Interaction of Potato Autophagy-Related StATG8 Family Proteins with Pathogen Effector and WRKY Transcription Factor in the Nucleus
Abstract
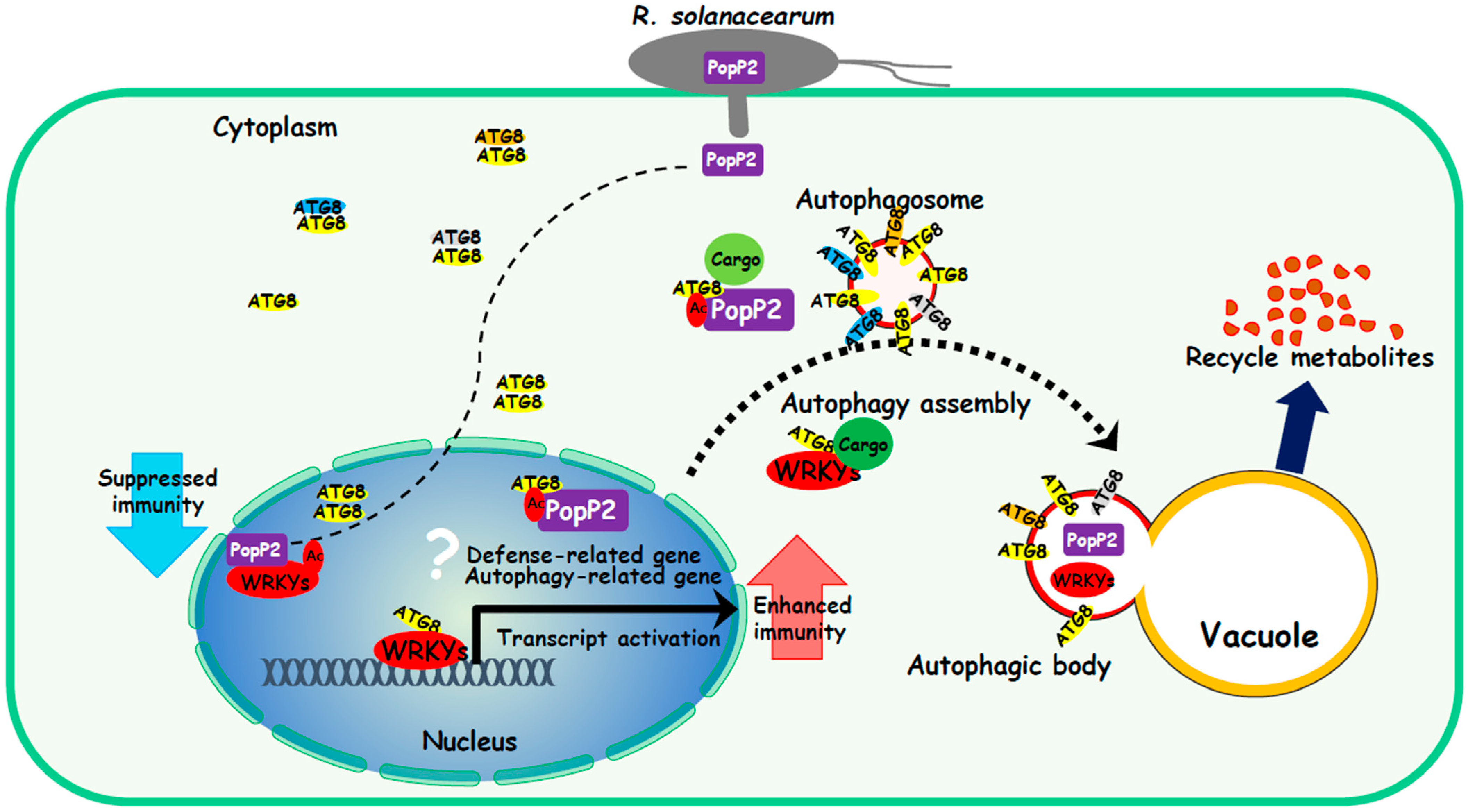
1. Introduction
2. Materials and Methods
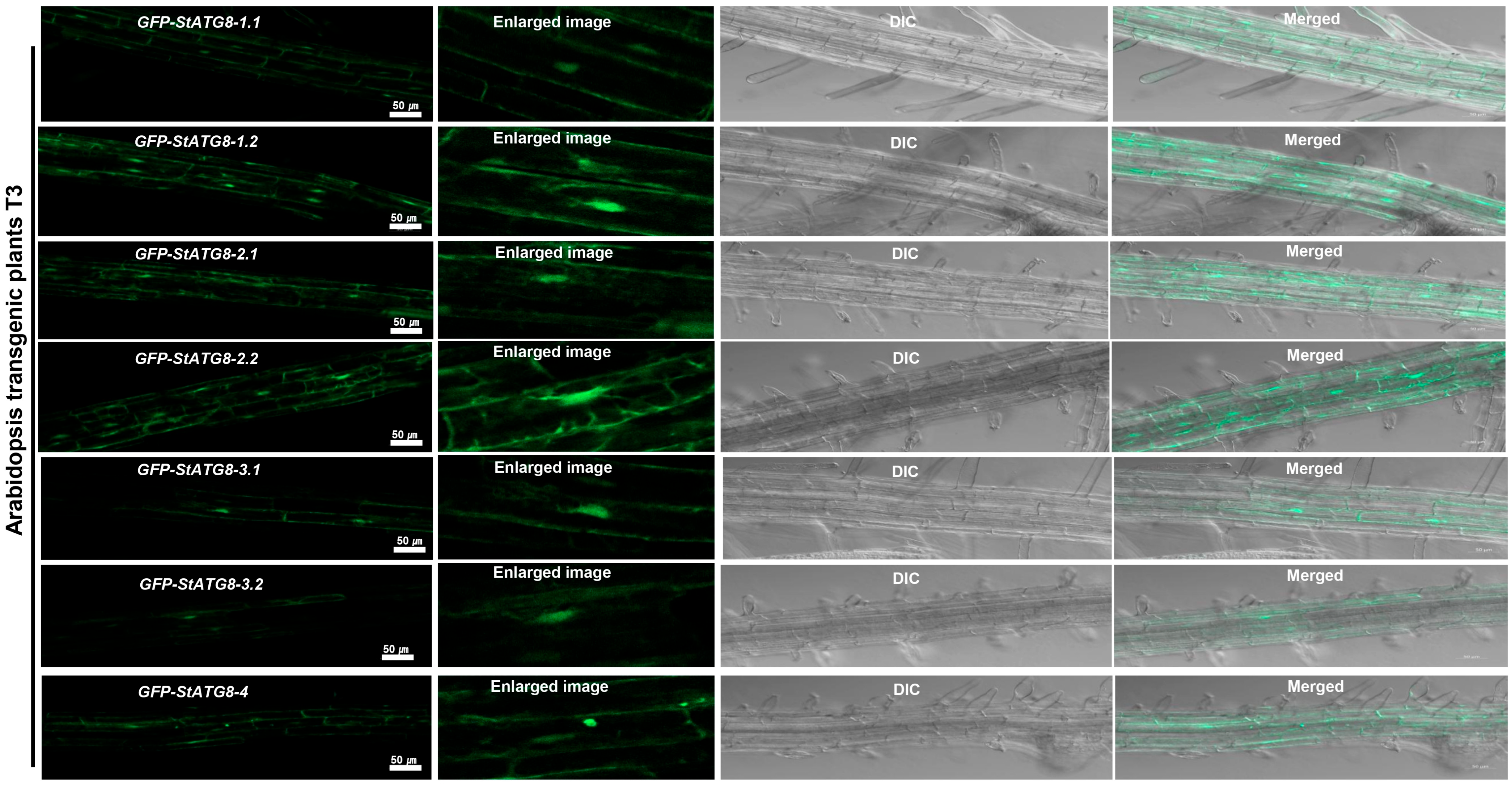
2.1. Plant Materials and Arabidopsis Transgenic Plants
2.2. BiFC Assay and Agro-Mediated Transient Gene Expression in N. benthamiana
2.3. Yeast Two-Hybrid Assay and Quantitative Liquid β-Gal Assays
2.4. Co-Immunoprecipitation (Co-IP) and Western Blot
2.5. Statistical Analyses
3. Results
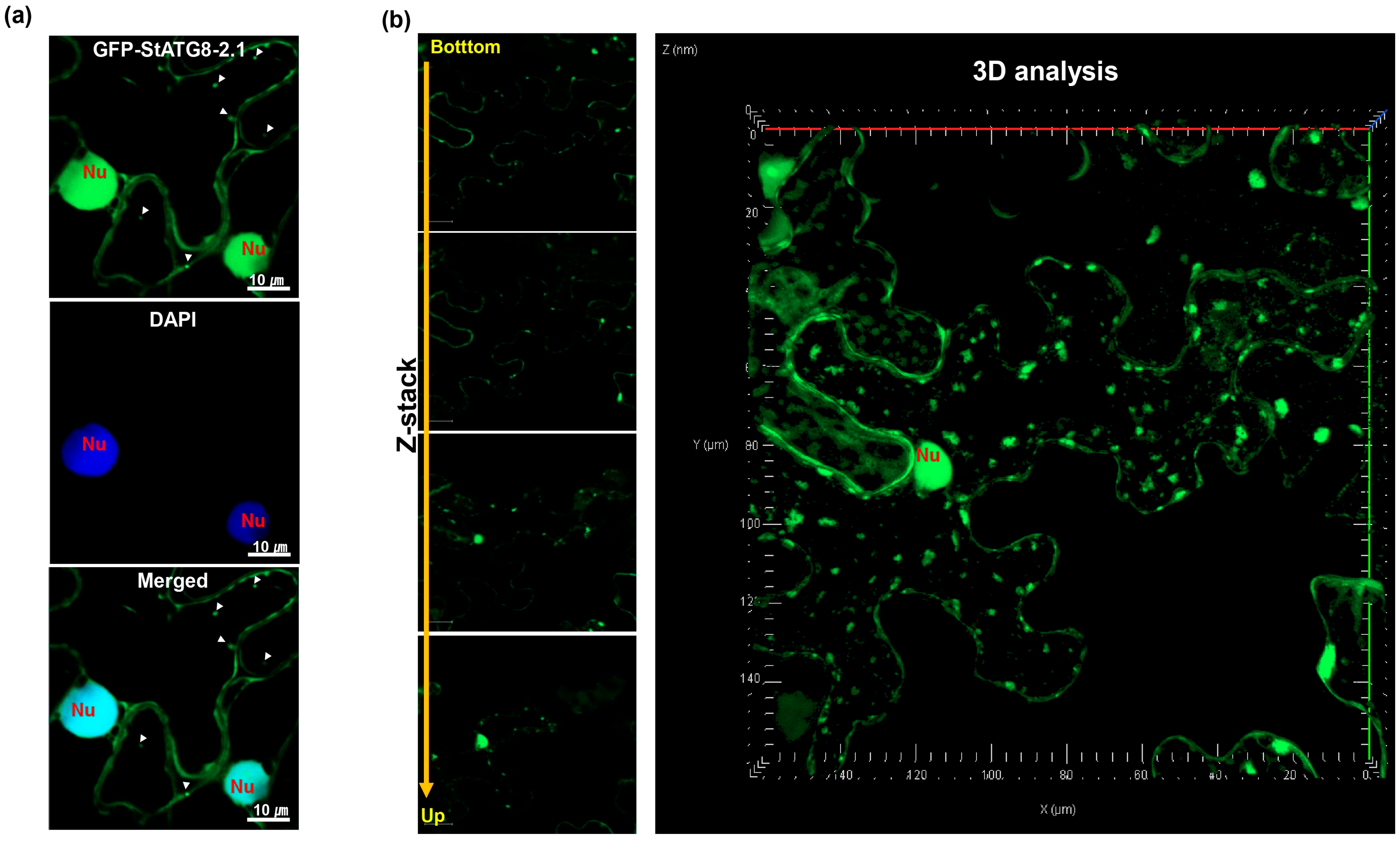
3.1. StATG8 Is Primarily Localized in the Cytoplasm and Nucleus of Primary Roots in Stable GFP-StATG8 Transgenic Plants
3.2. StATG8 Forms Homodimers
3.3. StATG8 Forms Heterodimers
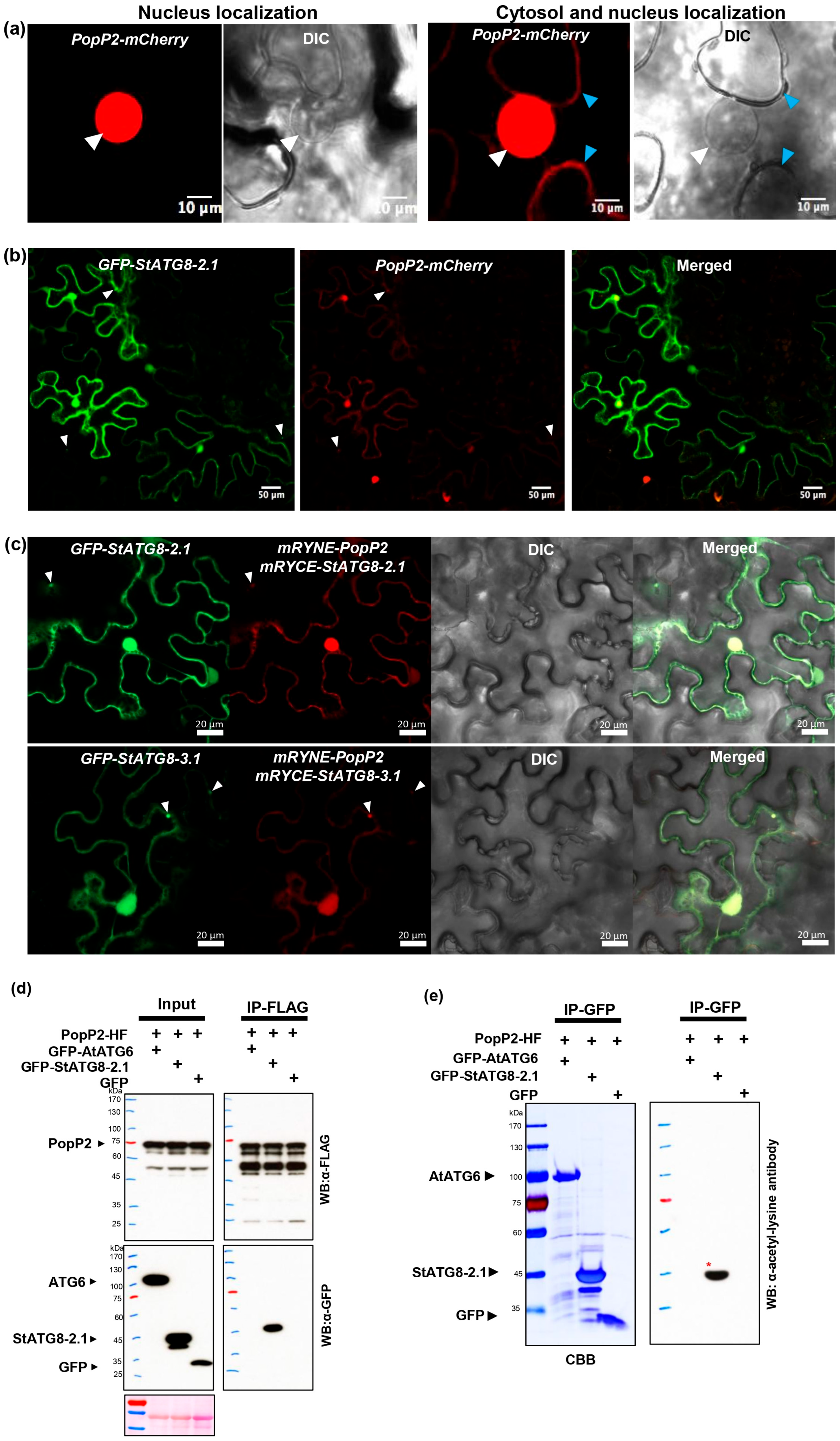
3.4. The Ralstonia Effector PopP2 Directly Associates with StATG8 and Relocalizes to Autophagosomes
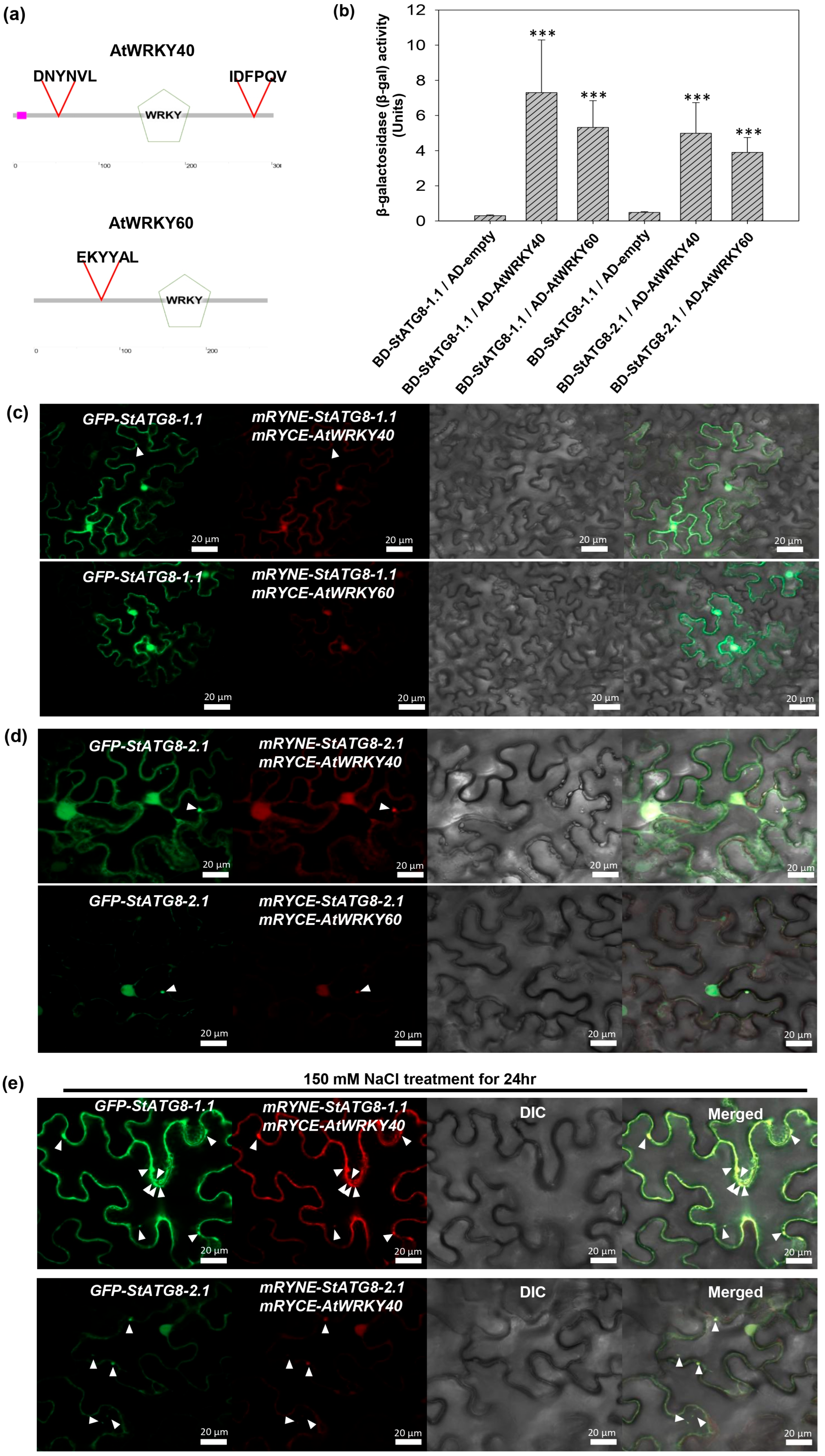
3.5. StATG8 Interacts with the WRKY Transcription Factor
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yagyu, M.; Yoshimoto, K. New insights into plant autophagy: Molecular mechanisms and roles in development and stress responses. J. Exp. Bot. 2023, 75, 1234–1251. [Google Scholar] [CrossRef]
- Eskelinen, E.L. Novel insights into autophagosome biogenesis revealed by cryo-electron tomography. FEBS Lett. 2024, 598, 9–16. [Google Scholar] [CrossRef]
- Juárez-Montiel, M.; Clark-Flores, D.; Tesillo-Moreno, P.; de la Vega-Camarillo, E.; Andrade-Pavón, D.; Hernández-García, J.A.; Hernández-Rodríguez, C.; Villa-Tanaca, L. Vacuolar proteases and autophagy in phytopathogenic fungi: A review. Front. Fungal Biol. 2022, 3, 948477. [Google Scholar] [CrossRef]
- Magen, S.; Seybold, H.; Laloum, D.; Avin-Wittenberg, T. Metabolism and autophagy in plants—A perfect match. FEBS Lett. 2022, 596, 2133–2151. [Google Scholar] [CrossRef]
- Zhou, Y.; Manghwar, H.; Hu, W.; Liu, F. Degradation Mechanism of Autophagy-Related Proteins and Research Progress. Int. J. Mol. Sci. 2022, 23, 7301. [Google Scholar] [CrossRef]
- Kuma, A.; Matsui, M.; Mizushima, N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: Caution in the interpretation of LC3 localization. Autophagy 2007, 3, 323–328. [Google Scholar] [CrossRef]
- Liu, R.; Zhang, R.; Yang, Y.; Liu, X.; Gong, Q. Monitoring Autophagy in Rice with GFP-ATG8 Marker Lines. Front. Plant Sci. 2022, 13, 866367. [Google Scholar] [CrossRef]
- Rogov, V.V.; Nezis, I.P.; Tsapras, P.; Zhang, H.; Dagdas, Y.; Noda, N.N.; Nakatogawa, H.; Wirth, M.; Mouilleron, S.; McEwan, D.G.; et al. Atg8 family proteins, LIR/AIM motifs and other interaction modes. Autophagy Rep. 2023, 2, 2188523. [Google Scholar] [CrossRef]
- Bassham, D.C. Methods for analysis of autophagy in plants. Methods 2015, 75, 181–188. [Google Scholar] [CrossRef]
- Zhang, W.; Nishimura, T.; Gahlot, D.; Saito, C.; Davis, C.; Jefferies, H.B.J.; Schreiber, A.; Thukral, L.; Tooze, S.A. Autophagosome membrane expansion is mediated by the N-terminus and cis-membrane association of human ATG8s. eLife 2023, 12, e89185. [Google Scholar] [CrossRef]
- Kellner, R.; De la Concepcion, J.C.; Maqbool, A.; Kamoun, S.; Dagdas, Y.F. ATG8 Expansion: A Driver of Selective Autophagy Diversification? Trends Plant Sci. 2017, 22, 204–214. [Google Scholar] [CrossRef]
- Marshall, R.S.; Hua, Z.; Mali, S.; McLoughlin, F.; Vierstra, R.D. ATG8-Binding UIM Proteins Define a New Class of Autophagy Adaptors and Receptors. Cell 2019, 177, 766–781.e24. [Google Scholar] [CrossRef]
- Li, F.; Zhang, M.; Zhang, C.; Zhou, X. Nuclear autophagy degrades a geminivirus nuclear protein to restrict viral infection in solanaceous plants. New Phytol. 2020, 225, 1746–1761. [Google Scholar] [CrossRef]
- Yan, Y.; Wang, P.; He, C.; Shi, H. MeWRKY20 and its interacting and activating autophagy-related protein 8 (MeATG8) regulate plant disease resistance in cassava. Biochem. Biophys. Res. Commun. 2017, 494, 20–26. [Google Scholar] [CrossRef]
- Song, I.; Hong, S.; Huh, S.U. Identification and Expression Analysis of the Solanum tuberosum StATG8 Family Associated with the WRKY Transcription Factor. Plants 2022, 11, 2858. [Google Scholar] [CrossRef]
- Zhang, B.; Huang, S.; Guo, S.; Meng, Y.; Tian, Y.; Zhou, Y.; Chen, H.; Li, X.; Zhou, J.; Chen, W. ATG6 Interacting with NPR1 Increases Arabidopsis thaliana Resistance to Pst DC3000/avrRps4 by Increasing Its Nuclear Accumulation and Stability; eLife Sciences Publications, Ltd.: Cambridge, UK, 2025. [Google Scholar]
- Wang, J.; Xu, C.; Sun, Q.; Xu, J.; Chai, Y.; Berg, G.; Cernava, T.; Ma, Z.; Chen, Y. Post-translational regulation of autophagy is involved in intra-microbiome suppression of fungal pathogens. Microbiome 2021, 9, 131. [Google Scholar] [CrossRef]
- Lin, L.Y.; Chow, H.X.; Chen, C.H.; Mitsuda, N.; Chou, W.C.; Liu, T.Y. Role of autophagy-related proteins ATG8f and ATG8h in the maintenance of autophagic activity in Arabidopsis roots under phosphate starvation. Front. Plant Sci. 2023, 14, 1018984. [Google Scholar] [CrossRef]
- Chen, Q.; Soulay, F.; Saudemont, B.; Elmayan, T.; Marmagne, A.; Masclaux-Daubresse, C. Overexpression of ATG8 in Arabidopsis Stimulates Autophagic Activity and Increases Nitrogen Remobilization Efficiency and Grain Filling. Plant Cell Physiol. 2018, 60, 343–352. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Zess, E.K.; Jensen, C.; Cruz-Mireles, N.; De la Concepcion, J.C.; Sklenar, J.; Stephani, M.; Imre, R.; Roitinger, E.; Hughes, R.; Belhaj, K.; et al. N-terminal β-strand underpins biochemical specialization of an ATG8 isoform. PLoS Biol. 2019, 17, e3000373. [Google Scholar] [CrossRef]
- Ibrahim, T.; Khandare, V.; Mirkin, F.G.; Tumtas, Y.; Bubeck, D.; Bozkurt, T.O. AlphaFold2-multimer guided high-accuracy prediction of typical and atypical ATG8-binding motifs. PLoS Biol. 2023, 21, e3001962. [Google Scholar] [CrossRef]
- Picchianti, L.; Sánchez de Medina Hernández, V.; Zhan, N.; Irwin, N.A.T.; Groh, R.; Stephani, M.; Hornegger, H.; Beveridge, R.; Sawa-Makarska, J.; Lendl, T.; et al. Shuffled ATG8 interacting motifs form an ancestral bridge between UFMylation and autophagy. EMBO J. 2023, 42, e112053. [Google Scholar] [CrossRef]
- Huh, S.U. Novel Functions of Arabidopsis Pumilio RNA-Binding Protein 6 in Salt Stress. Agronomy 2022, 12, 2410. [Google Scholar] [CrossRef]
- Offenborn, J.N.; Waadt, R.; Kudla, J. Visualization and translocation of ternary Calcineurin-A/Calcineurin-B/Calmodulin-2 protein complexes by dual-color trimolecular fluorescence complementation. New Phytol. 2015, 208, 269–279. [Google Scholar] [CrossRef]
- Hong, S.; Huh, S.U. Members of the Capsicum annuum CaTrxh Family Respond to High Temperature and Exhibit Dynamic Hetero/Homo Interactions. Int. J. Mol. Sci. 2024, 25, 1729. [Google Scholar] [CrossRef]
- Sarris, P.F.; Duxbury, Z.; Huh, S.U.; Ma, Y.; Segonzac, C.; Sklenar, J.; Derbyshire, P.; Cevik, V.; Rallapalli, G.; Saucet, S.B.; et al. A Plant Immune Receptor Detects Pathogen Effectors that Target WRKY Transcription Factors. Cell 2015, 161, 1089–1100. [Google Scholar] [CrossRef]
- Ma, Y.; Guo, H.; Hu, L.; Martinez, P.P.; Moschou, P.N.; Cevik, V.; Ding, P.; Duxbury, Z.; Sarris, P.F.; Jones, J.D.G. Distinct modes of derepression of an Arabidopsis immune receptor complex by two different bacterial effectors. Proc. Natl. Acad. Sci. USA 2018, 115, 10218–10227. [Google Scholar] [CrossRef]
- Huh, S.U. Optimization of immune receptor-related hypersensitive cell death response assay using agrobacterium-mediated transient expression in tobacco plants. Plant Methods 2022, 18, 57. [Google Scholar] [CrossRef]
- Huh, S.U.; Cevik, V.; Ding, P.; Duxbury, Z.; Ma, Y.; Tomlinson, L.; Sarris, P.F.; Jones, J.D.G. Protein-protein interactions in the RPS4/RRS1 immune receptor complex. PLoS Pathog. 2017, 13, e1006376. [Google Scholar] [CrossRef]
- Huang, R.; Xu, Y.; Wan, W.; Shou, X.; Qian, J.; You, Z.; Liu, B.; Chang, C.; Zhou, T.; Lippincott-Schwartz, J.; et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol. Cell 2015, 57, 456–466. [Google Scholar] [CrossRef]
- Jacomin, A.C.; Samavedam, S.; Promponas, V.; Nezis, I.P. iLIR database: A web resource for LIR motif-containing proteins in eukaryotes. Autophagy 2016, 12, 1945–1953. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Huh, S.U. PopP2 interacts with PAD4 in an acetyltransferase activity-dependent manner and affects plant immunity. Plant Signal Behav. 2021, 16, 2017631. [Google Scholar] [CrossRef]
- Xia, Y.; Zou, R.; Escouboué, M.; Zhong, L.; Zhu, C.; Pouzet, C.; Wu, X.; Wang, Y.; Lv, G.; Zhou, H.; et al. Secondary-structure switch regulates the substrate binding of a YopJ family acetyltransferase. Nat. Commun. 2021, 12, 5969. [Google Scholar] [CrossRef]
- Tasset, C.; Bernoux, M.; Jauneau, A.; Pouzet, C.; Brière, C.; Kieffer-Jacquinod, S.; Rivas, S.; Marco, Y.; Deslandes, L. Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1-R-mediated immunity in Arabidopsis. PLoS Pathog. 2010, 6, e1001202. [Google Scholar] [CrossRef] [PubMed]
- Mukhi, N.; Brown, H.; Gorenkin, D.; Ding, P.; Bentham, A.R.; Stevenson, C.E.M.; Jones, J.D.G.; Banfield, M.J. Perception of structurally distinct effectors by the integrated WRKY domain of a plant immune receptor. Proc. Natl. Acad. Sci. USA 2021, 118, e2113996118. [Google Scholar] [CrossRef]
- Bernoux, M.; Timmers, T.; Jauneau, A.; Brière, C.; de Wit, P.J.; Marco, Y.; Deslandes, L. RD19, an Arabidopsis cysteine protease required for RRS1-R-mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell 2008, 20, 2252–2264. [Google Scholar] [CrossRef]
- Huh, S.U. Bacteria manipulate autophagy through acetylation in both fungi and plants. Crop Des. 2024, 3, 100058. [Google Scholar] [CrossRef]
- Kournoutis, A.; Johansen, T. LC3B is a cofactor for LMX1B-mediated transcription of autophagy genes in dopaminergic neurons. J. Cell Biol. 2023, 222, e202303008. [Google Scholar] [CrossRef]
- Jiménez-Moreno, N.; Kollareddy, M.; Stathakos, P.; Moss, J.J.; Antón, Z.; Shoemark, D.K.; Sessions, R.B.; Witzgall, R.; Caldwell, M.; Lane, J.D. ATG8-dependent LMX1B-autophagy crosstalk shapes human midbrain dopaminergic neuronal resilience. J. Cell Biol. 2023, 222, e201910133. [Google Scholar] [CrossRef]
- Wang, L.; Klionsky, D.J.; Shen, H.M. The emerging mechanisms and functions of microautophagy. Nat. Rev. Mol. Cell Biol. 2023, 24, 186–203. [Google Scholar] [CrossRef]
- Vargas, J.N.S.; Hamasaki, M.; Kawabata, T.; Youle, R.J.; Yoshimori, T. The mechanisms and roles of selective autophagy in mammals. Nat. Rev. Mol. Cell Biol. 2023, 24, 167–185. [Google Scholar] [CrossRef]
- Yang, Z.; Klionsky, D.J. An overview of the molecular mechanism of autophagy. Curr. Top. Microbiol. Immunol. 2009, 335, 1–32. [Google Scholar]
- Macharia, M.W.; Tan, W.Y.Z.; Das, P.P.; Naqvi, N.I.; Wong, S.M. Proximity-dependent biotinylation screening identifies NbHYPK as a novel interacting partner of ATG8 in plants. BMC Plant Biol. 2019, 19, 326. [Google Scholar] [CrossRef]
- Tsapras, P.; Nezis, I.P. A yeast two-hybrid screening identifies novel Atg8a interactors in Drosophila. Autophagy 2022, 18, 1211–1212. [Google Scholar] [CrossRef]
- Wang, Q.; Shakoor, N.; Boyher, A.; Veley, K.M.; Berry, J.C.; Mockler, T.C.; Bart, R.S. Escalation in the host-pathogen arms race: A host resistance response corresponds to a heightened bacterial virulence response. PLoS Pathog. 2021, 17, e1009175. [Google Scholar] [CrossRef]
- Liu, G.; Zeng, H.; Li, X.; Wei, Y.; Shi, H. Functional Analysis of MaWRKY24 in Transcriptional Activation of Autophagy-Related Gene 8f/g and Plant Disease Susceptibility to Soil-Borne Fusarium oxysporum f. sp. cubense. Pathogens 2019, 8, 264. [Google Scholar] [CrossRef]
- Wang, P.; Nolan, T.M.; Yin, Y.; Bassham, D.C. Identification of transcription factors that regulate ATG8 expression and autophagy in Arabidopsis. Autophagy 2020, 16, 123–139. [Google Scholar] [CrossRef]
- Wang, Y.; Cai, S.; Yin, L.; Shi, K.; Xia, X.; Zhou, Y.; Yu, J.; Zhou, J. Tomato HsfA1a plays a critical role in plant drought tolerance by activating ATG genes and inducing autophagy. Autophagy 2015, 11, 2033–2047. [Google Scholar] [CrossRef]
- Zhu, T.; Zou, L.; Li, Y.; Yao, X.; Xu, F.; Deng, X.; Zhang, D.; Lin, H. Mitochondrial alternative oxidase-dependent autophagy involved in ethylene-mediated drought tolerance in Solanum lycopersicum. Plant Biotechnol. J. 2018, 16, 2063–2076. [Google Scholar] [CrossRef]
- Yang, M.; Wang, L.; Chen, C.; Guo, X.; Lin, C.; Huang, W.; Chen, L. Genome-wide analysis of autophagy-related genes in Medicago truncatula highlights their roles in seed development and response to drought stress. Sci. Rep. 2021, 11, 22933. [Google Scholar] [CrossRef]
- Nakatogawa, H.; Ichimura, Y.; Ohsumi, Y. Atg8, a Ubiquitin-like Protein Required for Autophagosome Formation, Mediates Membrane Tethering and Hemifusion. Cell 2007, 130, 165–178. [Google Scholar] [CrossRef]
- Iriondo, M.N.; Etxaniz, A.; Varela, Y.R.; Ballesteros, U.; Hervás, J.H.; Montes, L.R.; Goñi, F.M.; Alonso, A. LC3 subfamily in cardiolipin-mediated mitophagy: A comparison of the LC3A, LC3B and LC3C homologs. Autophagy 2022, 18, 2985–3003. [Google Scholar] [CrossRef]
- Jatana, N.; Ascher, D.B.; Pires, D.E.V.; Gokhale, R.S.; Thukral, L. Human LC3 and GABARAP subfamily members achieve functional specificity via specific structural modulations. Autophagy 2020, 16, 239–255. [Google Scholar] [CrossRef] [PubMed]
- Schaaf, M.B.; Keulers, T.G.; Vooijs, M.A.; Rouschop, K.M. LC3/GABARAP family proteins: Autophagy-(un)related functions. FASEB J. 2016, 30, 3961–3978. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huh, S.U. Interaction of Potato Autophagy-Related StATG8 Family Proteins with Pathogen Effector and WRKY Transcription Factor in the Nucleus. Microorganisms 2025, 13, 1589. https://doi.org/10.3390/microorganisms13071589
Huh SU. Interaction of Potato Autophagy-Related StATG8 Family Proteins with Pathogen Effector and WRKY Transcription Factor in the Nucleus. Microorganisms. 2025; 13(7):1589. https://doi.org/10.3390/microorganisms13071589
Chicago/Turabian StyleHuh, Sung Un. 2025. "Interaction of Potato Autophagy-Related StATG8 Family Proteins with Pathogen Effector and WRKY Transcription Factor in the Nucleus" Microorganisms 13, no. 7: 1589. https://doi.org/10.3390/microorganisms13071589
APA StyleHuh, S. U. (2025). Interaction of Potato Autophagy-Related StATG8 Family Proteins with Pathogen Effector and WRKY Transcription Factor in the Nucleus. Microorganisms, 13(7), 1589. https://doi.org/10.3390/microorganisms13071589





