promoting mucosal adhesion. This makes them ideal for oral delivery in humans and animals, as they shield phages from gastric acidity and enable targeted intestinal release [143]. However, most studies remain at the in vitro or proof-of-concept stage, and more in vivo validation is required.
Hydrogel matrices, such as alginate–CaCO3 microcapsules, provide sustained phage release and have demonstrated efficacy in veterinary models by maintaining anti-Salmonella activity in poultry [139]. Their adaptability suggests potential application for Burkholderia-specific phages targeting both respiratory and gastrointestinal infections, though direct evidence for these specific phages is limited to date.
Other nanocarrier systems, such as liposomes, polymeric nanoparticles, nanofibers, and whey protein isolate-based films, expand phage therapy’s utility in clinical and agricultural settings [144,145,146,147]. These systems improve phage stability, controlled release, and adhesion to biological or environmental surfaces. Liposome encapsulation, for instance, protects phages in respiratory infections but requires optimization to address immune clearance and limited systemic circulation [148]. Notably, whey protein isolate-based films, especially when reinforced with chitosan nanofibers or nano-chitin, form biodegradable and biocompatible matrices ideal for encapsulating phages [149,150]. These composite systems support long-term storage, pH-responsive release, and enhanced adhesion, making them promising candidates for bioactive seed coatings and durable phage packaging in agricultural applications (Table 3).

Table 3.
Nanotechnology-enhanced phage delivery strategies.
6. Conclusions and Perspectives
The convergence of Burkholderia pathogens infecting plants, animals, and humans highlights their significance within the One Health framework. Some Burkholderia species exhibit cross-kingdom infectivity and share resistance mechanisms, such as efflux pumps, quorum sensing-regulated virulence, and biofilm formation. The zoonotic potential of Bpc species and the increasing clinical detection of Bcc strains from environmental and animal reservoirs emphasize the interconnectedness of ecosystems [5,45].
Lytic Burkholderia phages offer the foundation for phage therapy targeting Burkholderia-associated diseases in humans, animals, and plants. Genetically distinct Burkholderia phages belonging to different genera or even different families target the same Burkholderia species or multiple Burkholderia species causing the same disease, providing nature resources for phage cocktails with reduced risk of immune escape and resistance emergence. While temperate phages may be used after modification, either through the selection of lytic derivatives or synthetic design, advances in synthetic biology allow for the engineering of phages with defined host ranges, enabling a balance between therapeutic efficacy and biosafety against multidrug-resistant infections. Moreover, phage–antibiotic synergy can also stimulate increased phage activity and reduce the risk of resistance emergence, making it a valuable complement to phage cocktail strategy and synthetic biology in combating multidrug-resistant Burkholderia infections.
Effective phage therapy requires targeted delivery, environmental stability, and sustained activity. Appropriate delivery strategies can overcome biological and environmental barriers specific to each Burkholderia–host system. Lessons from clinical and veterinary applications, such as mucoadhesive polymers for gastrointestinal use and liposome encapsulation for respiratory infections, can be adapted for agricultural purposes. Encapsulation methods like alginate microbeads and alginate/chitosan composites protect phages from environmental stress and allow for controlled pH-responsive release. These formulations are compatible with diverse agricultural delivery modes, including foliar sprays, soil drenches, and seed coatings. Moreover, spray-dried phage powders and electrospun nanofiber matrices enable the production of stable, field-ready products. The natural mucoadhesive and biodegradable properties of alginate and chitosan enhance phage targeting and prolong antibacterial activity. Whey protein isolate-based films, especially those reinforced with chitosan nanofibers or nano-chitins, offer biodegradable and biocompatible matrices for long-term storage and sustained bioactivity.
Together, these nanotechnology-enabled delivery systems bridge the gap between laboratory research and real-world implementation of phage therapy. The development of controlled host range phage cocktails, refined application-specific delivery systems, field trials, and regulatory frameworks holds promises for establishing robust, sustainable, and scalable phage-based biocontrol strategies across various Burkholderia–host systems. This integrated approach aligns closely with the One Health perspective, offering an eco-friendly alternative to antibiotics and chemical pesticides for managing Burkholderia-associated diseases in clinical and agricultural contexts.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms13081873/s1, Table S1. Pathogenic Burkholderia species. Figure S1. Comparison of morphology-based and genome-based phage classification. Conventional morphology-based phage taxonomy classifies phages by capsid shape and tail type using electron microscopy. Modern genome-based phage taxonomy classifies phages by genome sequencing, overall DNA and protein similarity, and phylogenetic analyses based on core genes and proteins. This transition improves classification in capturing phage diversity and evolutionary relationships. References [158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177] are cited in the Supplementary Materials.
Author Contributions
Conceptualization, B.W., D.D., Y.W., J.B., J.L., B.L. and Q.A.; methodology, B.W. and J.Z.; validation, B.W. and Q.A.; formal analysis, B.W. and J.Z.; resources, L.C., J.B. and J.L.; writing—original draft preparation, B.W., J.Z., L.C., M.I. and C.L.; writing—review and editing, B.L., J.L. and Q.A.; visualization, B.W., M.I. and Q.A.; supervision, B.L. and Q.A.; project administration, L.C., J.B., D.D., Y.W. and J.L.; funding acquisition, D.D., Y.W., J.B., B.L. and J.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ningbo Natural Science Foundation (2022J197), the Shanghai Agricultural Science and Technology Innovation Project (T2023101), and Industrial Technology Projects of Department of Agriculture and Rural Affairs of Zhejiang Province (2025-5) of China.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Bach, E.; Sant’Anna, F.H.; dos Santos Seger, G.D.; Passaglia, L.M.P. Pangenome inventory of Burkholderia sensu lato, Burkholderia sensu stricto, and the Burkholderia cepacia complex reveals the uniqueness of Burkholderia catarinensis. Genomics 2022, 114, 398–408. [Google Scholar] [CrossRef]
- Mullins, A.J.; Mahenthiralingam, E. The hidden genomic diversity, specialized metabolite capacity, and revised taxonomy of Burkholderia sensu lato. Front. Microbiol. 2021, 12, 726847. [Google Scholar] [CrossRef]
- Elshafie, H.S.; Camele, I. An overview of metabolic activity, beneficial and pathogenic aspects of Burkholderia spp. Metabolites 2021, 11, 321. [Google Scholar] [CrossRef]
- Morya, R.; Salvachúa, D.; Thakur, I.S. Burkholderia: An untapped but promising bacterial genus for the conversion of aromatic compounds. Trends Biotechnol. 2020, 38, 963–975. [Google Scholar] [CrossRef]
- Limmathurotsakul, D.; Golding, N.; Dance, D.A.; Messina, J.P.; Pigott, D.M.; Moyes, C.L.; Rolim, D.B.; Bertherat, E.; Day, N.P.; Peacock, S.J.; et al. Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nat. Microbiol. 2016, 1, 15008. [Google Scholar] [CrossRef]
- Weinberg, J.B.; Alexander, B.D.; Majure, J.M.; Williams, L.W.; Kim, J.Y.; Vandamme, P.; LiPuma, J.J. Burkholderia glumae infection in an infant with chronic granulomatous disease. J. Clin. Microbiol. 2007, 45, 662–665. [Google Scholar] [CrossRef] [PubMed]
- Segonds, C.; Clavel-Batut, P.; Thouverez, M.; Grenet, D.; Le Coustumier, A.; Plésiat, P.; Chabanon, G. Microbiological and epidemiological features of clinical respiratory isolates of Burkholderia gladioli. J. Clin. Microbiol. 2009, 47, 1510–1516. [Google Scholar] [CrossRef] [PubMed]
- Rajendraprasad, S.; Creech, Z.A.; Truong, G.T.D.; Nguyen, T.; Addula, M.; Mendoza, N.; Velagapudi, M. Fatal case of Burkholderia gladioli pneumonia in a patient with COVID-19. Ochsner J. 2022, 22, 349–352. [Google Scholar] [CrossRef]
- Yao, L.; Qian, C.; Guo, J.; Zhang, H.; Li, Z.; Xie, D.; Xia, L.; Wu, Q.; Hong, M. Traceability and characteristic investigation of Burkholderia gladioli bloodstream infection in patients with hematologic malignancies. Blood 2023, 142, 5569. [Google Scholar] [CrossRef]
- Zhou, J.; Ren, H.; Hu, M.; Zhou, J.; Li, B.; Kong, N.; Zhang, Q.; Jin, Y.; Liang, L.; Yue, J. Characterization of Burkholderia cepacia complex core genome and the underlying recombination and positive selection. Front. Genet. 2020, 11, 506. [Google Scholar] [CrossRef]
- Vial, L.; Chapalain, A.; Groleau, M.C.; Déziel, E. The various lifestyles of the Burkholderia cepacia complex species: A tribute to adaptation. Environ. Microbiol. 2011, 13, 1–12. [Google Scholar] [CrossRef]
- Mannaa, M.; Park, I.; Seo, Y.-S. Genomic features and insights into the taxonomy, virulence, and benevolence of plant-associated Burkholderia species. Int. J. Mol. Sci. 2018, 20, 121. [Google Scholar] [CrossRef]
- Podnecky, N.L.; Rhodes, K.A.; Schweizer, H.P. Efflux pump-mediated drug resistance in Burkholderia. Front. Microbiol. 2015, 6, 305. [Google Scholar] [CrossRef]
- Rhodes, K.A.; Schweizer, H.P. Antibiotic resistance in Burkholderia species. Drug Resist. Updat. 2016, 28, 82–90. [Google Scholar] [CrossRef]
- Lauman, P.; Dennis, J.J. Advances in phage therapy: Targeting the Burkholderia cepacia complex. Viruses 2021, 13, 1331. [Google Scholar] [CrossRef]
- Merril, C.R.; Scholl, D.; Adhya, S.L. The prospect for bacteriophage therapy in Western medicine. Nat. Rev. Drug Discov. 2003, 2, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Guang-Han, O.; Leang-Chung, C.; Vellasamy, K.M.; Mariappan, V.; Li-Yen, C.; Vadivelu, J. Experimental phage therapy for Burkholderia pseudomallei infection. PLoS ONE 2016, 11, e0158213. [Google Scholar] [CrossRef]
- Semler, D.D.; Lynch, K.H.; Dennis, J.J. The promise of bacteriophage therapy for Burkholderia cepacia complex respiratory infections. Front. Cell. Infect. Microbiol. 2012, 1, 27. [Google Scholar] [CrossRef] [PubMed]
- Mehmood Khan, F.; Manohar, P.; Singh Gondil, V.; Mehra, N.; Kayode Oyejobi, G.; Odiwuor, N.; Ahmad, T.; Huang, G. The applications of animal models in phage therapy: An update. Hum. Vaccines Immunother. 2023, 19, 2175519. [Google Scholar] [CrossRef] [PubMed]
- Jungkhun, N.; Farias, A.R.; Barphagha, I.; Patarapuwadol, S.; Ham, J.H. Isolation and characterization of bacteriophages infecting Burkholderia glumae, the major causal agent of bacterial panicle blight in rice. Plant Dis. 2021, 105, 2551–2559. [Google Scholar] [CrossRef]
- Supina, B.S.; McCutcheon, J.G.; Peskett, S.R.; Stothard, P.; Dennis, J.J. A flagella-dependent Burkholderia jumbo phage controls rice seedling rot and steers Burkholderia glumae toward reduced virulence in rice seedlings. mBio 2025, 16, e02814-24. [Google Scholar] [CrossRef]
- Ryan, E.M.; Gorman, S.P.; Donnelly, R.F.; Gilmore, B.F. Recent advances in bacteriophage therapy: How delivery routes, formulation, concentration and timing influence the success of phage therapy. J. Pharm. Pharmacol. 2011, 63, 1253–1264. [Google Scholar] [CrossRef]
- Holtappels, D.; Fortuna, K.; Lavigne, R.; Wagemans, J. The future of phage biocontrol in integrated plant protection for sustainable crop production. Curr. Opin. Biotechnol. 2021, 68, 60–71. [Google Scholar] [CrossRef]
- Ke, D.; Luo, J.; Liu, P.; Shou, L.; Ijaz, M.; Ahmed, T.; Shahid, M.S.; An, Q.; Mustać, I.; Ondrasek, G.; et al. Advancements in Bacteriophages for the Fire Blight Pathogen Erwinia amylovora. Viruses 2024, 16, 1619. [Google Scholar] [CrossRef] [PubMed]
- Shang, W.; Xiong, Q.; Xie, Z.; Cheng, J.; Yu, B.; Zhang, H.; Su, Y.; Zhao, J. Functional, eco-friendly, and starch-based nanocarriers with sustained release of carvacrol for persistent control of tomato gray mold. Crop Health 2023, 1, 13. [Google Scholar] [CrossRef]
- Paczesny, J.; Bielec, K. Application of bacteriophages in nanotechnology. Nanomaterials 2020, 10, 1944. [Google Scholar] [CrossRef] [PubMed]
- Mahenthiralingam, E.; Urban, T.A.; Goldberg, J.B. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 2005, 3, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Drevinek, P.; Mahenthiralingam, E. Burkholderia cenocepacia in cystic fibrosis: Epidemiology and molecular mechanisms of virulence. Clin. Microbiol. Infect. 2010, 16, 821–830. [Google Scholar] [CrossRef]
- Roux, D.; Weatherholt, M.; Clark, B.; Gadjeva, M.; Renaud, D.; Scott, D.; Skurnik, D.; Priebe, G.P.; Pier, G.; Gerard, C.; et al. Immune recognition of the epidemic cystic fibrosis pathogen Burkholderia dolosa. Infect. Immun. 2017, 85, e00765-16. [Google Scholar] [CrossRef]
- Pham, A.; Volmer, J.G.; Chambers, D.C.; Smith, D.J.; Reid, D.W.; Burr, L.; Wells, T.J. Genomic analyses of Burkholderia respiratory isolates indicates two evolutionarily distinct B. anthina clades. Front. Microbiol. 2023, 14, 1274280. [Google Scholar] [CrossRef]
- Hudson, M.J. Outbreak of Burkholderia stabilis infections associated with contaminated nonsterile, multiuse ultrasound gel—10 states, May–September 2021. MMWR Morb. Mortal. Wkly Rep. 2022, 71, 1517–1521. [Google Scholar] [CrossRef]
- Seth-Smith, H.M.; Casanova, C.; Sommerstein, R.; Meinel, D.M.; Abdelbary, M.M.; Blanc, D.S.; Droz, S.; Führer, U.; Lienhard, R.; Lang, C.; et al. Phenotypic and genomic analyses of Burkholderia stabilis clinical contamination, Switzerland. Emerg. Infect. Dis. 2019, 25, 1084. [Google Scholar] [CrossRef] [PubMed]
- Nunvar, J.; Kalferstova, L.; Bloodworth, R.A.; Kolar, M.; Degrossi, J.; Lubovich, S.; Cardona, S.T.; Drevinek, P. Understanding the pathogenicity of Burkholderia contaminans, an emerging pathogen in cystic fibrosis. PLoS ONE 2016, 11, e0160975. [Google Scholar] [CrossRef]
- Moehring, R.W.; Lewis, S.S.; Isaacs, P.J.; Schell, W.A.; Thomann, W.R.; Althaus, M.M.; Hazen, K.C.; Dicks, K.V.; LiPuma, J.J.; Chen, L.F.; et al. Outbreak of bacteremia due to Burkholderia contaminans linked to intravenous fentanyl from an institutional compounding pharmacy. JAMA Intern. Med. 2014, 174, 606–612. [Google Scholar] [CrossRef]
- Fujii, Y.; Suwa, A.; Tsuyuki, Y.; Koyama, K.; Nio-Kobayashi, J.; Yoshii, K. The First case of a cat infected with Burkholderia pseudomultivorans, a member of the Burkholderia cepacia complex. Vet. Sci. 2024, 11, 559. [Google Scholar] [CrossRef]
- Peeters, C.; Zlosnik, J.E.; Spilker, T.; Hird, T.J.; LiPuma, J.J.; Vandamme, P. Burkholderia pseudomultivorans sp. nov. a novel Burkholderia cepacia complex species from human respiratory samples and the rhizosphere. Syst. Appl. Microbiol. 2013, 36, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Campos, C.G.; Byrd, M.S.; Cotter, P.A. Functional characterization of Burkholderia pseudomallei trimeric autotransporters. Infect. Immun. 2013, 81, 2788–2799. [Google Scholar] [CrossRef] [PubMed]
- Dance, D. Melioidosis: The tip of the iceberg? Clin. Microbiol. Rev. 1991, 4, 52–60. [Google Scholar] [CrossRef]
- Kettle, A.N.; Wernery, U. Glanders and the risk for its introduction through the international movement of horses. Equine Vet. J. 2016, 48, 654–658. [Google Scholar] [CrossRef]
- Khan, I.; Wieler, L.; Melzer, F.; Elschner, M.; Muhammad, G.; Ali, S.; Sprague, L.; Neubauer, H.; Saqib, M. Glanders in animals: A review on epidemiology, clinical presentation, diagnosis and countermeasures. Transbound. Emerg. Dis. 2013, 60, 204–221. [Google Scholar] [CrossRef]
- Srinivasan, A.; Kraus, C.N.; DeShazer, D.; Becker, P.M.; Dick, J.D.; Spacek, L.; Bartlett, J.G.; Byrne, W.R.; Thomas, D.L. Glanders in a military research microbiologist. N. Engl. J. Med. 2001, 345, 256–258. [Google Scholar] [CrossRef] [PubMed]
- Brett, P.J.; DeShazer, D.; Woods, D.E. Note: Burkholderia thailandensis sp. nov. a Burkholderia pseudomallei-like species. Int. J. Syst. Evol. Microbiol. 1998, 48, 317–320. [Google Scholar] [CrossRef]
- Glass, M.B.; Gee, J.E.; Steigerwalt, A.G.; Cavuoti, D.; Barton, T.; Hardy, R.D.; Godoy, D.; Spratt, B.G.; Clark, T.A.; Wilkins, P.P. Pneumonia and septicemia caused by Burkholderia thailandensis in the United States. J. Clin. Microbiol. 2006, 44, 4601–4604. [Google Scholar] [CrossRef]
- Lertpatanasuwan, N.; Sermsri, K.; Petkaseam, A.; Trakulsomboon, S.; Thamlikitkul, V.; Suputtamongkol, Y. Arabinose-positive Burkholderia pseudomallei infection in humans: Case report. Clin. Infect. Dis. 1999, 28, 927–928. [Google Scholar] [CrossRef]
- Patro, S.; Sharma, V.; Choudhary, A.; Varuneil, Y.; Pathi, B.K.; Pattnaik, S.S.; Pathi, B. Clinical and microbiological insights into Burkholderia infections: A retrospective study from a tertiary care hospital. Cureus 2025, 17, e76742. [Google Scholar] [CrossRef]
- Leitão, J.H.; Sousa, S.A.; Ferreira, A.S.; Ramos, C.G.; Silva, I.N.; Moreira, L.M. Pathogenicity, virulence factors, and strategies to fight against Burkholderia cepacia complex pathogens and related species. Appl. Microbiol. Biotechnol. 2010, 87, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Mahenthiralingam, E.; Baldwin, A.; Dowson, C.G. Burkholderia cepacia complex bacteria: Opportunistic pathogens with important natural biology. J. Appl. Microbiol. 2008, 104, 1539–1551. [Google Scholar] [CrossRef]
- Uehlinger, S.; Schwager, S.; Bernier, S.P.; Riedel, K.; Nguyen, D.T.; Sokol, P.A.; Eberl, L. Identification of specific and universal virulence factors in Burkholderia cenocepacia strains by using multiple infection hosts. Infect. Immun. 2009, 77, 4102–4110. [Google Scholar] [CrossRef]
- Nguyen, Q.H.; Nguyen, C.L.; Nguyen, T.S.; Do, B.N.; Tran, T.T.T.; Le, T.T.H.; Bui, T.T.; Le, H.S.; Van Quyen, D.; Hayer, J.; et al. Genomic insights into an extensively drug-resistant and hypervirulent Burkholderia dolosa N149 isolate of a novel sequence type (ST2237) from a Vietnamese patient hospitalised for stroke. J. Glob. Antimicrob. Resist. 2024, 37, 44–47. [Google Scholar] [CrossRef]
- Bernier, S.P.; Son, S.; Surette, M.G. The Mla pathway plays an essential role in the intrinsic resistance of Burkholderia cepacia complex species to antimicrobials and host innate components. J. Bacteriol. 2018, 200, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Deng, P.; Wang, X.; Baird, S.M.; Showmaker, K.C.; Smith, L.; Peterson, D.G.; Lu, S. Comparative genome-wide analysis reveals that Burkholderia contaminans MS 14 possesses multiple antimicrobial biosynthesis genes but not major genetic loci required for pathogenesis. Microbiologyopen 2016, 5, 353–369. [Google Scholar] [CrossRef]
- Galyov, E.E.; Brett, P.J.; DeShazer, D. Molecular insights into Burkholderia pseudomallei and Burkholderia mallei pathogenesis. Annu. Rev. Microbiol. 2010, 64, 495–517. [Google Scholar] [CrossRef]
- Wiersinga, W.J.; Van der Poll, T.; White, N.J.; Day, N.P.; Peacock, S.J. Melioidosis: Insights into the pathogenicity of Burkholderia pseudomallei. Nat. Rev. Microbiol. 2006, 4, 272–282. [Google Scholar] [CrossRef]
- Bzdyl, N.M.; Moran, C.L.; Bendo, J.; Sarkar-Tyson, M. Pathogenicity and virulence of Burkholderia pseudomallei. Virulence 2022, 13, 2139063. [Google Scholar] [CrossRef] [PubMed]
- Ham, J.H.; Melanson, R.A.; Rush, M.C. Burkholderia glumae: Next major pathogen of rice? Mol. Plant Pathol. 2011, 12, 329–339. [Google Scholar] [CrossRef]
- Jeong, Y.; Kim, J.; Kim, S.; Kang, Y.; Nagamatsu, T.; Hwang, I. Toxoflavin produced by Burkholderia glumae causing rice grain rot is responsible for inducing bacterial wilt in many field crops. Plant Dis. 2003, 87, 890–895. [Google Scholar] [CrossRef]
- Cui, Z.; Wang, S.; Kakar, K.U.; Xie, G.; Li, B.; Chen, G.; Zhu, B. Genome sequence and adaptation analysis of the human and rice pathogenic strain Burkholderia glumae AU6208. Pathogens 2021, 10, 87. [Google Scholar] [CrossRef] [PubMed]
- Brizendine, K.; Baddley, J.; Pappas, P.; Leon, K.; Rodriguez, J. Fatal Burkholderia gladioli infection misidentified as Empedobacter brevis in a lung transplant recipient with cystic fibrosis. Transpl. Infect. Dis. 2012, 14, E13–E18. [Google Scholar] [CrossRef] [PubMed]
- Graves, M.; Robin, T.; Chipman, A.M.; Wong, J.; Khashe, S.; Janda, J.M. Four additional cases of Burkholderia gladioli infection with microbiological correlates and review. Clin. Infect. Dis. 1997, 25, 838–842. [Google Scholar] [CrossRef]
- Nandakumar, R.; Shahjahan, A.; Yuan, X.; Dickstein, E.; Groth, D.; Clark, C.; Cartwright, R.; Rush, M. Burkholderia glumae and B. gladioli cause bacterial panicle blight in rice in the southern United States. Plant Dis. 2009, 93, 896–905. [Google Scholar] [CrossRef]
- Ura, H.; Furuya, N.; Iiyama, K.; Hidaka, M.; Tsuchiya, K.; Matsuyama, N. Burkholderia gladioli associated with symptoms of bacterial grain rot and leaf-sheath browning of rice plants. J. Gen. Plant Pathol. 2006, 72, 98–103. [Google Scholar] [CrossRef]
- Wang, M.; Wei, P.; Cao, M.; Zhu, L.; Lu, Y. First report of rice seedling blight caused by Burkholderia plantarii in north and southeast China. Plant Dis. 2016, 100, 645. [Google Scholar] [CrossRef]
- Matsumoto, H.; Fan, X.; Wang, Y.; Kusstatscher, P.; Duan, J.; Wu, S.; Chen, S.; Qiao, K.; Wang, Y.; Ma, B.; et al. Bacterial seed endophyte shapes disease resistance in rice. Nat. Plants 2021, 7, 60–72. [Google Scholar] [CrossRef]
- Burkholder, W.H. Sour skin, a bacterial rot of onion bulbs. Phytopathology 1950, 40, 115–117. [Google Scholar]
- Morales-Ruíz, L.-M.; Rodríguez-Cisneros, M.; Kerber-Díaz, J.-C.; Rojas-Rojas, F.-U.; Ibarra, J.A.; Estrada-de Los Santos, P. Burkholderia orbicola sp. nov. a novel species within the Burkholderia cepacia complex. Arch. Microbiol. 2022, 204, 178. [Google Scholar] [CrossRef] [PubMed]
- Velez, L.S.; Aburjaile, F.F.; Farias, A.R.; Baia, A.D.; Oliveira, W.J.; Silva, A.M.; Benko-Iseppon, A.M.; Azevedo, V.; Brenig, B.; Ham, J.H.; et al. Burkholderia semiarida sp. nov. and Burkholderia sola sp. nov. two novel B. cepacia complex species causing onion sour skin. Syst. Appl. Microbiol. 2023, 46, 126415. [Google Scholar] [CrossRef]
- Kim, N.; Lee, D.; Lee, S.-B.; Lim, G.-H.; Kim, S.-W.; Kim, T.-J.; Park, D.-S.; Seo, Y.-S. Understanding Burkholderia glumae BGR1 virulence through the application of toxoflavin-degrading enzyme, TxeA. Plants 2023, 12, 3934. [Google Scholar] [CrossRef] [PubMed]
- Lelis, T.; Peng, J.; Barphagha, I.; Chen, R.; Ham, J.H. The virulence function and regulation of the metalloprotease gene prtA in the plant-pathogenic bacterium Burkholderia glumae. Mol. Plant-Microbe Interact. 2019, 32, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Marunga, J.; Goo, E.; Kang, Y.; Hwang, I. Mutations in the two-component GluS-GluR regulatory system confer resistance to β-lactam antibiotics in Burkholderia glumae. Front. Microbiol. 2021, 12, 721444. [Google Scholar] [CrossRef]
- Lee, J.; Park, J.; Kim, S.; Park, I.; Seo, Y.S. Differential regulation of toxoflavin production and its role in the enhanced virulence of Burkholderia gladioli. Mol. Plant Pathol. 2016, 17, 65–76. [Google Scholar] [CrossRef]
- Paudel, S.; Franco, Y.; Zhao, M.; Dutta, B.; Kvitko, B.H. Distinct virulence mechanisms of Burkholderia gladioli in onion foliar and bulb scale tissues. Mol. Plant-Microbe Interact. 2025, 38, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Zeiser, E.T.; Becka, S.A.; Wilson, B.M.; Barnes, M.D.; LiPuma, J.J.; Papp-Wallace, K.M. “Switching partners”: Piperacillin-avibactam is a highly potent combination against multidrug-resistant Burkholderia cepacia complex and Burkholderia gladioli cystic fibrosis isolates. J. Clin. Microbiol. 2019, 57, e00181-19. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, E.B.; Santos, L.R.d.; Egito, A.A.d.; Santos, M.G.d.; Mantovani, C.; Rieger, J.d.S.G.; Abrantes, G.A.d.S.; Suniga, P.A.P.; Favacho, J.d.M.; Pinto, I.B.; et al. Assessment of the virulence of the Burkholderia mallei strain BAC 86/19 in BALB/c mice. Microorganisms 2023, 11, 2597. [Google Scholar] [CrossRef]
- Memišević, V.; Zavaljevski, N.; Pieper, R.; Rajagopala, S.V.; Kwon, K.; Townsend, K.; Yu, C.; Yu, X.; DeShazer, D.; Reifman, J.; et al. Novel Burkholderia mallei virulence factors linked to specific host-pathogen protein interactions. Mol. Cell. Proteom. 2013, 12, 3036–3051. [Google Scholar] [CrossRef]
- Saikh, K.U.; Mott, T.M. Innate immune response to Burkholderia mallei. Curr. Opin. Infect. Dis. 2017, 30, 297–302. [Google Scholar] [CrossRef]
- Li, J.; Zhong, Q.; Li, J.; Chong, H.-M.; Wang, L.-X.; Xing, Y.; Lu, W.-P. Genomic features and virulence characteristics of a rare Burkholderia thailandensis strain causing human infection. J. Med. Microbiol. 2023, 72, 001688. [Google Scholar] [CrossRef]
- Thapa, S.S.; Al-Tohamy, A.; Grove, A. The global regulator MftR controls virulence and siderophore production in Burkholderia thailandensis. J. Bacteriol. 2022, 204, e00237-22. [Google Scholar] [CrossRef]
- Kovacs-Simon, A.; Hemsley, C.; Scott, A.; Prior, J.; Titball, R. Burkholderia thailandensis strain E555 is a surrogate for the investigation of Burkholderia pseudomallei replication and survival in macrophages. BMC Microbiol. 2019, 19, 97. [Google Scholar] [CrossRef]
- Hyman, P. Phages for phage therapy: Isolation, characterization, and host range breadth. Pharmaceuticals 2019, 12, 35. [Google Scholar] [CrossRef]
- Khan Mirzaei, M.; Nilsson, A.S. Isolation of phages for phage therapy: A comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE 2015, 10, e0118557. [Google Scholar] [CrossRef] [PubMed]
- Adachi, N.; Tsukamoto, S.; Inoue, Y.; Azegami, K. Control of bacterial seedling rot and seedling blight of rice by bacteriophage. Plant Dis. 2012, 96, 1033–1036. [Google Scholar] [CrossRef] [PubMed]
- Kanaizuka, A.; Sasaki, R.; Miyashita, S.; Ando, S.; Ito, K.; Fukuhara, T.; Takahashi, H. Isolation of Burkholderia jumbo phages and their utilization as biocontrol agents to suppress rice seedling rot disease. J. Gen. Plant Pathol. 2023, 89, 24–34. [Google Scholar] [CrossRef]
- Sasaki, R.; Miyashita, S.; Ando, S.; Ito, K.; Fukuhara, T.; Takahashi, H. Isolation and characterization of a novel jumbo phage from leaf litter compost and its suppressive effect on rice seedling rot diseases. Viruses 2021, 13, 591. [Google Scholar] [CrossRef] [PubMed]
- Gill, J.J.; Summer, E.J.; Russell, W.K.; Cologna, S.M.; Carlile, T.M.; Fuller, A.C.; Kitsopoulos, K.; Mebane, L.M.; Parkinson, B.N.; Sullivan, D.; et al. Genomes and characterization of phages Bcep22 and BcepIL02, founders of a novel phage type in Burkholderia cenocepacia. J. Bacteriol. 2011, 193, 5300–5313. [Google Scholar] [CrossRef]
- Roszniowski, B.; Latka, A.; Maciejewska, B.; Vandenheuvel, D.; Olszak, T.; Briers, Y.; Holt, G.S.; Valvano, M.A.; Lavigne, R.; Smith, D.L.; et al. The temperate Burkholderia phage AP3 of the Peduovirinae shows efficient antimicrobial activity against B. cenocepacia of the IIIA lineage. Appl. Microbiol. Biotechnol. 2017, 101, 1203–1216. [Google Scholar] [CrossRef]
- Woods, D.E.; Jeddeloh, J.A.; Fritz, D.L.; DeShazer, D. Burkholderia thailandensis E125 harbors a temperate bacteriophage specific for Burkholderia mallei. J. Bacteriol. 2002, 184, 4003–4017. [Google Scholar] [CrossRef]
- DeShazer, D. Genomic diversity of Burkholderia pseudomallei clinical isolates: Subtractive hybridization reveals a Burkholderia mallei-specific prophage in B. pseudomallei 1026b. J. Bacteriol. 2004, 186, 3938–3950. [Google Scholar] [CrossRef]
- Gatedee, J.; Kritsiriwuthinan, K.; Galyov, E.E.; Shan, J.; Dubinina, E.; Intarak, N.; Clokie, M.R.; Korbsrisate, S. Isolation and characterization of a novel podovirus which infects Burkholderia pseudomallei. Virol. J. 2011, 8, 366. [Google Scholar] [CrossRef]
- Ackermann, H.-W. Bacteriophage electron microscopy. Adv. Virus Res. 2012, 82, 1–32. [Google Scholar]
- Lynch, K.H.; Stothard, P.; Dennis, J.J. Genomic analysis and relatedness of P2-like phages of the Burkholderia cepacia complex. BMC Genom. 2010, 11, 599. [Google Scholar] [CrossRef]
- Ronning, C.M.; Losada, L.; Brinkac, L.; Inman, J.; Ulrich, R.L.; Schell, M.; Nierman, W.C.; DeShazer, D. Genetic and phenotypic diversity in Burkholderia: Contributions by prophage and phage-like elements. BMC Microbiol. 2010, 10, 202. [Google Scholar] [CrossRef]
- Shan, J.; Korbsrisate, S.; Withatanung, P.; Adler, N.L.; Clokie, M.R.; Galyov, E.E. Temperature dependent bacteriophages of a tropical bacterial pathogen. Front. Microbiol. 2014, 5, 599. [Google Scholar] [CrossRef] [PubMed]
- Letarov, A.V.; Letarova, M.A.; Ivanov, P.A.; Belalov, I.S.; Clokie, M.R.; Galyov, E.E. Genetic analysis of the cold-sensitive growth phenotype of Burkholderia pseudomallei/thailandensis bacteriophage AMP1. Sci. Rep. 2022, 12, 4288. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, R.; Pires, D.P.; Costa, A.R.; Azeredo, J. Phage therapy: Going temperate? Trends Microbiol. 2019, 27, 368–378. [Google Scholar] [CrossRef] [PubMed]
- Seed, K.D.; Dennis, J.J. Isolation and characterization of bacteriophages of the Burkholderia cepacia complex. FEMS Microbiol. Lett. 2005, 251, 273–280. [Google Scholar] [CrossRef]
- Islam, M.Z.; Fokine, A.; Mahalingam, M.; Zhang, Z.; Garcia-Doval, C.; van Raaij, M.J.; Rossmann, M.G.; Rao, V.B. Molecular anatomy of the receptor binding module of a bacteriophage long tail fiber. PLoS Pathog. 2019, 15, e1008193. [Google Scholar] [CrossRef]
- Taslem Mourosi, J.; Awe, A.; Guo, W.; Batra, H.; Ganesh, H.; Wu, X.; Zhu, J. Understanding bacteriophage tail fiber interaction with host surface receptor: The key “blueprint” for reprogramming phage host range. Int. J. Mol. Sci. 2022, 23, 12146. [Google Scholar] [CrossRef]
- Le, S.; He, X.; Tan, Y.; Huang, G.; Zhang, L.; Lux, R.; Shi, W.; Hu, F. Mapping the tail fiber as the receptor binding protein responsible for differential host specificity of Pseudomonas aeruginosa bacteriophages PaP1 and JG004. PLoS ONE 2013, 8, e68562. [Google Scholar] [CrossRef]
- He, P.; Cao, F.; Qu, Q.; Geng, H.; Yang, X.; Xu, T.; Wang, R.; Jia, X.; Lu, M.; Zeng, P.; et al. Host range expansion of Acinetobacter phage vB_Ab4_Hep4 driven by a spontaneous tail tubular mutation. Front. Cell. Infect. Microbiol. 2024, 14, 1301089. [Google Scholar] [CrossRef]
- Zhang, J.; Ning, H.; Lin, H.; She, J.; Wang, L.; Jing, Y.; Wang, J. Expansion of the plaquing host range and improvement of the absorption rate of a T5-like Salmonella phage by altering the long tail fibers. Appl. Environ. Microbiol. 2022, 88, e00895-22. [Google Scholar] [CrossRef]
- Turner, D.; Kropinski, A.M.; Adriaenssens, E.M. A roadmap for genome-based phage taxonomy. Viruses 2021, 13, 506. [Google Scholar] [CrossRef]
- Turner, D.; Shkoporov, A.N.; Lood, C.; Millard, A.D.; Dutilh, B.E.; Alfenas-Zerbini, P.; Van Zyl, L.J.; Aziz, R.K.; Oksanen, H.M.; Poranen, M.M.; et al. Abolishment of morphology-based taxa and change to binomial species names: 2022 taxonomy update of the ICTV bacterial viruses subcommittee. Arch. Virol. 2023, 168, 74. [Google Scholar] [CrossRef] [PubMed]
- Quinones-Olvera, N.; Owen, S.V.; McCully, L.M.; Marin, M.G.; Rand, E.A.; Fan, A.C.; Martins Dosumu, O.J.; Paul, K.; Sanchez Castaño, C.E.; Petherbridge, R.; et al. Diverse and abundant phages exploit conjugative plasmids. Nat. Commun. 2024, 15, 3197. [Google Scholar] [CrossRef]
- Park, K.E. The Genomes of Bacteriophages NY12 and Sauron. Bachelor’s Thesis, Texas A&M University, College Station, TX, USA, 2014. [Google Scholar]
- Porat, S.B.; Gelman, D.; Yerushalmy, O.; Alkalay-Oren, S.; Coppenhagen-Glazer, S.; Cohen-Cymberknoh, M.; Kerem, E.; Amirav, I.; Nir-Paz, R.; Hazan, R. Expanding clinical phage microbiology: Simulating phage inhalation for respiratory tract infections. ERJ Open Res. 2021, 7, 00367–02021. [Google Scholar] [CrossRef]
- Lynch, K.H.; Stothard, P.; Dennis, J.J. Comparative analysis of two phenotypically-similar but genomically-distinct Burkholderia cenocepacia-specific bacteriophages. BMC Genom. 2012, 13, 223. [Google Scholar] [CrossRef]
- Rezene, S.; Yao, G.; Le, T.; Burrowes, B.; Gonzalez, C.; Liu, M.; Gill, J. Complete genome sequence of Burkholderia cenocepacia phage Paku. Microbiol. Resour. Announc. 2022, 11, e01220-21. [Google Scholar] [CrossRef]
- Yu, Z.; Yao, G.; Vizoso-Pinto, M.G.; Sun, L.; Young, R.; Gonzalez, C.; Liu, M. Complete genome sequence of Burkholderia gladioli Phage Maja. Microbiol. Resour. Announc. 2021, 10, 10–1128. [Google Scholar] [CrossRef]
- Summer, E.J.; Gill, J.J.; Upton, C.; Gonzalez, C.F.; Young, R. Role of phages in the pathogenesis of Burkholderia, or ‘Where are the toxin genes in Burkholderia phages?’. Curr. Opin. Microbiol. 2007, 10, 410–417. [Google Scholar] [CrossRef]
- Wang, T.; Cheng, B.; Jiao, R.; Zhang, X.; Zhang, D.; Cheng, X.; Ling, N.; Ye, Y. Characterization of a novel high-efficiency cracking Burkholderia gladiolus phage vB_BglM_WTB and its application in black fungus. Int. J. Food Microbiol. 2024, 414, 110615. [Google Scholar] [CrossRef] [PubMed]
- Ahern, S.J.; Das, M.; Bhowmick, T.S.; Young, R.; Gonzalez, C.F. Characterization of novel virulent broad-host-range phages of Xylella fastidiosa and Xanthomonas. J. Bacteriol. 2014, 196, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Lauman, P.; Dennis, J.J. Prophylactic phage biocontrol prevents Burkholderia gladioli infection in a quantitative ex planta model of bacterial virulence. Appl. Environ. Microbiol. 2024, 90, e01317-24. [Google Scholar] [CrossRef]
- Goudie, A.D.; Lynch, K.H.; Seed, K.D.; Stothard, P.; Shrivastava, S.; Wishart, D.S.; Dennis, J.J. Genomic sequence and activity of KS10, a transposable phage of the Burkholderia cepacia complex. BMC Genom. 2008, 9, 615. [Google Scholar] [CrossRef] [PubMed]
- Kvitko, B.H.; Cox, C.R.; DeShazer, D.; Johnson, S.L.; Voorhees, K.J.; Schweizer, H.P. φX216, a P2-like bacteriophage with broad Burkholderia pseudomallei and B. mallei strain infectivity. BMC Microbiol. 2012, 12, 289. [Google Scholar] [CrossRef] [PubMed]
- Muangsombut, V.; Withatanung, P.; Chantratita, N.; Chareonsudjai, S.; Lim, J.; Galyov, E.E.; Ottiwet, O.; Sengyee, S.; Janesomboon, S.; Loessner, M.J.; et al. Rapid clinical screening of Burkholderia pseudomallei colonies by a bacteriophage tail fiber-based latex agglutination assay. Appl. Environ. Microbiol. 2021, 87, e03019-20. [Google Scholar] [CrossRef]
- Khrongsee, P.; Kaewrakmuk, J.; Alami-Rose, M.; Subramaniam, K.; Waltzek, T.B.; Schweizer, H.P.; Tuanyok, A. Exploring Burkholderia pseudomallei-specific bacteriophages: Overcoming O-antigen specificity and adaptive mutation in phage tail fiber. Front. Bacteriol. 2024, 3, 1433593. [Google Scholar] [CrossRef]
- Sasaki, R.; Miyashita, S.; Ando, S.; Ito, K.; Fukuhara, T.; Kormelink, R.; Takahashi, H. Complete genomic sequence of a novel phytopathogenic Burkholderia phage isolated from fallen leaf compost. Arch. Virol. 2021, 166, 313–316. [Google Scholar] [CrossRef]
- Godoy, B.; Yao, G.; Le, T.; Vizoso-Pinto, M.G.; Gill, J.; Gonzalez, C.; Liu, M. Complete genome sequence of Burkholderia gladioli Myophage Mana. Microbiol. Resour. Announc. 2021, 10, e00402-21. [Google Scholar] [CrossRef]
- Yordpratum, U.; Tattawasart, U.; Wongratanacheewin, S.; Sermswan, R.W. Novel lytic bacteriophages from soil that lyse Burkholderia pseudomallei. FEMS Microbiol. Lett. 2011, 314, 81–88. [Google Scholar] [CrossRef]
- Lynch, K.H.; Stothard, P.; Dennis, J.J. Characterization of DC1, a broad-host-range Bcep22-like podovirus. Appl. Environ. Microbiol. 2012, 78, 889–891. [Google Scholar] [CrossRef]
- Garcia, J.; Yao, G.; Vizoso-Pinto, M.G.; Clark, J.; Le, T.; Gonzalez, C.; Gill, J.; Liu, M. Complete genome sequence of Burkholderia cenocepacia phage mica. Microbiol. Resour. Announc. 2021, 10, e01407-20. [Google Scholar] [CrossRef]
- Summer, E.J.; Gonzalez, C.F.; Bomer, M.; Carlile, T.; Embry, A.; Kucherka, A.M.; Lee, J.; Mebane, L.; Morrison, W.C.; Mark, L.; et al. Divergence and mosaicism among virulent soil phages of the Burkholderia cepacia complex. J. Bacteriol. 2006, 188, 255–268. [Google Scholar] [CrossRef]
- Hammerl, J.A.; Volkmar, S.; Jacob, D.; Klein, I.; Jäckel, C.; Hertwig, S. The Burkholderia thailandensis phages ΦE058 and ΦE067 represent distinct prototypes of a new subgroup of temperate Burkholderia myoviruses. Front. Microbiol. 2020, 11, 1120. [Google Scholar] [CrossRef]
- Gafford-Gaby, D.; Yao, G.; Le, T.; Clark, J.; Gonzalez, C.; Gill, J.; Liu, M. Complete genome sequence of Burkholderia cenocepacia phage Magia. Microbiol. Resour. Announc. 2021, 10, e01473-20. [Google Scholar] [CrossRef]
- Mera, L. Bcep176 and Bglu421-Two Novel Phages Contributing to the Understanding of Pathogenicity and Diversity in Burkholderiacae. Bachelor’s Thesis, Texas A&M University, College Station, TX, USA, 2006. [Google Scholar]
- Lynch, K.H.; Seed, K.D.; Stothard, P.; Dennis, J.J. Inactivation of Burkholderia cepacia complex phage KS9 gp41 identifies the phage repressor and generates lytic virions. J. Virol. 2010, 84, 1276–1288. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. VICTOR: Genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics 2017, 33, 3396–3404. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Desper, R.; Gascuel, O. FastME, 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [PubMed]
- Göker, M.; García-Blázquez, G.; Voglmayr, H.; Tellería, M.T.; Martín, M.P. Molecular taxonomy of phytopathogenic fungi: A case study in Peronospora. PLoS ONE 2009, 4, e6319. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A.; et al. Complete genome sequence of DSM 30083 (T), the type strain (U5/41(T)) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genom. Sci. 2014, 9, 2. [Google Scholar] [CrossRef]
- Walmagh, M.; Boczkowska, B.; Grymonprez, B.; Briers, Y.; Drulis-Kawa, Z.; Lavigne, R. Characterization of five novel endolysins from Gram-negative infecting bacteriophages. Appl. Microbiol. Biotechnol. 2013, 97, 4369–4375. [Google Scholar] [CrossRef]
- Comeau, A.M.; Tétart, F.; Trojet, S.N.; Prère, M.-F.; Krisch, H. Phage-antibiotic synergy (PAS): β-lactam and quinolone antibiotics stimulate virulent phage growth. PLoS ONE 2007, 2, e799. [Google Scholar] [CrossRef]
- Ruest, M.K.; Supina, B.S.; Dennis, J.J. Bacteriophage steering of Burkholderia cenocepacia toward reduced virulence and increased antibiotic sensitivity. J. Bacteriol. 2023, 205, e00196-23. [Google Scholar] [CrossRef] [PubMed]
- Kraus, S.; Fletcher, M.L.; Łapińska, U.; Chawla, K.; Baker, E.; Attrill, E.L.; O’Neill, P.; Farbos, A.; Jeffries, A.; Galyov, E.E.; et al. Phage-induced efflux down-regulation boosts antibiotic efficacy. PLoS Pathog. 2024, 20, e1012361. [Google Scholar] [CrossRef]
- Egido, J.E.; Costa, A.R.; Aparicio-Maldonado, C.; Haas, P.-J.; Brouns, S.J. Mechanisms and clinical importance of bacteriophage resistance. FEMS Microbiol. Rev. 2022, 46, fuab048. [Google Scholar] [CrossRef]
- Dicks, L.M.; Vermeulen, W. Bacteriophage–Host interactions and the therapeutic potential of bacteriophages. Viruses 2024, 16, 478. [Google Scholar] [CrossRef]
- Kamal, F.; Dennis, J.J. Burkholderia cepacia complex phage-antibiotic synergy (PAS): Antibiotics stimulate lytic phage activity. Appl. Environ. Microbiol. 2015, 81, 1132–1138. [Google Scholar] [CrossRef]
- Canning, J.S.; Laucirica, D.R.; Ling, K.-M.; Nicol, M.P.; Stick, S.M.; Kicic, A. Phage therapy to treat cystic fibrosis Burkholderia cepacia complex lung infections: Perspectives and challenges. Front. Microbiol. 2024, 15, 1476041. [Google Scholar] [CrossRef] [PubMed]
- Colom, J.; Cano-Sarabia, M.; Otero, J.; Aríñez-Soriano, J.; Cortés, P.; Maspoch, D.; Llagostera, M. Microencapsulation with alginate/CaCO3: A strategy for improved phage therapy. Sci. Rep. 2017, 7, 41441. [Google Scholar] [CrossRef] [PubMed]
- Jain, L.; Kumar, V.; Jain, S.K.; Kaushal, P.; Ghosh, P.K. Isolation of bacteriophages infecting Xanthomonas oryzae pv. oryzae and genomic characterization of novel phage vB_XooS_NR08 for biocontrol of bacterial leaf blight of rice. Front. Microbiol. 2023, 14, 1084025. [Google Scholar]
- Vila, M.M.; Balcão, L.M.; Balcão, V.M. Phage delivery strategies for biocontrolling human, animal, and plant bacterial infections: State of the art. Pharmaceutics 2024, 16, 374. [Google Scholar] [CrossRef]
- Erdrich, S.H.; Schurr, U.; Frunzke, J.; Arsova, B. Seed coating with phages for sustainable plant biocontrol of plant pathogens and influence of the seed coat mucilage. Microb. Biotechnol. 2024, 17, e14507. [Google Scholar] [CrossRef]
- Li, S.; Zhang, H.; Chen, K.; Jin, M.; Vu, S.H.; Jung, S.; He, N.; Zheng, Z.; Lee, M.-S. Application of chitosan/alginate nanoparticle in oral drug delivery systems: Prospects and challenges. Drug Deliv. 2022, 29, 1142–1149. [Google Scholar] [CrossRef] [PubMed]
- Yadollahi, Z.; Motiei, M.; Kazantseva, N.; Císař, J.; Sáha, P. Whey protein isolate-chitosan PolyElectrolyte nanoparticles as a drug delivery system. Molecules 2023, 28, 1724. [Google Scholar] [CrossRef] [PubMed]
- Cinquerrui, S.; Mancuso, F.; Vladisavljević, G.T.; Bakker, S.E.; Malik, D.J. Nanoencapsulation of bacteriophages in liposomes prepared using microfluidic hydrodynamic flow focusing. Front. Microbiol. 2018, 9, 2172. [Google Scholar] [CrossRef] [PubMed]
- Sangwan, A.; Singh, N. Advanced nanostrategies for biomolecule delivery in plant disease management. J. Agric. Food Chem. 2024, 73, 66–84. [Google Scholar] [CrossRef]
- Choińska-Pulit, A.; Mituła, P.; Śliwka, P.; Łaba, W.; Skaradzińska, A. Bacteriophage encapsulation: Trends and potential applications. Trends Food Sci. Technol. 2015, 45, 212–221. [Google Scholar] [CrossRef]
- Wang, X.; Xie, Z.; Zhao, J.; Zhu, Z.; Yang, C.; Liu, Y. Prospects of inhaled phage therapy for combatting pulmonary infections. Front. Cell. Infect. Microbiol. 2021, 11, 758392. [Google Scholar] [CrossRef]
- Luo, M.; Ma, L.; Guo, Y.; Zhu, C.; Chen, J.; Zhang, B.; Zhu, J.; Jellicoe, M.; He, S.; Zou, Y.; et al. Preparation and characterization of microcapsules and tablets for probiotic encapsulation via whey protein isolate-nanochitin complex coacervation. Int. J. Biol. Macromol. 2025, 285, 138225. [Google Scholar] [CrossRef]
- Mohammadi, M.; Mirabzadeh, S.; Shahvalizadeh, R.; Hamishehkar, H. Development of novel active packaging films based on whey protein isolate incorporated with chitosan nanofiber and nano-formulated cinnamon oil. Int. J. Biol. Macromol. 2020, 149, 11–20. [Google Scholar] [CrossRef]
- Vonasek, E.; Le, P.; Nitin, N. Encapsulation of bacteriophages in whey protein films for extended storage and release. Food Hydrocoll. 2014, 37, 7–13. [Google Scholar] [CrossRef]
- Lu, Y.; Cheng, D.; Niu, B.; Wang, X.; Wu, X.; Wang, A. Properties of poly (lactic-co-glycolic acid) and progress of poly (lactic-co-glycolic acid)-based biodegradable materials in biomedical research. Pharmaceuticals 2023, 16, 454. [Google Scholar] [CrossRef]
- Hines, D.J.; Kaplan, D.L. Poly (lactic-co-glycolic) acid-controlled-release systems: Experimental and modeling insights. Crit. Rev. Ther. Drug Carr. Syst. 2013, 30, 257–276. [Google Scholar] [CrossRef]
- Kim, S.-G.; Giri, S.S.; Jo, S.-J.; Kang, J.-W.; Lee, S.-B.; Jung, W.-J.; Lee, Y.-M.; Kim, H.-J.; Kim, J.-H.; Park, S.-C. Prolongation of fate of bacteriophages in vivo by polylactic-co-glycolic-acid/alginate-composite encapsulation. Antibiotics 2022, 11, 1264. [Google Scholar] [CrossRef]
- Golshahi, L.; Lynch, K.; Dennis, J.; Finlay, W. In vitro lung delivery of bacteriophages KS4-M and ΦKZ using dry powder inhalers for treatment of Burkholderia cepacia complex and Pseudomonas aeruginosa infections in cystic fibrosis. J. Appl. Microbiol. 2011, 110, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, T.; Luo, J.; Noman, M.; Ijaz, M.; Wang, X.; Masood, H.A.; Manzoor, N.; Wang, Y.; Li, B. Microbe-mediated nanoparticle intervention for the management of plant diseases. Crop Health 2023, 1, 3. [Google Scholar] [CrossRef]
- Noman, M.; Ahmed, T.; Wang, J.; Ijaz, M.; Shahid, M.; Islam, M.S.; Azizullah; Manzoor, I.; Li, D.; Song, F. Nano-enabled crop resilience against pathogens: Potential, mechanisms and strategies. Crop Health 2023, 1, 15. [Google Scholar] [CrossRef]
- Thomassen, M.; Demko, C.; Klinger, J.; Stern, R. Pseudomonas cepacia colonization among patients with cystic fibrosis: A new opportunist. Am. Rev. Respir. Dis. 1985, 131, 791–796. [Google Scholar]
- Zhang, Y.; Liu, F.; Wang, B.; Qiu, D.; Liu, J.; Wu, H.; Cheng, C.; Bei, X.; Lü, P. First report of Burkholderia cepacia causing finger-tip rot on banana fruit in the Guangxi province of China. Plant Dis. 2022, 106, 1979. [Google Scholar] [CrossRef]
- Cain, C.L.; Cole, S.D.; Bradley II, C.W.; Canfield, M.S.; Mauldin, E.A. Clinical and histopathological features of Burkholderia cepacia complex dermatitis in dogs: A series of four cases. Vet. Dermatol. 2018, 29, 457-e156. [Google Scholar] [CrossRef]
- Epithelia, W.-D.H.A. Role of Actin Filament Network in Burkholderia multivorans Invasion in Well-Differentiated Human Airway Epithelia. Infect. Immun. 2003, 71, 6607–6609. [Google Scholar] [CrossRef]
- Peralta, D.P.; Chang, A.Y.; Ariza-Hutchinson, A.; Ho, C.A. Burkholderia multivorans: A rare yet emerging cause of bacterial meningitis. IDCases 2018, 11, 61–63. [Google Scholar] [CrossRef]
- Kar, M.; Dubey, A.; Sahu, C.; Patel, S.S. Burkholderia vietnamiensis causing bacteremia in patients suffering from B-Cell acute lymphocytic leukemia: A case series and review of literature. J. Lab. Physicians 2023, 16, 134–139. [Google Scholar] [CrossRef]
- Flores-Vega, V.R.; Lara-Zavala, B.A.; Jarillo-Quijada, M.D.; Fernández-Vázquez, J.L.; Alcántar-Curiel, M.D.; Vargas-Roldán, S.Y.; Ares, M.A.; de la Cruz, M.A.; Morfín-Otero, R.; Rodríguez-Noriega, E. Burkholderia vietnamiensis causing infections in noncystic fibrosis patients in a tertiary care hospital in Mexico. Diagn. Microbiol. Infect. Dis. 2023, 105, 115866. [Google Scholar] [CrossRef]
- Kalish, L.A.; Waltz, D.A.; Dovey, M.; Potter-Bynoe, G.; McAdam, A.J.; LiPuma, J.J.; Gerard, C.; Goldmann, D. Impact of Burkholderia dolosa on lung function and survival in cystic fibrosis. Am. J. Respir. Crit. Care Med. 2006, 173, 421–425. [Google Scholar] [CrossRef]
- Vial, L.; Groleau, M.-C.; Lamarche, M.G.; Filion, G.; Castonguay-Vanier, J.; Dekimpe, V.; Daigle, F.; Charette, S.J.; Déziel, E. Phase variation has a role in Burkholderia ambifaria niche adaptation. ISME J. 2010, 4, 49–60. [Google Scholar] [CrossRef]
- Alshiekheid, M.A.; Dou, A.M.; Algahtani, M.; Al-Megrin, W.A.I.; Alhawday, Y.A.; Alradhi, A.E.; Bukhari, K.; Alharbi, B.F.; Algefary, A.N.; Alhunayhani, B.A. Bioinformatics and immunoinformatics assisted multiepitope vaccine construct against Burkholderia anthina. Saudi Pharm. J. 2024, 32, 101917. [Google Scholar] [CrossRef]
- Manno, G.; Dalmastri, C.; Tabacchioni, S.; Vandamme, P.; Lorini, R.; Minicucci, L.; Romano, L.; Giannattasio, A.; Chiarini, L.; Bevivino, A. Epidemiology and clinical course of Burkholderia cepacia complex infections, particularly those caused by different Burkholderia cenocepacia strains, among patients attending an Italian cystic fibrosis center. J. Clin. Microbiol. 2004, 42, 1491–1497. [Google Scholar] [CrossRef]
- Savi, D.; De Biase, R.V.; Amaddeo, A.; Anile, M.; Venuta, F.; Ruberto, F.; Simmonds, N.; Cimino, G.; Quattrucci, S. Burkholderia pyrrocinia in cystic fibrosis lung transplantation: A case report. Transplant. Proc. 2014, 46, 295–297. [Google Scholar] [CrossRef] [PubMed]
- Martina, P.; Bettiol, M.; Vescina, C.; Montanaro, P.; Mannino, M.C.; Prieto, C.I.; Vay, C.; Naumann, D.; Schmitt, J.; Yantorno, O. Genetic diversity of Burkholderia contaminans isolates from cystic fibrosis patients in Argentina. J. Clin. Microbiol. 2013, 51, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, C.P.; Barreto, C.; Pereira, L.; Lito, L.; Melo Cristino, J.; Sa-Correia, I. Incidence of Burkholderia contaminans at a cystic fibrosis centre with an unusually high representation of Burkholderia cepacia during 15 years of epidemiological surveillance. J. Med. Microbiol. 2015, 64, 927–935. [Google Scholar] [CrossRef]
- Leong, L.E.; Lagana, D.; Carter, G.P.; Wang, Q.; Smith, K.; Stinear, T.P.; Shaw, D.; Sintchenko, V.; Wesselingh, S.L.; Bastian, I. Burkholderia lata infections from intrinsically contaminated chlorhexidine mouthwash, Australia, 2016. Emerging Infect. Dis. 2018, 24, 2109. [Google Scholar] [CrossRef] [PubMed]
- Dance, D.A. Ecology of Burkholderia pseudomallei and the interactions between environmental Burkholderia spp. and human–animal hosts. Acta Trop. 2000, 74, 159–168. [Google Scholar] [CrossRef]
- Lazar Adler, N.R.; Govan, B.; Cullinane, M.; Harper, M.; Adler, B.; Boyce, J.D. The molecular and cellular basis of pathogenesis in melioidosis: How does Burkholderia pseudomallei cause disease? FEMS Microbiol. Rev. 2009, 33, 1079–1099. [Google Scholar] [CrossRef] [PubMed]
- Glass, M.B.; Steigerwalt, A.G.; Jordan, J.G.; Wilkins, P.P.; Gee, J.E. Burkholderia oklahomensis sp. nov. a Burkholderia pseudomallei-like species formerly known as the Oklahoma strain of Pseudomonas pseudomallei. Int. J. Syst. Evol. Microbiol. 2006, 56, 2171–2176. [Google Scholar] [CrossRef] [PubMed]
- Vandamme, P.; Peeters, C.; De Smet, B.; Price, E.P.; Sarovich, D.S.; Henry, D.A.; Hird, T.J.; Zlosnik, J.E.; Mayo, M.; Warner, J. Comparative genomics of Burkholderia singularis sp. nov. a low G+ C content, free-living bacterium that defies taxonomic dissection of the genus Burkholderia. Front. Microbiol. 2017, 8, 1679. [Google Scholar] [CrossRef] [PubMed]
- Azegami, K.; Nishiyama, K.; Kato, H. Effect of iron limitation on “Pseudomonas plantarii” growth and tropolone and protein production. Appl. Environ. Microbiol. 1988, 54, 844–847. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).