Healthy Ageing and Gut Microbiota: A Study on Longevity in Adults
Abstract
1. Background
2. Materials and Methods
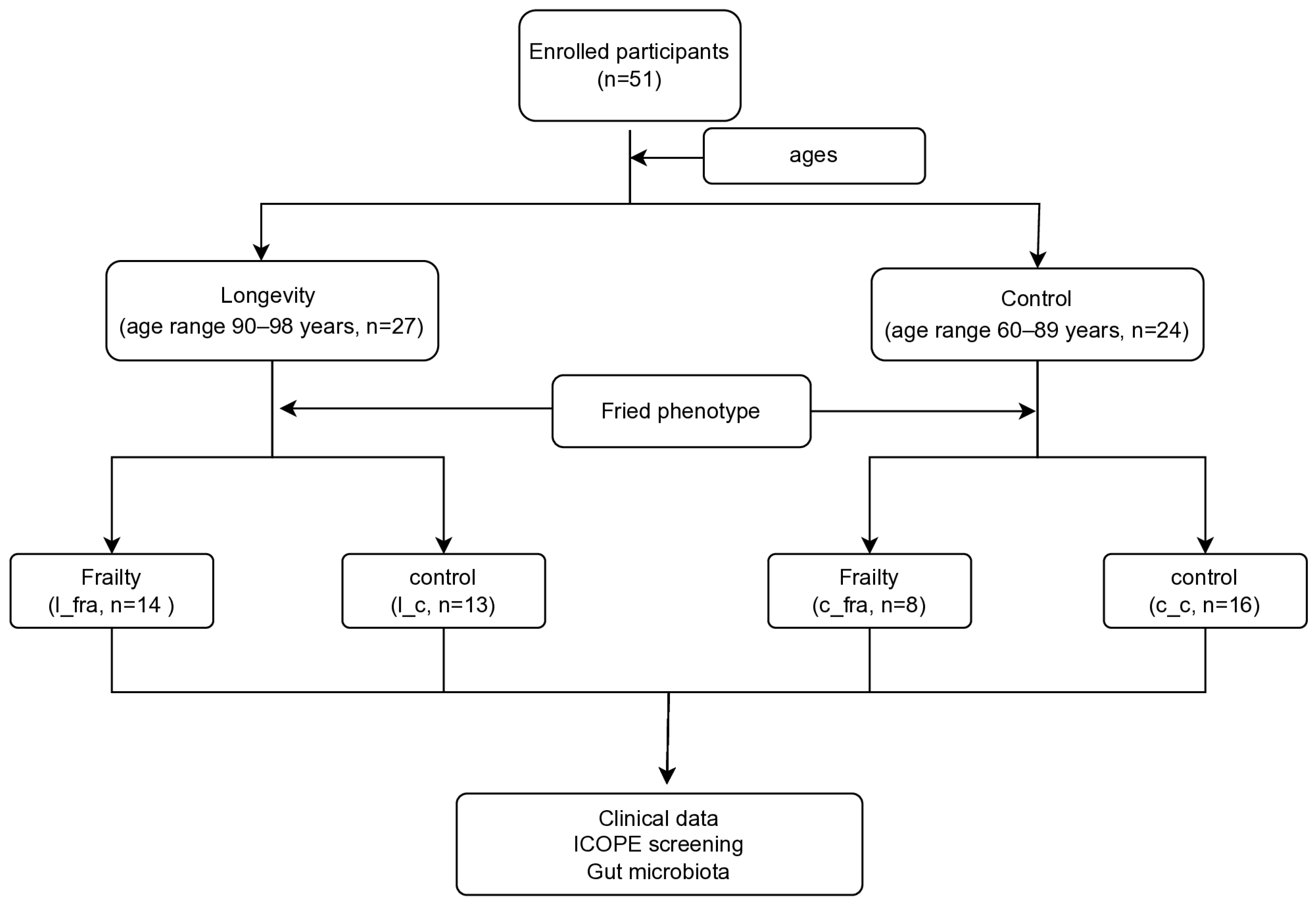
2.1. Study Participants
2.2. Sample Collection
2.3. Sequencing and Bioinformatics
2.4. Longevity-Associated Microbial Index
2.5. Functional Profiles of Microbial Communities with HMP Unified Metabolic Analysis Network 2 (HUMAnN2)
2.6. Statistical Analysis
3. Results
3.1. Demographic and Clinical Data of the Study Population
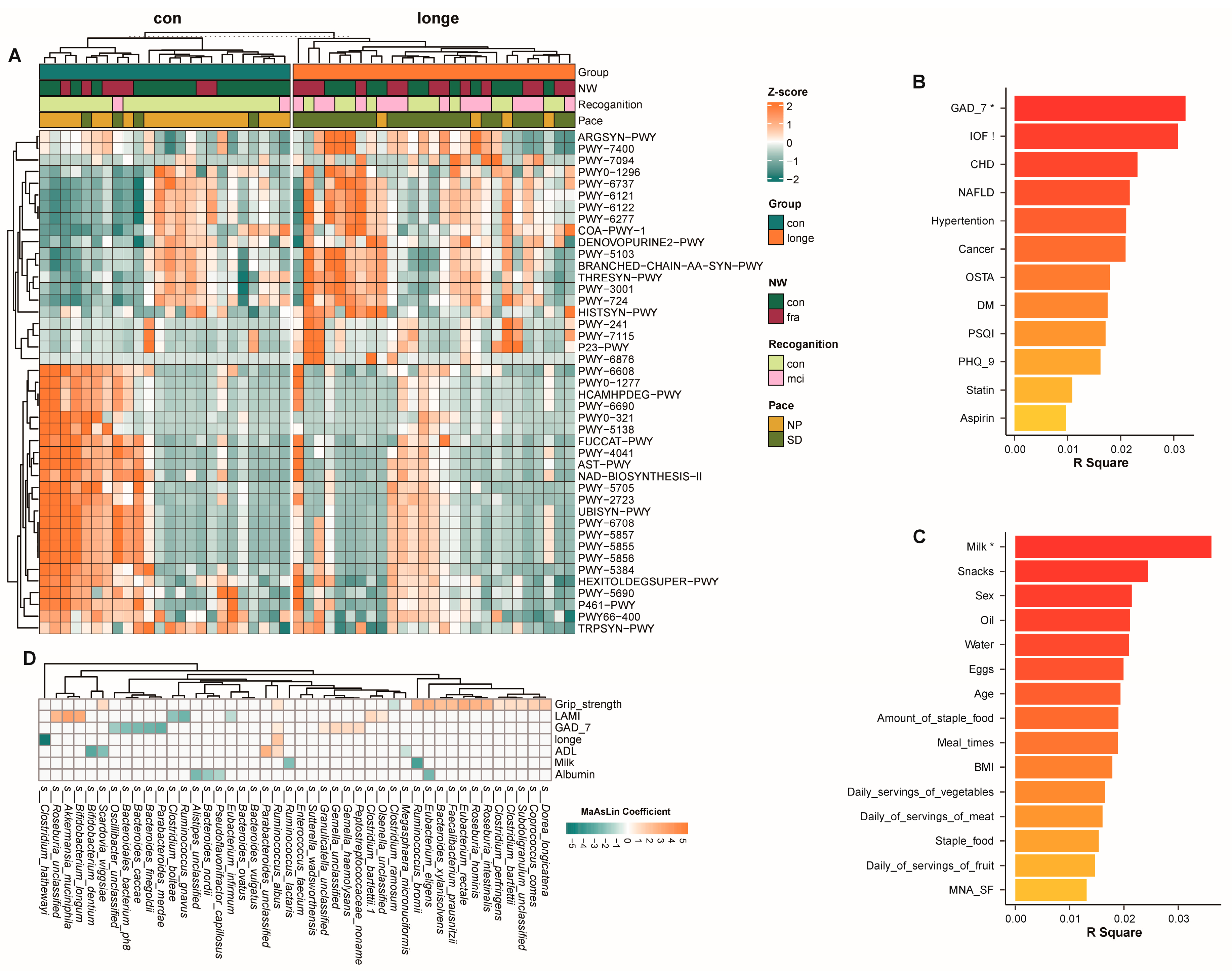
3.2. Bacterial Diversity, Abundance, and Functionality
3.3. Gut Microbiota and Clinical Data Based on Age and Frailty
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DM | diabetes mellitus |
| WHO | World Health Organisation |
| ICOPE | Integrated Care for Older People |
| MMSE | Mini-Mental State Examination |
| SPPB | Short Physical Performance Battery |
| MNA_SF | Mini-Nutritional Assessment Short Form |
| GAD-7 | Brief 7-Item Self-Report Questionnaire for Generalised Anxiety Disorder |
| PHQ-9 | Patient Health Questionnaire-9 |
| MCI | mild cognitive impairment |
| IC | impaired intrinsic ability |
| NAFLD | non-alcoholic fatty liver disease |
| HTN | hypertension |
| FFQ | Food Frequency Questionnaire |
| LAMI | The longevity-associated microbial index |
| CHD | coronary heart disease |
| BCAA | branched amino acid |
References
- Partridge, L.; Deelen, J.; Slagboom, P.E. Facing up to the global challenges of ageing. Nature 2018, 561, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Rudnicka, E.; Napierała, P.; Podfigurna, A.; Męczekalski, B.; Smolarczyk, R.; Grymowicz, M. The World Health Organization (WHO) approach to healthy ageing. Maturitas 2020, 139, 6–11. [Google Scholar] [CrossRef]
- Beard, J.R.; Officer, A.; de Carvalho, I.A.; Sadana, R.; Pot, A.M.; Michel, J.-P.; Lloyd-Sherlock, P.; Epping-Jordan, J.E.; Peeters, G.M.E.E.G.; Mahanani, W.R.; et al. The World report on ageing and health: A policy framework for healthy ageing. Lancet 2016, 387, 2145–2154. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Hua, Y.; Zeng, B.; Ning, R.; Li, Y.; Zhao, J. Gut microbiota signatures of longevity. Curr. Biol. 2016, 26, R832–R833. [Google Scholar] [CrossRef]
- Palmas, V.; Pisanu, S.; Madau, V.; Casula, E.; Deledda, A.; Cusano, R.; Uva, P.; Loviselli, A.; Velluzzi, F.; Manzin, A. Gut Microbiota Markers and Dietary Habits Associated with Extreme Longevity in Healthy Sardinian Centenarians. Nutrients 2022, 14, 2436. [Google Scholar] [CrossRef] [PubMed]
- Biagi, E.; Franceschi, C.; Rampelli, S.; Severgnini, M.; Ostan, R.; Turroni, S.; Consolandi, C.; Quercia, S.; Scurti, M.; Monti, D.; et al. Gut Microbiota and Extreme Longevity. Curr. Biol. 2016, 26, 1480–1485. [Google Scholar] [CrossRef] [PubMed]
- Ko, C.-Y.; Liu, Q.-Q.; Su, H.-Z.; Zhang, H.-P.; Fan, J.-M.; Yang, J.-H.; Hu, A.-K.; Liu, Y.-Q.; Chou, D.; Zeng, Y.-M. Gut microbiota in obstructive sleep apnea-hypopnea syndrome: Disease-related dysbiosis and metabolic comorbidities. Clin. Sci. 2019, 133, 905–917. [Google Scholar] [CrossRef]
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’Connor, E.M.; Cusack, S.; Harris, H.M.B.; Coakley, M.; Lakshminarayanan, B.; O’Sullivan, O.; et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012, 488, 178–184. [Google Scholar] [CrossRef]
- Sonnenburg, E.D.; Sonnenburg, J.L. The ancestral and industrialized gut microbiota and implications for human health. Nat. Rev. Microbiol. 2019, 17, 383–390. [Google Scholar] [CrossRef]
- Pang, S.; Chen, X.; Lu, Z.; Meng, L.; Huang, Y.; Yu, X.; Huang, L.; Ye, P.; Chen, X.; Liang, J.; et al. Longevity of centenarians is reflected by the gut microbiome with youth-associated signatures. Nat. Aging 2023, 3, 436–449. [Google Scholar] [CrossRef]
- Dent, E.; Martin, F.C.; Bergman, H.; Woo, J.; Romero-Ortuno, R.; Walston, J.D. Management of frailty: Opportunities, challenges, and future directions. Lancet 2019, 394, 1376–1386. [Google Scholar] [CrossRef] [PubMed]
- Integrated Care for Older People: Guidelines on Community-Level Interventions to Manage Declines in Intrinsic Capacity. Geneva2017. Available online: https://www.who.int/publications/i/item/9789241550109 (accessed on 1 January 2017).
- Fried, L.P.; Tangen, C.M.; Walston, J.; Newman, A.B.; Hirsch, C.; Gottdiener, J.; Seeman, T.; Tracy, R.; Kop, W.J.; Burke, G.; et al. Frailty in older adults: Evidence for a phenotype. J. Gerontol. A Biol. Sci. Med. Sci. 2001, 56, M146–M157. [Google Scholar] [CrossRef]
- Liu, S.; Yu, X.; Wang, X.; Li, J.; Jiang, S.; Kang, L.; Liu, X. Intrinsic Capacity predicts adverse outcomes using Integrated Care for Older People screening tool in a senior community in Beijing. Arch. Gerontol. Geriatr. 2021, 94, 104358. [Google Scholar] [CrossRef] [PubMed]
- Folstein, M.F.; Folstein, S.E.; McHugh, P.R. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res. 1975, 12, 189–198. [Google Scholar] [CrossRef]
- Mangione, K.K.; Craik, R.L.; McCormick, A.A.; Blevins, H.L.; White, M.B.; Sullivan-Marx, E.M.; Tomlinson, J.D. Detectable changes in physical performance measures in elderly African Americans. Phys. Ther. 2010, 90, 921–927. [Google Scholar] [CrossRef] [PubMed]
- Rubenstein, L.Z.; Harker, J.O.; Salvà, A.; Guigoz, Y.; Vellas, B. Screening for undernutrition in geriatric practice: Developing the short-form mini-nutritional assessment (MNA-SF). J. Gerontol. A Biol. Sci. Med. Sci. 2001, 56, M366–M372. [Google Scholar] [CrossRef]
- Spitzer, R.L.; Kroenke, K.; Williams, J.B.W.; Löwe, B. A brief measure for assessing generalized anxiety disorder: The GAD-7. Arch. Intern. Med. 2006, 166, 1092–1097. [Google Scholar] [CrossRef]
- Kroenke, K.; Spitzer, R.L.; Williams, J.B. The PHQ-9: Validity of a brief depression severity measure. J. Gen. Intern. Med. 2001, 16, 606–613. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Peng, Y.; Leung, H.C.M.; Yiu, S.M.; Chin, F.Y.L. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef]
- Zhu, W.; Lomsadze, A.; Borodovsky, M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010, 38, e132. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Fernandes, A.D.; Reid, J.N.; Macklaim, J.M.; McMurrough, T.A.; Edgell, D.R.; Gloor, G.B. Unifying the analysis of high-throughput sequencing datasets: Characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome 2014, 2, 15. [Google Scholar] [CrossRef]
- Mallick, H.; Rahnavard, A.; McIver, L.J.; Ma, S.; Zhang, Y.; Nguyen, L.H.; Tickle, T.L.; Weingart, G.; Ren, B.; Schwager, E.H.; et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput. Biol. 2021, 17, e1009442. [Google Scholar] [CrossRef]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’HAra, R.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Vegan: Community Ecology Package, R Package Version 2.0-10; CRAN; 2013. Available online: https://github.com/vegandevs/vegan (accessed on 1 January 2017).
- Tatsumi, S.; Iritani, R.; Cadotte, M.W. Partitioning the temporal changes in abundance-based beta diversity into loss and gain components. Methods Ecol. Evol. 2022, 13, 2042–2048. [Google Scholar] [CrossRef]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, J.-N.; Sun, B.-H.; Liu, Q.; Ma, J.; Zhang, Q.; Liu, Y.-X.; Chen, N.; Chen, F. The role of genotype and diet in shaping gut microbiome in a genetic vitamin A deficient mouse model. J. Genet. Genom. 2022, 49, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Franzosa, E.A.; McIver, L.J.; Rahnavard, G.; Thompson, L.R.; Schirmer, M.; Weingart, G.; Lipson, K.S.; Knight, R.; Caporaso, J.G.; Segata, N.; et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 2018, 15, 962–968. [Google Scholar] [CrossRef]
- Stevens, C.; Norris, S.; Arbeeva, L.; Carter, S.; Enomoto, M.; Nelson, A.E.; Lascelles, B.D.X. Gut Microbiome and Osteoarthritis: Insights from the Naturally Occurring Canine Model of Osteoarthritis. Arthritis Rheumatol. 2024, 76, 1758–1763. [Google Scholar] [CrossRef]
- Gutiérrez-Robledo, L.M.; García-Chanes, R.E.; González-Bautista, E.; Rosas-Carrasco, O. Validation of Two Intrinsic Capacity Scales and Its Relationship with Frailty and Other Outcomes in Mexican Community-Dwelling Older Adults. J. Nutr. Health Aging 2021, 25, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Odamaki, T.; Kato, K.; Sugahara, H.; Hashikura, N.; Takahashi, S.; Xiao, J.-Z.; Abe, F.; Osawa, R. Age-related changes in gut microbiota composition from newborn to centenarian: A cross-sectional study. BMC Microbiol. 2016, 16, 90. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, Q.; Cheng, L.; Buch, H.; Zhang, F. Akkermansia muciniphila is a promising probiotic. Microb. Biotechnol. 2019, 12, 1109–1125. [Google Scholar] [CrossRef]
- Cani, P.D.; Depommier, C.; Derrien, M.; Everard, A.; de Vos, W.M. Akkermansia muciniphila: Paradigm for next-generation beneficial microorganisms. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 625–637. [Google Scholar] [CrossRef]
- Nie, C.; He, T.; Zhang, W.; Zhang, G.; Ma, X. Branched Chain Amino Acids: Beyond Nutrition Metabolism. Int. J. Mol. Sci. 2018, 19, 954. [Google Scholar] [CrossRef]
- Pei, T.; Zhu, D.; Yang, S.; Hu, R.; Wang, F.; Zhang, J.; Yan, S.; Ju, L.; He, Z.; Han, Z.; et al. Bacteroides plebeius improves muscle wasting in chronic kidney disease by modulating the gut-renal muscle axis. J. Cell. Mol. Med. 2022, 26, 6066–6078. [Google Scholar] [CrossRef]
- Shimizu, J.; Kubota, T.; Takada, E.; Takai, K.; Fujiwara, N.; Arimitsu, N.; Murayama, M.A.; Ueda, Y.; Wakisaka, S.; Suzuki, T.; et al. Propionate-producing bacteria in the intestine may associate with skewed responses of IL10-producing regulatory T cells in patients with relapsing polychondritis. PLoS ONE 2018, 13, e0203657. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, J.; Wang, H.; Lu, W.; Lee, Y.K.; Zhao, J.; Zhang, H. Machine learning framework for gut microbiome biomarkers discovery and modulation analysis in large-scale obese population. BMC Genom. 2022, 23, 850. [Google Scholar] [CrossRef] [PubMed]
- Denamur, E.; Clermont, O.; Bonacorsi, S.; Gordon, D. The population genetics of pathogenic Escherichia coli. Nat. Rev. Microbiol. 2021, 19, 37–54. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Sun, Q.; Zheng, H.; Zhang, Y.; Wang, Y.; Liu, S.; Duan, L. Roseburia hominis Alleviates Neuroinflammation via Short-Chain Fatty Acids through Histone Deacetylase Inhibition. Mol. Nutr. Food Res. 2022, 66, e2200164. [Google Scholar] [CrossRef] [PubMed]
- Machiels, K.; Joossens, M.; Sabino, J.; De Preter, V.; Arijs, I.; Eeckhaut, V.; Ballet, V.; Claes, K.; Van Immerseel, F.; Verbeke, K.; et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014, 63, 1275–1283. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]



| Intrinsic Capacity | Tests | Scores |
|---|---|---|
| Cognition | MMSE | Scores equal or greater than 27 (1)/less than 27 (0) |
| Locomotion Chair rise test | Rise from a chair five times without using arms within 14 s | Yes (1)/No (0) |
| Vitality | ||
| Weight loss | Have you unintentionally lost more than 3 kg over the last 3 months? | Yes (0)/No (1) |
| Appetite loss | Have you experienced loss of appetite? | Yes (0)/No (1) |
| Sensory | ||
| Visual impairment | Do you have any problems with your eyes: difficulties in seeing far, reading, or eye disease? | Yes (0)/No (1) |
| Hearing loss | Do you have any problems in hearing: difficulties in hearing whispers? | Yes (0)/No (1) |
| Psychosocial | ||
| Depressive symptoms | PHQ-9 score | Scores greater than 4 (0)/scores equal to or less than 4 (1) |
| Anxiety symptoms | GAD-7 score | Scores greater than 4 (0)/scores equal to or less than 4 (1) |
| Longe Group vs. CON Group | Longe Group | CON Group | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Longe Group (≥90, n = 27) | CON Group (60–89, n = 24) | p Value | l_fra (n = 14) | l_c (n = 13) | p | C_fra (n = 8) | C-c (n = 16) | p Value | |
| Age | 91.59 ± 1.89 | 79.08 ± 6.83 | <0.001 | 91.85 ± 1.66 | 91.30 ± 2.14 | 0.406 | 80.75 ± 5.65 | 78.25 ± 7.37 | 0.410 |
| Female | 8 (29.6%) | 14 (58.3%) | 0.039 | 5 (35.7%) | 3 (23.1%) | 0.678 | 1 (12.5%) | 13(81.2%) | 0.002 |
| DM | 6 (22.2%) | 10 (41.7%) | 0.135 | 3 (21.4%) | 3 (23.1%) | 1.000 | 6 (75.0%) | 4(25.0%) | 0.032 |
| HTN | 18 (66.7%) | 18 (75.0%) | 0.514 | 11 (78.6%) | 7 (53.8%) | 0.236 | 7 (87.5%) | 11(68.8%) | 0.621 |
| CHD | 8 (30.8%) | 4 (16.7%) | 0.243 | 6 (42.9%) | 2 (16.7) | 0.216 | 3 (37.5%) | 1(6.2%) | 0.091 |
| NAFLD | 2 (7.4%) | 6 (25.0%) | 0.127 | 1 (7.1%) | 1 (7.1%) | 1.000 | 1 (12.5%) | 5(31.2%) | 0.621 |
| Statin | 14 (51.9%) | 16 (66.7%) | 0.283 | 11 (78.6%) | 3 (23.1%) | 0.004 | 6 (75.0%) | 10(62.5%) | 0.667 |
| Metformin | 0 (0.0%) | 5 (20.8%) | 0.018 | - | - | - | 3 (37.5%) | 2(12.5%) | 0.289 |
| Aspirin | 8 (29.6%) | 9 (37.5%) | 0.552 | 6 (42.9%) | 2 (15.4%) | 0.209 | 4 (50.0%) | 5(31.2%) | 0.412 |
| Albumin | 35.36 ± 3.65 | 39.06 ± 4.09 | 0.008 | 34.74 ± 3.85 | 37.83 ± 0.40 | 0.200 | 36.90 ± 4.93 | 40.29 ± 3.06 | 0.059 |
| Haemoglobin | 121.40 ± 19.48 | 126.27 ± 17.33 | 0.430 | 118.83 ± 19.93 | 131.67 ± 16.56 | 0.325 | 124.00 ± 28.33 | 127.57 ± 6.92 | 0.736 |
| BMI | 24.70 ± 3.50 | 24.12 ± 3.09 | 0.537 | 25.25 ± 4.38 | 24.10 ± 2.21 | 0.406 | 23.82 ± 2.52 | 24.27 ± 3.40 | 0.747 |
| Grip strength | 19.56 ± 7.27 | 20.17 ± 6.44 | 0.756 | 15.00 ± 5.21 | 24.48 ± 5.88 | <0.001 | 19.41 ± 9.64 | 20.54 ± 4.45 | 0.760 |
| Time_4.57m | 10.02 ± 5.66 | 4.80 ± 2.45 | <0.001 | 6.61 ± 3.62 | 13.18 ± 5.43 | 0.001 | 6.33 ± 3.70 | 4.04 ± 0.97 | 0.126 |
| CRP | 8.82 ± 18.42 | 3.68 ± 9.19 | 0.331 | 10.80 ± 20.26 | 0.90 ± 0.46 | 0.425 | 6.69 ± 15.21 | 1.96 ± 1.76 | 0.410 |
| ESR | 15.80 ± 17.22 | 16.87 ± 24.06 | 0.883 | 18.42 ± 18.40 | 5.33 ± 3.06 | 0.254 | 26.50 ± 36.96 | 11.36 ± 10.48 | 0.292 |
| Creatinine | 95.33 ± 29.93 | 76.50 ± 18.31 | 0.023 | 96.50 ± 33.44 | 90.67 ± 9.02 | 0.775 | 84.87 ± 17.33 | 71.71 ± 17.67 | 0.106 |
| BUN | 6.50 ± 2.50 | 7.07 ± 1.70 | 0.404 | 6.59 ± 2.77 | 6.14 ± 1.15 | 0.794 | 7.15 ± 1.66 | 7.04 ± 1.78 | 0.882 |
| MMSE | 24.78 ± 4.50 | 28.17 ± 1.20 | 0.001 | 22.57 ± 4.78 | 27.15 ± 2.70 | 0.006 | 27.62 ± 1.06 | 28.44 ± 1.21 | 0.121 |
| MNA-SF | 12.85 ± 2.68 | 16.33 ± 0.96 | <0.001 | 11.50 ± 2.93 | 14.31 ± 1.38 | 0.004 | 16.50 ± 0.76 | 16.25 ± 1.06 | 0.561 |
| GAD-7 | 2.04 ± 3.43 | 2.54 ± 3.02 | 0.582 | 3.21 ± 4.21 | 0.77 ± 1.74 | 0.061 | 3.13 ± 3.18 | 2.25 ± 3.00 | 0.516 |
| PHQ-9 | 2.67 ± 3.82 | 3.13 ± 3.89 | 0.674 | 4.36 ± 4.40 | 0.85 ± 1.95 | 0.014 | 4.13 ± 3.80 | 2.63 ± 3.96 | 0.385 |
| PSQI | 8.67 ± 4.22 | 9.12 ± 3.65 | 0.682 | 11.29 ± 3.73 | 5.85 ± 2.64 | <0.010 | 10.00 ± 3.70 | 8.69 ± 3.66 | 0.419 |
| OSTA | −5.32 ± 2.27 | −3.42 ± 2.18 | 0.004 | −5.19 ± 2.69 | −5.47 ± 1.81 | 0.754 | −3.14 ± 2.68 | −3.56 ± 1.96 | 0.662 |
| Intrinsic capacity | 5.44 ± 2.15 | 6.71 ± 1.46 | 0.017 | 4.00 ± 1.66 | 7.00 ± 1.41 | <0.010 | 7.15 ± 1.04 | 4.50±1.29 | <0.010 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, L.; Xu, J.; Xue, Q.; Wei, Y.; Wang, J. Healthy Ageing and Gut Microbiota: A Study on Longevity in Adults. Microorganisms 2025, 13, 1657. https://doi.org/10.3390/microorganisms13071657
Deng L, Xu J, Xue Q, Wei Y, Wang J. Healthy Ageing and Gut Microbiota: A Study on Longevity in Adults. Microorganisms. 2025; 13(7):1657. https://doi.org/10.3390/microorganisms13071657
Chicago/Turabian StyleDeng, Lihua, Jun Xu, Qian Xue, Yanan Wei, and Jingtong Wang. 2025. "Healthy Ageing and Gut Microbiota: A Study on Longevity in Adults" Microorganisms 13, no. 7: 1657. https://doi.org/10.3390/microorganisms13071657
APA StyleDeng, L., Xu, J., Xue, Q., Wei, Y., & Wang, J. (2025). Healthy Ageing and Gut Microbiota: A Study on Longevity in Adults. Microorganisms, 13(7), 1657. https://doi.org/10.3390/microorganisms13071657






