Isolation, Identification, and Antibiotic Resistance, CRISPR System Analysis of Escherichia coli from Forest Musk Deer in Western China
Abstract
1. Introduction
2. Materials and Methods
2.1. Test Strains
2.2. DNA Extraction and 16S rRNA Gene Analysis
2.3. Antibiotic Susceptibility Testing
2.4. Antibiotic Resistance Gene Detection
2.5. CRISPR Sequence Analysis
2.6. Data Processing and Analysis
3. Results
3.1. Antibiotic Resistance Phenotype Analysis
3.2. Distribution of Resistance Genes in E. coli
3.3. Relationship Between ARGs and AMRs
3.4. Characteristics Analysis of CRISPR System in Isolated Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MDR | multi-drug resistance |
| CRISPR-Cas | Clustered Regularly Interspaced Short Palindromic Repeats-associated protein |
| WHO | World Health Organization |
| ARGs | Antibiotic resistance genes |
| DR | direct repeats |
| IUCN | International Union for Conservation of Nature |
| CLSI | Clinical and Laboratory Standards Institute |
| r | Pearson correlation coefficient |
| WGS | whole-genome sequencing |
References
- Mendelson, M.; Laxminarayan, R.; Limmathurotsakul, D.; Kariuki, S.; Gyansa-Lutterodt, M.; Charani, E.; Singh, S.; Walia, K.; Gales, A.C.; Mpundu, M. Antimicrobial resistance and the great divide: Inequity in priorities and agendas between the Global North and the Global South threatens global mitigation of antimicrobial resistance. Lancet Glob. Health 2024, 12, e516–e521. [Google Scholar] [CrossRef]
- Hernando-Amado, S.; Coque, T.M.; Baquero, F.; Martínez, J.L. Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nat. Microbiol. 2019, 4, 1432–1442. [Google Scholar] [CrossRef]
- Smit, C.C.H.; Lambert, M.; Rogers, K.; Djordjevic, S.P.; Van Oijen, A.M.; Keighley, C.; Taxis, K.; Robertson, H.; Pont, L.G. One Health Determinants of Escherichia coli Antimicrobial Resistance in Humans in the Community: An Umbrella Review. Int. J. Mol. Sci. 2023, 24, 17204. [Google Scholar] [CrossRef]
- Li, B.; Yang, Y.; Ma, L.; Ju, F.; Guo, F.; Tiedje, J.M.; Zhang, T. Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J. 2015, 9, 2490–2502. [Google Scholar] [CrossRef]
- Li, F.; Li, M.; Nie, L.; Zuo, J.; Fan, W.; Lian, L.; Hu, J.; Chen, S.; Jiang, W.; Han, X.; et al. Molecular Epidemiology and Antibiotic Resistance Associated with Avian Pathogenic Escherichia coli in Shanxi Province, China, from 2021 to 2023. Microorganisms 2025, 13, 541. [Google Scholar] [CrossRef]
- Blake, K.S.; Xue, Y.P.; Gillespie, V.J.; Fishbein, S.R.; Tolia, N.H.; Wencewicz, T.A.; Dantas, G. The tetracycline resistome is shaped by selection for specific resistance mechanisms by each antibiotic generation. Nat. Commun. 2025, 16, 1452. [Google Scholar] [CrossRef]
- Sommer, M.O.A.; Munck, C.; Toft-Kehler, R.V.; Andersson, D.I. Prediction of antibiotic resistance: Time for a new preclinical paradigm? Nat. Rev. Microbiol. 2017, 15, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Madec, J.Y.; Lupo, A.; Schink, A.K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial Resistance in Escherichia coli. Microbiol. Spectr. 2018, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Yuan, Q.; Mathieu, J.; Stadler, L.; Senehi, N.; Sun, R.; Alvarez, P.J. Antibiotic resistance genes from livestock waste: Occurrence, dissemination, and treatment. NPJ Clean Water 2020, 3, 4. [Google Scholar] [CrossRef]
- Dong, H.; Cui, Y.; Zhang, D. CRISPR/Cas Technologies and Their Applications in Escherichia coli. Front. Bioeng. Biotechnol. 2021, 9, 762676. [Google Scholar] [CrossRef]
- Tong, Y.; Jørgensen, T.S.; Whitford, C.M.; Webe, T.; Lee, S.Y. A versatile genetic engineering toolkit for E. coli based on CRISPR-prime editing. Nat. Commun. 2021, 12, 5206. [Google Scholar] [CrossRef]
- Pandova, M.; Kizheva, Y.; Hristova, P. Relationship Between CRISPR-Cas Systems and Acquisition of Tetracycline Resistance in Non-Clinical Enterococcus Populations in Bulgaria. Antibiotics 2025, 14, 145. [Google Scholar] [CrossRef] [PubMed]
- Palmer, K.L.; Gilmore, M.S. Multidrug-resistant enterococci lack CRISPR-cas. mBio 2010, 1, e00227-10. [Google Scholar] [CrossRef] [PubMed]
- Gholizadeh, P.; Aghazadeh, M.; Ghotaslou, R.; Rezaee, M.A.; Pirzadeh, T.; Cui, L.; Watanabe, S.; Feizi, H.; Kadkhoda, H.; Kafil, H.S. Role of CRISPR-Cas system on antibiotic resistance patterns of Enterococcus faecalis. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 49. [Google Scholar] [CrossRef] [PubMed]
- Jie, H.; Li, F.; Liu, Q.; Zheng, T.; Tan, H.; Feng, X.; Zhao, G.; Zeng, D.; Li, D.; Xu, Z.; et al. Elucidating metabolites and biosynthetic pathways during musk maturation: Insights from forest musk deer. Front. Pharmacol. 2025, 16, 1503138. [Google Scholar] [CrossRef]
- Gong, R.; Song, S.; Ai, Y.; Wang, S.; Dong, X.; Ren, Z.; Xie, H.; Jiang, B.; Zhao, L. Exploring the growing forest musk deer (Moschus berezovskii) dietary protein requirement based on gut microbiome. Front. Microbiol. 2023, 14, 1124163. [Google Scholar] [CrossRef]
- Wu, X.; Yang, W.; Cheng, J.; Luo, Y.; Fu, W.; Zhou, L.; Wu, J.; Wang, Y.; Zhong, Z.; Yang, Z.; et al. Molecular cloning, prokaryotic expression and its application potential evaluation of interferon (IFN)-ω of forest musk deer. Sci. Rep. 2023, 13, 10625. [Google Scholar] [CrossRef]
- Church, D.L.; Cerutti, L.; Gürtler, A.; Griener, T.; Zelazny, A.; Emler, S. Performance and Application of 16S rRNA Gene Cycle Sequencing for Routine Identification of Bacteria in the Clinical Microbiology Laboratory. Clin. Microbiol. Rev. 2020, 33, 19. [Google Scholar] [CrossRef]
- Salam, M.A.; Al-Amin, M.Y.; Pawar, J.S.; Akhter, N.; Lucy, I.B. Conventional methods and future trends in antimicrobial susceptibility testing. Saudi J. Biol. Sci. 2023, 30, 103582. [Google Scholar] [CrossRef]
- Humphries, R.; Bobenchik, A.M.; Hindler, J.A.; Schuetz, A.N. Overview of Changes to the Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing, M100, 31st Edition. J. Clin. Microbiol. 2021, 59, e0021321. [Google Scholar] [CrossRef]
- Liu, H.; Pan, S.; Cheng, Y.; Luo, L.; Zhou, L.; Fan, S.; Wang, L.; Jiang, S.; Zhou, Z.; Liu, H.; et al. Distribution and associations for antimicrobial resistance and antibiotic resistance genes of Escherichia coli from musk deer (Moschus berezovskii) in Sichuan, China. PLoS ONE 2023, 18, e0289028. [Google Scholar] [CrossRef]
- Zhu, Z.; Jiang, S.; Qi, M.; Liu, H.; Zhang, S.; Liu, H.; Zhou, Z.; Wang, L.; Wang, C.; Luo, Y.; et al. Prevalence and characterization of antibiotic resistance genes and integrons in Escherichia coli isolates from captive non-human primates of 13 zoos in China. Sci. Total Environ. 2021, 798, 149268. [Google Scholar] [CrossRef] [PubMed]
- Schober, P.; Boer, C.; Schwarte, L.A. Correlation Coefficients: Appropriate Use and Interpretation. Anesth. Analg. 2018, 126, 1763–1768. [Google Scholar] [CrossRef] [PubMed]
- Kunin, V.; Sorek, R.; Hugenholtz, P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007, 8, R61. [Google Scholar] [CrossRef] [PubMed]
- Nkansa-Gyamfi, N.A.; Kazibwe, J.; Traore, D.A.; Nji, E. Prevalence of multidrug-, extensive drug-, and pandrug-resistant commensal Escherichia coli isolated from healthy humans in community settings in low- and middle-income countries: A systematic review and meta-analysis. Glob. Health Action 2019, 12 (Suppl. S1), 1815272. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Rozwadowski, M.; Gawel, D. Molecular Factors and Mechanisms Driving Multidrug Resistance in Uropathogenic Escherichia coli-An Update. Genes 2022, 13, 1397. [Google Scholar] [CrossRef]
- He, T.; Wang, R.; Liu, D.; Walsh, T.R.; Zhang, R.; Lv, Y.; Ke, Y.; Ji, Q.; Wei, R.; Liu, Z.; et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 2019, 4, 1450–1456. [Google Scholar] [CrossRef]
- Ćwiek, K.; Woźniak-Biel, A.; Karwańska, M.; Siedlecka, M.; Lammens, C.; Rebelo, A.R.; Hendriksen, R.S.; Kuczkowski, M.; Chmielewska-Władyka, M.; Wieliczko, A. Phenotypic and genotypic characterization of mcr-1-positive multidrug-resistant Escherichia coli ST93, ST117, ST156, ST10, and ST744 isolated from poultry in Poland. Braz. J. Microbiol. 2021, 52, 1597–1609. [Google Scholar] [CrossRef]
- El Maghraby, H.M.; El-Sayed, H.A.; Hussein, S.; El Azawy, D.S.; Attia, O.; Orabi, E.E.; Fahmy, Y.A. Detection of phylogrouping, adhesin, and extended spectrum β-lactamases genes in hospital acquired uropathogenic Escherichia coli isolates. Mol. Biol. Rep. 2024, 51, 143. [Google Scholar] [CrossRef]
- Peng, S.; Zheng, H.; Herrero-Fresno, A.; Olsen, J.E.; Dalsgaard, A.; Ding, Z. Co-occurrence of antimicrobial and metal resistance genes in pig feces and agricultural fields fertilized with slurry. Sci. Total Environ. 2021, 792, 148259. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, Y.; Mulchandani, R.; Van Boeckel, T.P. Global surveillance of antimicrobial resistance in food animals using priority drugs maps. Nat. Commun. 2024, 15, 763. [Google Scholar] [CrossRef] [PubMed]
- Thaker, M.; Spanogiannopoulos, P.; Wright, G.D. The tetracycline resistome. Cell. Mol. Life Sci. 2010, 67, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Roberts, K.D.; Zhu, Y.; Azad, M.A.K.; Han, M.L.; Wang, J.; Wang, L.; Yu, H.H.; Horne, A.S.; Pinson, J.-A.; Rudd, D.; et al. A synthetic lipopeptide targeting top-priority multidrug-resistant Gram-negative pathogens. Nat. Commun. 2022, 13, 1625. [Google Scholar] [CrossRef] [PubMed]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef]
- Pfeifer, N.M.; Weber, M.; Wiegand, E.; Barth, S.A.; Berens, C.; Menge, C. Escherichia coli resistant to the highest priority critically important fluoroquinolone or 3rd and 4th generation cephalosporin antibiotics persist in pigsties. Appl. Environ. Microbiol. 2025, 91, e0138624. [Google Scholar] [CrossRef]
- El Badawy, A.; Hamza, D.; Ahmed, Z.; Sabry, M.A. Seasonal and environmental drivers of antibiotic resistance and virulence in Escherichia coli from aquaculture and their public health implications. Sci. Rep. 2025, 15, 16100. [Google Scholar] [CrossRef]
- Tyrrell, C.; Burgess, C.M.; Brennan, F.P.; Muenzenmaier, D.; Drissner, D.; Leigh, R.J.; Walsh, F. Genomic analysis of antimicrobial resistant Escherichia coli isolated from manure and manured agricultural grasslands. npj Antimicrob. Resist. 2025, 3, 8. [Google Scholar] [CrossRef]
- Zhou, J.; Zhou, J.; Chen, M.; Lü, P.; Jiang, C. Prevalence of β-lactam antibiotic resistance of Escherichia coli isolated from a neonatal intensive care unit. BMC Pediatr. 2025, 25, 95. [Google Scholar] [CrossRef]
- Sun, J.; Chen, C.; Cui, C.Y.; Zhang, Y.; Liu, X.; Cui, Z.-H.; Ma, X.Y.; Feng, Y.; Fang, L.X.; Lian, X.-L. Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat. Microbiol. 2019, 4, 1457–1464. [Google Scholar] [CrossRef]
- Brodersen, D.E.; Clemons, W.M.; Carter, A.P.; Morgan-Warren, R.J.; Wimberly, B.T.; Ramakrishnan, V. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 2000, 103, 1143–1154. [Google Scholar] [CrossRef]
- Sun, C.; Cui, M.; Zhang, S.; Wang, H.; Song, L.; Zhang, C.; Zhao, Q.; Liu, D.; Wang, Y.; Shen, J.; et al. Plasmid-mediated tigecycline-resistant gene tet(X4) in Escherichia coli from food-producing animals, China, 2008–2018. Emerg. Microbes Infect. 2019, 8, 1524–1527. [Google Scholar] [CrossRef]
- Fan, X.Y.; Jiang, Y.; Wu, H.; Liu, J.; Gu, Q.Y.; Wang, Z.Y.; Sun, L.; Jiao, X.; Li, Q.; Wang, J. Distribution and spread of tigecycline resistance gene tet(X4) in Escherichia coli from different sources. Front. Cell. Infect. Microbiol. 2024, 14, 1399732. [Google Scholar] [CrossRef]
- Cheng, Y.; Li, Y.; Yang, M.; He, Y.; Shi, X.; Zhang, Z.; Zhong, Y.; Zhang, Y.; Si, H. Emergence of novel tigecycline resistance gene tet(X5) variant in multidrug-resistant Acinetobacter indicus of swine farming environments. Vet. Microbiol. 2023, 284, 109837. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Cai, W.; Jiang, L.; Wang, Z.; Liu, Y. Large-Scale Analysis of Fitness Cost of tet(X4)-Positive Plasmids in Escherichia coli. Front. Cell. Infect. Microbiol. 2022, 12, 798802. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lu, M.J.; Wang, Z.Y.; Jiang, Y.; Wu, H.; Pan, Z.M.; Jiao, X. Tigecycline-resistant Escherichia coli ST761 carrying tet(X4) in a pig farm, China. Front. Microbiol. 2022, 13, 967313. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Zhou, M.; Cong, Z.; Xiao, X.; Liu, L.; Chen, S.; Jiang, W.; Wu, Y.; Liu, R. A host defense peptide-mimicking prodrug activated by drug-resistant Gram-negative bacterial infections. Sci. Transl. Med. 2025, 17, eadl4870. [Google Scholar] [CrossRef]
- Liu, X.; Wu, Y.; Mao, C.; Shen, J.; Zhu, K. Host-acting antibacterial compounds combat cytosolic bacteria. Trends Microbiol. 2022, 30, 761–777. [Google Scholar] [CrossRef]
- Stacy, A.; Andrade-Oliveira, V.; McCulloch, J.A.; Hild, B.; Oh, J.H.; Perez-Chaparro, P.J.; Sim, C.K.; Lim, A.I.; Link, V.M.; Enamorado, M.; et al. Infection trains the host for microbiota-enhanced resistance to pathogens. Cell 2021, 184, 615–627. [Google Scholar] [CrossRef]
- Stine, W.; Akiyama, T.; Weiss, D.; Kim, M. Lineage-dependent variations in single-cell antibiotic susceptibility reveal the selective inheritance of phenotypic resistance in bacteria. Nat. Commun. 2025, 16, 4655. [Google Scholar] [CrossRef]
- Deekshit, V.K.; Srikumar, S. ‘To be, or not to be’-The dilemma of ‘silent’ antimicrobial resistance genes in bacteria. J. Appl. Microbiol. 2022, 133, 2902–2914. [Google Scholar] [CrossRef]
- Hughes, D.; Andersson, D.I. Environmental and genetic modulation of the phenotypic expression of antibiotic resistance. FEMS Microbiol. Rev. 2017, 41, 374–391. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Snijders, A.P.; Chakravorty, R.; Ahmed, M.; Tarek, A.M.; Hossain, M.A. Comparative network clustering of direct repeats (DRs) and cas genes confirms the possibility of the horizontal transfer of CRISPR locus among bacteria. Mol. Phylogenet. Evol. 2010, 56, 878–887. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.M.; Wolf, Y.I.; Yakunin, A.F. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Toro, M.; Cao, G.; Ju, W.; Allard, M.; Barrangou, R.; Zhao, S.; Brown, E.; Meng, J. Association of clustered regularly interspaced short palindromic repeat (CRISPR) elements with specific serotypes and virulence potential of shiga toxin-producing Escherichia coli. Appl. Environ. Microbiol. 2014, 80, 1411–1420. [Google Scholar] [CrossRef]
- Yin, S.; Jensen, M.A.; Bai, J.; Debroy, C.; Barrangou, R.; Dudley, E.G. The evolutionary divergence of Shiga toxin-producing Escherichia coli is reflected in clustered regularly interspaced short palindromic repeat (CRISPR) spacer composition. Appl. Environ. Microbiol. 2013, 79, 5710–5720. [Google Scholar] [CrossRef]


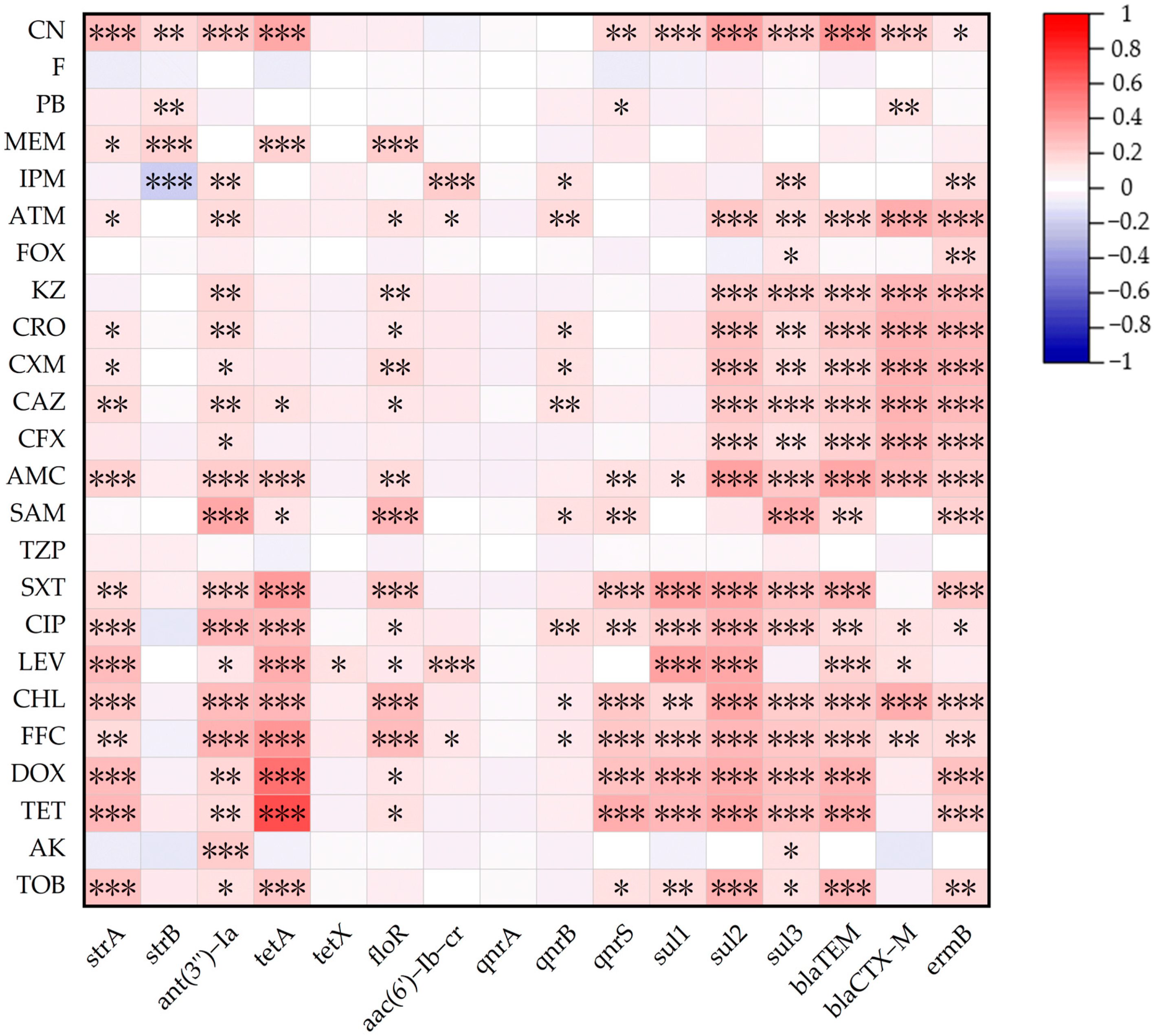


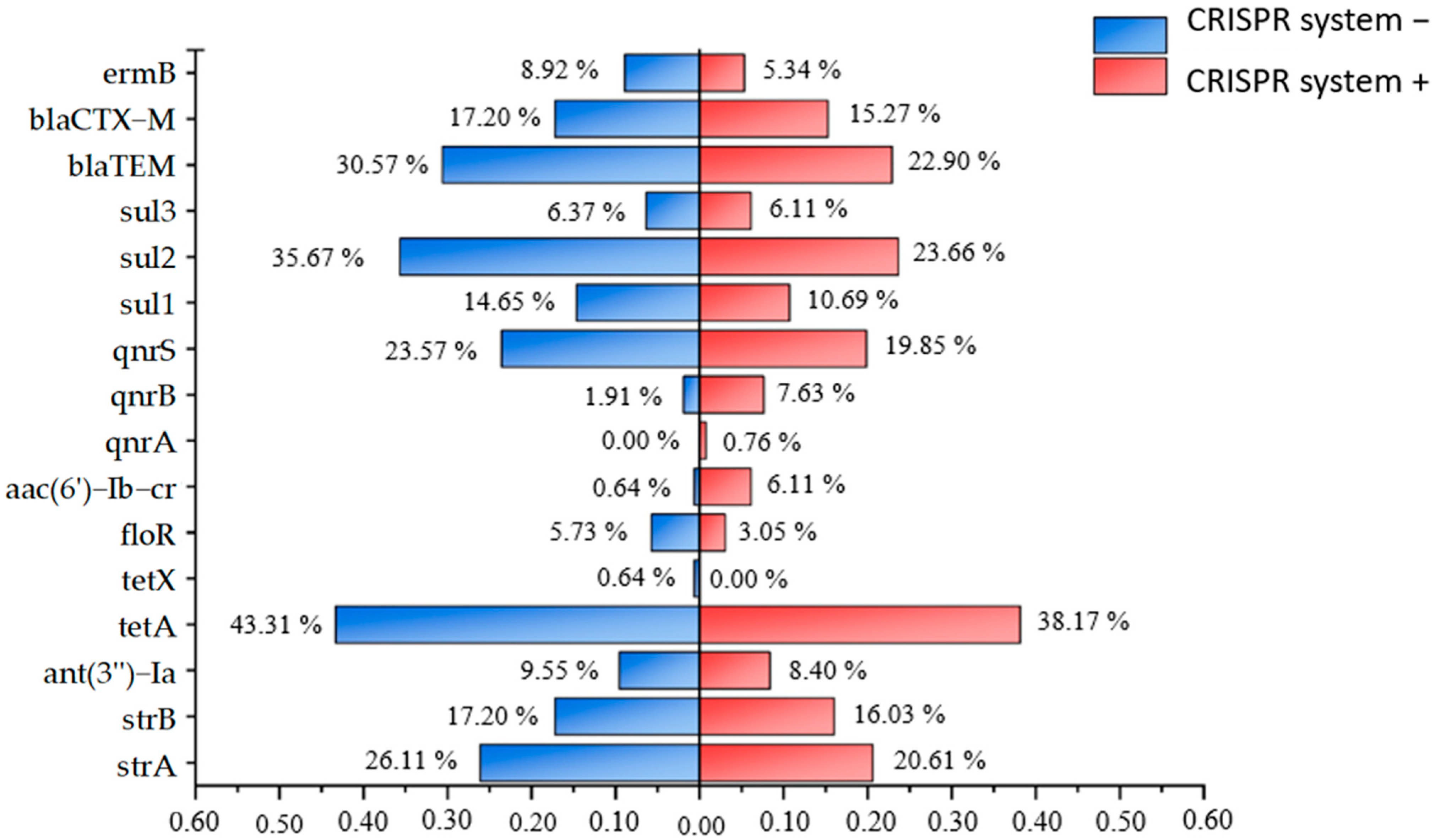
| Category of Antimicrobial | No. of Resistant Isolates (%) | Antibiotic | No. of Resistant Isolates (%) |
|---|---|---|---|
| aminoglycosides | 119 (41.32) | gentamicin (CN) | 90 (31.25) |
| tobramycin (TOB) | 53 (18.40) | ||
| amikacin (AK) | 33 (11.46) | ||
| tetracyclines | 145 (50.34) | tetracycline (TET) | 139 (48.26) |
| doxycycline (DOX) | 120 (41.67) | ||
| amide alcohols | 93 (32.29) | florfenicol (FFC) | 66 (22.92) |
| chloramphenicol (CHL) | 84 (29.17) | ||
| quinolones | 82 (28.47) | Levofloxacin (LEV) | 40 (13.89) |
| ciprofloxacin (CIP) | 51 (17.71) | ||
| sulfonamides | 105 (36.11) | trimethoprim–sulfamethoxazole (SXT) | 105 (36.46) |
| β-lactams | 212 (73.61) | piperacillin/tazobactam10:1 (TZP) | 12 (4.17) |
| ampicillin/sulbactam 1:1 (SAM) | 31 (10.76) | ||
| amoxicillin/clavulanic acid 2:1 (AMC) | 101 (35.07) | ||
| cefalexin (CFX) | 130 (45.14) | ||
| ceftazidime (CAZ) | 82 (28.47) | ||
| cefuroxime sodium (CXM) | 108 (37.5) | ||
| ceftriaxone (CRO) | 107 (37.15) | ||
| cephazolin (KZ) | 108 (37.5) | ||
| cefoxitin (FOX) | 9 (3.13) | ||
| aztreonam (ATM) | 99 (34.37) | ||
| imipenem (IPM) | 89 (30.9) | ||
| meropenem (MEM) | 15 (5.21) | ||
| polypeptides | 5 (1.74) | polymyxin b (PB) | 5 (1.74) |
| furans | 7 (2.43) | nitrofurantoin (F) | 7 (2.43) |
| Antibiotic | No. of Resistant Isolates in Sichuan (%) | No. of Resistant Isolates in Shanxi (%) | No. of Resistant Isolates in Yunnan (%) |
|---|---|---|---|
| Gentamicin (CN) | 45 (27.61) | 30 (42.86) | 15 (27.27) |
| Tobramycin (TOB) | 30 (18.40) | 16 (22.86) | 7 (12.73) |
| Amikacin (AK) | 32 (19.63) | 1 (1.43) | 0 (0.00) |
| Tetracycline (TET) | 79 (48.47) | 40 (57.14) | 20 (36.36) |
| Doxycycline (DOX) | 73 (44.79) | 32 (45.71) | 15 (27.27) |
| Florfenicol (FFC) | 43 (26.38) | 16 (22.86) | 7 (12.73) |
| Chloramphenicol (CHL) | 49 (30.06) | 28 (40.00) | 7 (12.73) |
| Levofloxacin (LEV) | 25 (15.34) | 12 (17.14) | 3 (5.45) |
| Ciprofloxacin (CIP) | 35 (21.47) | 13 (18.57) | 3 (5.45) |
| trimethoprim–sulfamethoxazole (SXT) | 60 (36.81) | 26 (37.14) | 19 (34.55) |
| piperacillin/tazobactam10:1 (TZP) | 9 (5.52) | 3 (4.29) | 0 (0.00) |
| ampicillin/sulbactam 1:1 (SAM) | 24 (14.72) | 7 (10.00) | 0 (0.00) |
| amoxicillin/clavulanic acid 2:1 (AMC) | 62 (38.04) | 24 (34.29) | 15 (27.27) |
| cefalexin (CFX) | 81 (49.69) | 26 (37.14) | 23 (41.82) |
| ceftazidime (CAZ) | 57 (34.79) | 21 (30.00) | 4 (7.27) |
| cefuroxime sodium (CXM) | 67 (41.10) | 24 (34.29) | 17 (30.91) |
| ceftriaxone (CRO) | 66 (40.49) | 26 (37.14) | 15 (27.27) |
| cephazolin (KZ) | 68 (41.72) | 23 (32.86) | 17 (30.91) |
| cefoxitin (FOX) | 5 (3.07) | 2 (2.86) | 2 (3.64) |
| aztreonam (ATM) | 68 (41.72) | 25 (35.71) | 6 (10.91) |
| imipenem (IPM) | 86 (52.76) | 3 (4.29) | 0 (0.00) |
| meropenem (MEM) | 7 (4.29) | 7 (10.00) | 1 (1.82) |
| polymyxin b (PB) | 2 (1.23) | 3 (4.29) | 0 (0.00) |
| nitrofurantoin (F) | 7 (4.29) | 0 (0.00) | 0 (0.00) |
| Category of Antimicrobial | No. of Resistant Isolates (%) | ARGs | No. of Positive Isolates (%) |
|---|---|---|---|
| aminoglycosides | 119 (41.32) | strA | 68 (23.61%) |
| strB | 48 (16.67%) | ||
| gyrB | 262 (90.97%) | ||
| ant(3′′)-Ia | 26 (9.03%) | ||
| aph(3′)-IIa | 0 (0%) | ||
| tetracyclines | 145 (50.34) | tetA | 118 (40.97%) |
| tetX | 1 (0.35%) | ||
| amide alcohols | 93 (32.29) | flor ermB | 13 (4.51%) 21 (7.29%) |
| quinolones | 82 (28.47) | aac(6′)-Ib-cr | 9 (3.13%) |
| qnrA | 1 (0.35%) | ||
| qnrB | 13 (4.51%) | ||
| qnrS | 63 (21.88%) | ||
| sulfonamides | 105 (36.11) | sul1 | 37 (12.85%) |
| sul2 | 87 (30.21%) | ||
| sul3 | 68 (23.61%) | ||
| β-lactams | 212 (73.61) | blaTEM | 78 (27.08%) |
| blaCTX-M | 47 (16.32%) | ||
| blaSHV | 0 (0%) |
| ARGs | Number of Positive Strains in Sichuan (%) | Number of Positive Strains in Shanxi (%) | Number of Positive Strains in Yunnan (%) |
|---|---|---|---|
| floR | 7 (4.29%) | 6 (8.57%) | 0 (0.00%) |
| blaTEM | 40 (24.54%) | 37 (52.86%) | 1 (1.82%) |
| sul1 | 20 (12.27%) | 16 (22.86%) | 1 (1.82%) |
| sul2 | 46 (28.22%) | 40 (57.14%) | 1 (1.82%) |
| strA | 24 (14.72%) | 39 (55.71%) | 5 (9.09%) |
| gyrB | 148 (90.80%) | 65 (92.86%) | 49 (89.09%) |
| qnrS | 29 (17.79%) | 26 (37.14%) | 8 (14.55%) |
| strB | 8 (4.91%) | 36 (51.43%) | 4 (7.27%) |
| blaCTX-M | 24 (14.72%) | 18 (25.71%) | 5 (9.09%) |
| ermB | 19 (11.66%) | 2 (2.86%) | 0 (0.00%) |
| sul3 | 15 (9.20%) | 1 (1.43%) | 2 (3.64%) |
| blaSHV | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) |
| qnrA | 1 (0.61%) | 0 (0.00%) | 0 (0.00%) |
| qnrB | 12 (7.36%) | 1 (1.43%) | 0 (0.00%) |
| aac(6′)-Ib-cr | 9 (5.52%) | 0 (0.00%) | 0 (0.00%) |
| ant(3′′)-Ia | 26 (15.95%) | 0 (0.00%) | 0 (0.00%) |
| aph(3′)-IIa | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) |
| tetA | 61 (37.42%) | 39 (55.71%) | 18 (32.73%) |
| tetX | 1 (0.61%) | 0 (0.00%) | 0 (0.00%) |
| Repeat Sequences Number | Repeat Sequence (5′–3′) | Minimum Free Energy Prediction |
|---|---|---|
| Sequence 1 | GTGTTCCCCGCGCCAGCGGGGATAAACCG | −15.20 kcal/mol |
| Sequence 2 | CGGTTTATCCCCGCTGGCGCGGGGAACAC | −14.20 kcal/mol |
| Sequence 3 | TGTGTTCCCCGCGCCAGCGGGGATAAACCG | −15.20 kcal/mol |
| Sequence 4 | GTGTTCCCCGCGCCAGCGGGGATAAACC | −15.20 kcal/mol |
| Sequence 5 | GTGTTCCCCGCGTCAGCGGGGATAAACCG | −15.20 kcal/mol |
| Sequence 6 | CGGTTTATCCCCGCTGGCGCGGGGAACACG | −14.20 kcal/mol |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, K.; Wu, X.; Ding, H.; Ma, B.; Li, Z.; Wang, Y.; Yang, Z.; Yao, X.; Luo, Y. Isolation, Identification, and Antibiotic Resistance, CRISPR System Analysis of Escherichia coli from Forest Musk Deer in Western China. Microorganisms 2025, 13, 1683. https://doi.org/10.3390/microorganisms13071683
Yang K, Wu X, Ding H, Ma B, Li Z, Wang Y, Yang Z, Yao X, Luo Y. Isolation, Identification, and Antibiotic Resistance, CRISPR System Analysis of Escherichia coli from Forest Musk Deer in Western China. Microorganisms. 2025; 13(7):1683. https://doi.org/10.3390/microorganisms13071683
Chicago/Turabian StyleYang, Kaiwei, Xi Wu, Hui Ding, Bingcun Ma, Zengting Li, Yin Wang, Zexiao Yang, Xueping Yao, and Yan Luo. 2025. "Isolation, Identification, and Antibiotic Resistance, CRISPR System Analysis of Escherichia coli from Forest Musk Deer in Western China" Microorganisms 13, no. 7: 1683. https://doi.org/10.3390/microorganisms13071683
APA StyleYang, K., Wu, X., Ding, H., Ma, B., Li, Z., Wang, Y., Yang, Z., Yao, X., & Luo, Y. (2025). Isolation, Identification, and Antibiotic Resistance, CRISPR System Analysis of Escherichia coli from Forest Musk Deer in Western China. Microorganisms, 13(7), 1683. https://doi.org/10.3390/microorganisms13071683




