Mutations in Genes with a Role in Cell Envelope Biosynthesis Render Gram-Negative Bacteria Highly Susceptible to the Anti-Infective Small Molecule D66
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Media, and Antibiotics
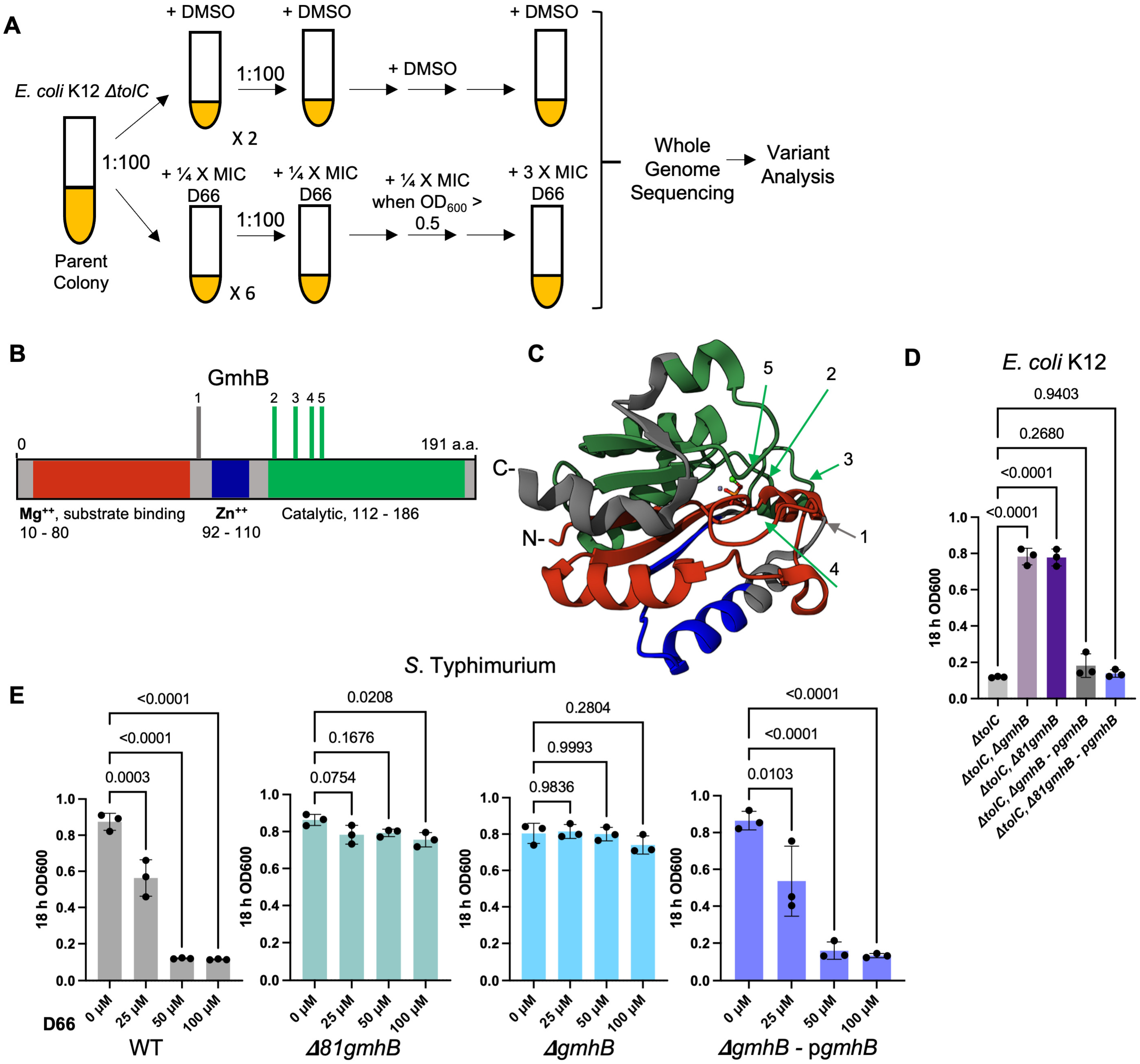
2.2. Evolution of D66-Resistant E. coli K12 ΔtolC Mutants and Genetic Analyses
2.3. Construction of E. coli K12 and S. Typhimurium SL1344 gmhB Mutant Strains
2.4. Plasmid Construction for Complementation of gmhB
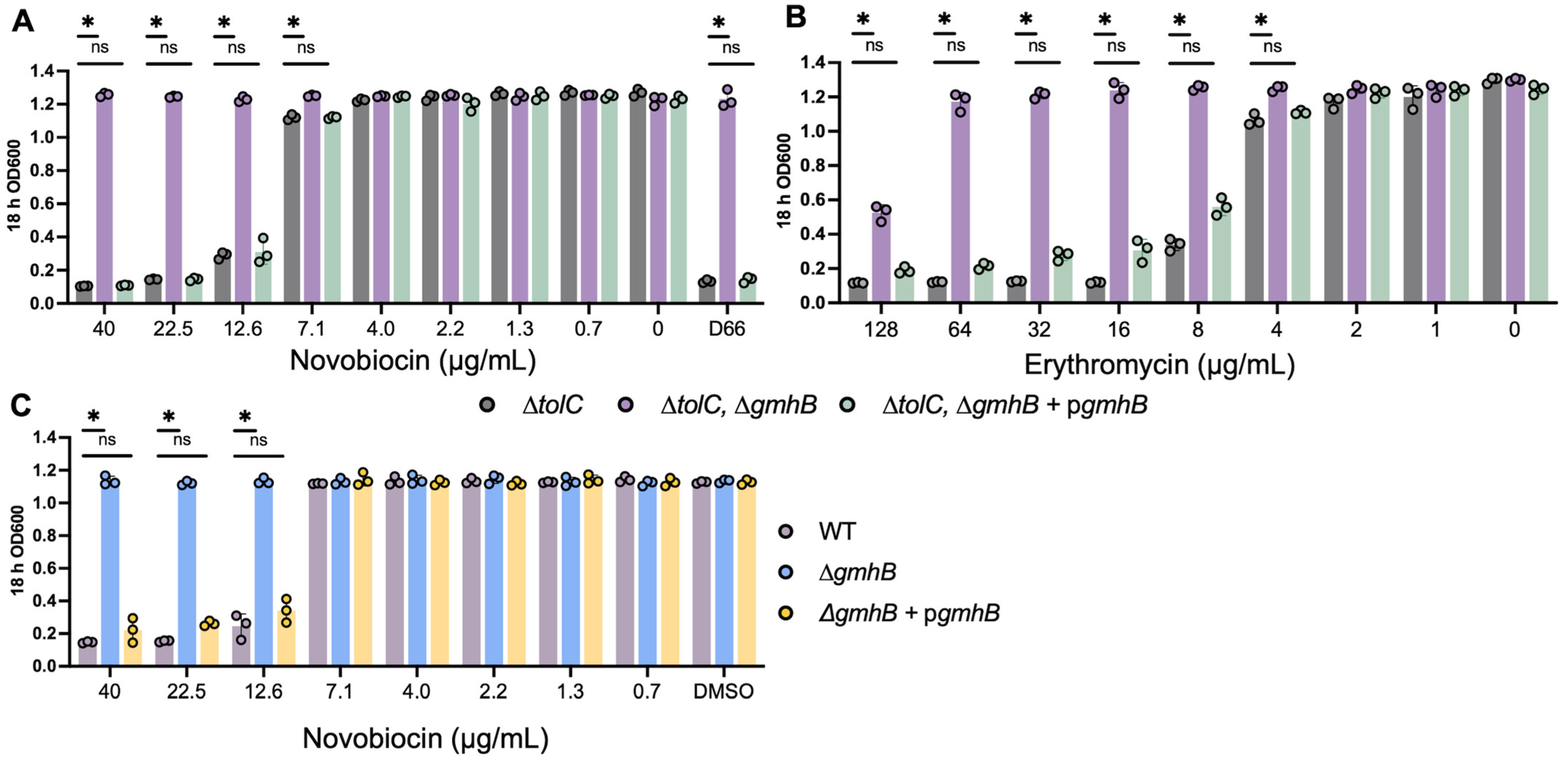
2.5. Sensitivity to Novobiocin and Erythromycin
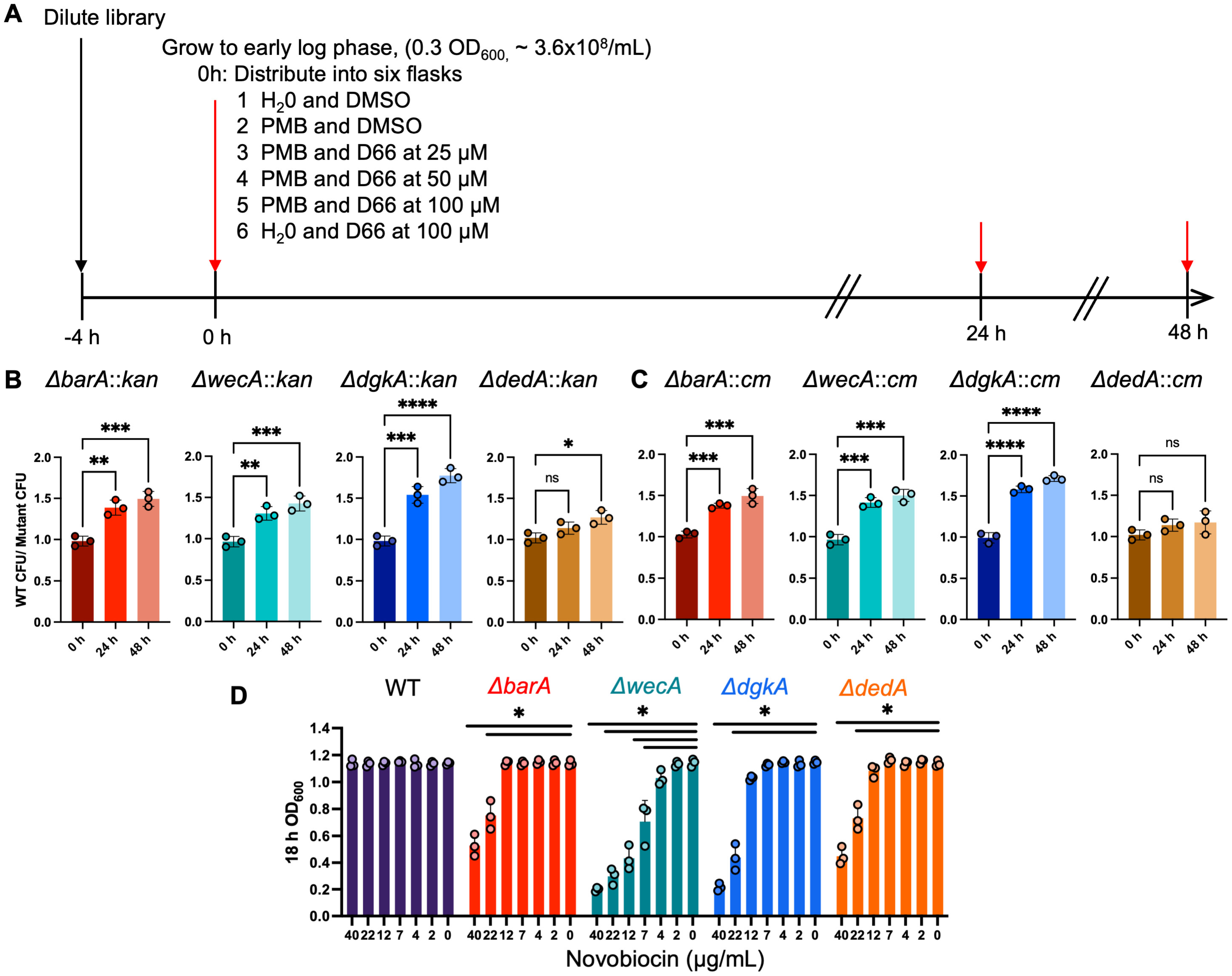
2.6. Selection of Transposon Mutants with Reduced Fitness in the Presence of D66
2.7. Validation of Transposon Mutants in Competitive Fitness Assays
2.8. Statistical Analyses
3. Results
3.1. Mutations in gmhB/yaeD Confer Heritable D66 Resistance in Broth
3.2. Strains Lacking gmhB Have a Less Permeable Cell Envelope
3.3. Competition for Growth Identifies Transposon Mutants with Reduced Fitness in the Presence of D66
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zgurskaya, H.I.; Rybenkov, V.V.; Krishnamoorthy, G.; Leus, I.V. Trans-Envelope Multidrug Efflux Pumps of Gram-Negative Bacteria and Their Synergism with the Outer Membrane Barrier. Res. Microbiol. 2018, 169, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Bertani, B.; Ruiz, N. Function and Biogenesis of Lipopolysaccharides. EcoSal Plus 2018, 8. [Google Scholar] [CrossRef]
- Clifton, L.A.; Skoda, M.W.A.; Le Brun, A.P.; Ciesielski, F.; Kuzmenko, I.; Holt, S.A.; Lakey, J.H. Effect of Divalent Cation Removal on the Structure of Gram-Negative Bacterial Outer Membrane Models. Langmuir 2015, 31, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Farhana, A.; Khan, Y.S. Biochemistry, Lipopolysaccharide. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Hews, C.L.; Cho, T.; Rowley, G.; Raivio, T.L. Maintaining Integrity Under Stress: Envelope Stress Response Regulation of Pathogenesis in Gram-Negative Bacteria. Front. Cell. Infect. Microbiol. 2019, 9, 313. [Google Scholar] [CrossRef]
- Li, X.-Z.; Plésiat, P.; Nikaido, H. The Challenge of Efflux-Mediated Antibiotic Resistance in Gram-Negative Bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef]
- Davis, K.P.; Morales, Y.; Ende, R.J.; Peters, R.; McCabe, A.L.; Mecsas, J.; Aldridge, B.B. Critical Role of Growth Medium for Detecting Drug Interactions in Gram-Negative Bacteria That Model in Vivo Responses. mBio 2024, 15, e0015924. [Google Scholar] [CrossRef]
- Nizet, V. The Accidental Orthodoxy of Drs. Mueller and Hinton. EBioMedicine 2017, 22, 26–27. [Google Scholar] [CrossRef] [PubMed]
- Lesic, B.; Lépine, F.; Déziel, E.; Zhang, J.; Zhang, Q.; Padfield, K.; Castonguay, M.-H.; Milot, S.; Stachel, S.; Tzika, A.A.; et al. Inhibitors of Pathogen Intercellular Signals as Selective Anti-Infective Compounds. PLoS Pathog. 2007, 3, 1229–1239. [Google Scholar] [CrossRef]
- Nikaido, H. Outer Membrane of Salmonella Typhimurium. Transmembrane Diffusion of Some Hydrophobic Substances. Biochim. Biophys. Acta 1976, 433, 118–132. [Google Scholar] [CrossRef]
- Augustus, A.M.; Celaya, T.; Husain, F.; Humbard, M.; Misra, R. Antibiotic-Sensitive TolC Mutants and Their Suppressors. J. Bacteriol. 2004, 186, 1851–1860. [Google Scholar] [CrossRef]
- Kumaraswamy, M.; Lin, L.; Olson, J.; Sun, C.-F.; Nonejuie, P.; Corriden, R.; Döhrmann, S.; Ali, S.R.; Amaro, D.; Rohde, M.; et al. Standard Susceptibility Testing Overlooks Potent Azithromycin Activity and Cationic Peptide Synergy against MDR Stenotrophomonas Maltophilia. J. Antimicrob. Chemother. 2016, 71, 1264–1269. [Google Scholar] [CrossRef] [PubMed]
- Ulloa, E.R.; Kousha, A.; Tsunemoto, H.; Pogliano, J.; Licitra, C.; LiPuma, J.J.; Sakoulas, G.; Nizet, V.; Kumaraswamy, M. Azithromycin Exerts Bactericidal Activity and Enhances Innate Immune Mediated Killing of MDR Achromobacter Xylosoxidans. Infect. Microbes Dis. 2020, 2, 10–17. [Google Scholar] [CrossRef]
- Heesterbeek, D.A.C.; Martin, N.I.; Velthuizen, A.; Duijst, M.; Ruyken, M.; Wubbolts, R.; Rooijakkers, S.H.M.; Bardoel, B.W. Complement-Dependent Outer Membrane Perturbation Sensitizes Gram-Negative Bacteria to Gram-Positive Specific Antibiotics. Sci. Rep. 2019, 9, 3074. [Google Scholar] [CrossRef]
- Sakoulas, G.; Kumaraswamy, M.; Kousha, A.; Nizet, V. Interaction of Antibiotics with Innate Host Defense Factors against Salmonella Enterica Serotype Newport. mSphere 2017, 2, e00410-17. [Google Scholar] [CrossRef] [PubMed]
- Reens, A.L.; Crooks, A.L.; Su, C.-C.; Nagy, T.A.; Reens, D.L.; Podoll, J.D.; Edwards, M.E.; Yu, E.W.; Detweiler, C.S. A Cell-Based Infection Assay Identifies Efflux Pump Modulators That Reduce Bacterial Intracellular Load. PLoS Pathog. 2018, 14, e1007115. [Google Scholar] [CrossRef]
- Ellis, M.J.; Tsai, C.N.; Johnson, J.W.; French, S.; Elhenawy, W.; Porwollik, S.; Andrews-Polymenis, H.; McClelland, M.; Magolan, J.; Coombes, B.K.; et al. A Macrophage-Based Screen Identifies Antibacterial Compounds Selective for Intracellular Salmonella Typhimurium. Nat. Commun. 2019, 10, 197. [Google Scholar] [CrossRef]
- Tsai, C.N.; Massicotte, M.-A.; MacNair, C.R.; Perry, J.N.; Brown, E.D.; Coombes, B.K. Screening under Infection-Relevant Conditions Reveals Chemical Sensitivity in Multidrug Resistant Invasive Non-Typhoidal Salmonella (iNTS). RSC Chem. Biol. 2023, 4, 600–612. [Google Scholar] [CrossRef]
- Xie, T.; Liu, G.; Ma, J.; Wang, Y.; Gao, R.; Geng, S.; Jiao, X.; Barrow, P. Nifuratel Reduces Salmonella Survival in Macrophages by Extracellular and Intracellular Antibacterial Activity. Microbiol. Spectr. 2023, 11, e0514722. [Google Scholar] [CrossRef]
- Nagy, T.A.; Crooks, A.L.; Quintana, J.L.J.; Detweiler, C.S. Clofazimine Reduces the Survival of Salmonella Enterica in Macrophages and Mice. ACS Infect. Dis. 2020, 6, 1238–1249. [Google Scholar] [CrossRef]
- Dombach, J.L.; Quintana, J.L.J.; Nagy, T.A.; Wan, C.; Crooks, A.L.; Yu, H.; Su, C.-C.; Yu, E.W.; Shen, J.; Detweiler, C.S. A Small Molecule That Mitigates Bacterial Infection Disrupts Gram-Negative Cell Membranes and Is Inhibited by Cholesterol and Neutral Lipids. PLoS Pathog. 2020, 16, e1009119. [Google Scholar] [CrossRef]
- Pearson eText for Brock Biology of Microorganisms, 15e—Exchange. Available online: https://exchange.pearson.com/products/e9088234-75f8-4b2c-9e87-e06c0c4aec3f/pearson-etext-for-brock-biology-of-microorganisms-15e?uuid=e9088234-75f8-4b2c-9e87-e06c0c4aec3f (accessed on 29 July 2024).
- Mohiuddin, S.G.; Ghosh, S.; Kavousi, P.; Orman, M.A. Proton Motive Force Inhibitors Are Detrimental to Methicillin-Resistant Staphylococcus Aureus Strains. Microbiol. Spectr. 2022, 10, e0202422. [Google Scholar] [CrossRef]
- Villanueva, J.A.; Crooks, A.L.; Nagy, T.A.; Quintana, J.L.J.; Dalebroux, Z.D.; Detweiler, C.S. Salmonella Enterica Infections Are Disrupted by Two Small Molecules That Accumulate within Phagosomes and Differentially Damage Bacterial Inner Membranes. mBio 2022, e0179022. [Google Scholar] [CrossRef]
- Dombach, J.L.; Quintana, J.L.; Allgood, S.C.; Nagy, T.A.; Gustafson, D.L.; Detweiler, C.S. A Small Molecule That Disrupts S. Typhimurium Membrane Voltage without Cell Lysis Reduces Bacterial Colonization of Mice. PLoS Pathog. 2022, 18, e1010606. [Google Scholar] [CrossRef] [PubMed]
- Heithoff, D.M.; Mahan, S.P.; Barnes, V.L.; Leyn, S.A.; George, C.X.; Zlamal, J.E.; Limwongyut, J.; Bazan, G.C.; Fried, J.C.; Fitzgibbons, L.N.; et al. A Broad-Spectrum Synthetic Antibiotic That Does Not Evoke Bacterial Resistance. EBioMedicine 2023, 89, 104461. [Google Scholar] [CrossRef] [PubMed]
- Dombach, J.L.; Christensen, G.L.; Allgood, S.C.; Quintana, J.L.J.; Detweiler, C.S. Inhibition of Multiple Staphylococcal Growth States by a Small Molecule That Disrupts Membrane Fluidity and Voltage. mSphere 2024, 9, e0077223. [Google Scholar] [CrossRef]
- Ofek, I.; Cohen, S.; Rahmani, R.; Kabha, K.; Tamarkin, D.; Herzig, Y.; Rubinstein, E. Antibacterial Synergism of Polymyxin B Nonapeptide and Hydrophobic Antibiotics in Experimental Gram-Negative Infections in Mice. Antimicrob. Agents Chemother. 1994, 38, 374–377. [Google Scholar] [CrossRef] [PubMed]
- Nishino, K.; Nikaido, E.; Yamaguchi, A. Regulation and Physiological Function of Multidrug Efflux Pumps in Escherichia coli and Salmonella. Biochim. Biophys. Acta 2009, 1794, 834–843. [Google Scholar] [CrossRef]
- Bertani, G. Studies on Lysogenesis. I. The Mode of Phage Liberation by Lysogenic Escherichia coli. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [CrossRef]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-Frame, Single-Gene Knockout Mutants: The Keio Collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef]
- Montaño, E.T.; Nideffer, J.F.; Sugie, J.; Enustun, E.; Shapiro, A.B.; Tsunemoto, H.; Derman, A.I.; Pogliano, K.; Pogliano, J. Bacterial Cytological Profiling Identifies Rhodanine-Containing PAINS Analogs as Specific Inhibitors of Escherichia coli Thymidylate Kinase In Vivo. J. Bacteriol. 2021, 203. [Google Scholar] [CrossRef]
- Hoiseth, S.K.; Stocker, B.A.D. Aromatic-Dependent Salmonella Typhimurium Are Non-Virulent and Effective as Live Vaccines. Nature 1981, 291, 238–239. [Google Scholar] [CrossRef] [PubMed]
- Santiviago, C.A.; Reynolds, M.M.; Porwollik, S.; Choi, S.-H.; Long, F.; Andrews-Polymenis, H.L.; McClelland, M. Analysis of Pools of Targeted Salmonella Deletion Mutants Identifies Novel Genes Affecting Fitness during Competitive Infection in Mice. PLoS Pathog. 2009, 5, e1000477. [Google Scholar] [CrossRef]
- Deatherage, D.E.; Barrick, J.E. Identification of Mutations in Laboratory-Evolved Microbes from next-Generation Sequencing Data Using Breseq. Methods Mol. Biol. 2014, 1151, 165–188. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-Step Inactivation of Chromosomal Genes in Escherichia coli K-12 Using PCR Products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
- Canals, R.; Xia, X.-Q.; Fronick, C.; Clifton, S.W.; Ahmer, B.M.M.; Andrews-Polymenis, H.L.; Porwollik, S.; McClelland, M. High-Throughput Comparison of Gene Fitness among Related Bacteria. BMC Genom. 2012, 13, 212. [Google Scholar] [CrossRef]
- Jayeola, V.; McClelland, M.; Porwollik, S.; Chu, W.; Farber, J.; Kathariou, S. Identification of Novel Genes Mediating Survival of Salmonella on Low-Moisture Foods via Transposon Sequencing Analysis. Front. Microbiol. 2020, 11, 726. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Porwollik, S.; Santiviago, C.A.; Cheng, P.; Long, F.; Desai, P.; Fredlund, J.; Srikumar, S.; Silva, C.A.; Chu, W.; Chen, X.; et al. Defined Single-Gene and Multi-Gene Deletion Mutant Collections in Salmonella Enterica Sv Typhimurium. PLoS ONE 2014, 9, e99820. [Google Scholar] [CrossRef]
- Stanley, S.A.; Grant, S.S.; Kawate, T.; Iwase, N.; Shimizu, M.; Wivagg, C.; Silvis, M.; Kazyanskaya, E.; Aquadro, J.; Golas, A.; et al. Identification of Novel Inhibitors of M. Tuberculosis Growth Using Whole Cell Based High-Throughput Screening. ACS Chem. Biol. 2012, 7, 1377–1384. [Google Scholar] [CrossRef]
- La Rosa, V.; Poce, G.; Canseco, J.O.; Buroni, S.; Pasca, M.R.; Biava, M.; Raju, R.M.; Porretta, G.C.; Alfonso, S.; Battilocchio, C.; et al. MmpL3 Is the Cellular Target of the Antitubercular Pyrrole Derivative BM212. Antimicrob. Agents Chemother. 2012, 56, 324–331. [Google Scholar] [CrossRef]
- Cho, H.J.; Misra, R. Mutational Activation of Antibiotic Resistant Mechanisms in the Absence of Major Drug Efflux Systems of Escherichia coli. J. Bacteriol. 2021, 203, e0010921. [Google Scholar] [CrossRef]
- Klenotic, P.A.; Yu, E.W. Structural Analysis of Resistance-Nodulation Cell Division Transporters. Microbiol. Mol. Biol. Rev. 2024, 88, e0019823. [Google Scholar] [CrossRef]
- Kneidinger, B.; Marolda, C.; Graninger, M.; Zamyatina, A.; McArthur, F.; Kosma, P.; Valvano, M.A.; Messner, P. Biosynthesis Pathway of ADP-L-Glycero-Beta-D-Manno-Heptose in Escherichia coli. J. Bacteriol. 2002, 184, 363–369. [Google Scholar] [CrossRef]
- Nobre, T.M.; Martynowycz, M.W.; Andreev, K.; Kuzmenko, I.; Nikaido, H.; Gidalevitz, D. Modification of Salmonella Lipopolysaccharides Prevents the Outer Membrane Penetration of Novobiocin. Biophys. J. 2015, 109, 2537–2545. [Google Scholar] [CrossRef]
- May, J.M.; Owens, T.W.; Mandler, M.D.; Simpson, B.W.; Lazarus, M.B.; Sherman, D.J.; Davis, R.M.; Okuda, S.; Massefski, W.; Ruiz, N.; et al. The Antibiotic Novobiocin Binds and Activates the ATPase That Powers Lipopolysaccharide Transport. J. Am. Chem. Soc. 2017, 139, 17221–17224. [Google Scholar] [CrossRef]
- Nikaido, H.; Vaara, M. Molecular Basis of Bacterial Outer Membrane Permeability. Microbiol. Rev. 1985, 49, 1–32. [Google Scholar] [CrossRef]
- Mandler, M.D.; Baidin, V.; Lee, J.; Pahil, K.S.; Owens, T.W.; Kahne, D. Novobiocin Enhances Polymyxin Activity by Stimulating Lipopolysaccharide Transport. J. Am. Chem. Soc. 2018, 140, 6749–6753. [Google Scholar] [CrossRef]
- Todor, H.; Herrera, N.; Gross, C. Three Bacterial DedA Subfamilies with Distinct Functions and Phylogenetic Distribution. bioRxiv 2023, 14, e00028-23. [Google Scholar] [CrossRef]
- Schmidt, G.; Mayer, H.; Mäkelä, P.H. Presence of Rfe Genes in Escherichia coli: Their Participation in Biosynthesis of O Antigen and Enterobacterial Common Antigen. J. Bacteriol. 1976, 127, 755–762. [Google Scholar] [CrossRef]
- Purcell, A.B.; Voss, B.J.; Trent, M.S. Diacylglycerol Kinase A Is Essential for Polymyxin Resistance Provided by EptA, MCR-1, and Other Lipid A Phosphoethanolamine Transferases. J. Bacteriol. 2022, 204, e00498-21. [Google Scholar] [CrossRef]
- Prouty, A.M.; Gunn, J.S. Salmonella Enterica Serovar Typhimurium Invasion Is Repressed in the Presence of Bile. Infect. Immun. 2000, 68, 6763–6769. [Google Scholar] [CrossRef]
- Coombes, B.K.; Brown, N.F.; Valdez, Y.; Brumell, J.H.; Finlay, B.B. Expression and Secretion of Salmonella Pathogenicity Island-2 Virulence Genes in Response to Acidification Exhibit Differential Requirements of a Functional Type III Secretion Apparatus and SsaL. J. Biol. Chem. 2004, 279, 49804–49815. [Google Scholar] [CrossRef]
- Beuzón, C.R.; Banks, G.; Deiwick, J.; Hensel, M.; Holden, D.W. pH-Dependent Secretion of SseB, a Product of the SPI-2 Type III Secretion System of Salmonella Typhimurium. Mol. Microbiol. 1999, 33, 806–816. [Google Scholar] [CrossRef] [PubMed]
- Teplitski, M.; Goodier, R.I.; Ahmer, B.M.M. Pathways Leading from BarA/SirA to Motility and Virulence Gene Expression in Salmonella. J. Bacteriol. 2003, 185, 7257–7265. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.Y.; Gui, G.; Wei, B.; Preston, J.F.; Oakford, L.; Yüksel, U.; Giedroc, D.P.; Romeo, T. The RNA Molecule CsrB Binds to the Global Regulatory Protein CsrA and Antagonizes Its Activity in Escherichia coli. J. Biol. Chem. 1997, 272, 17502–17510. [Google Scholar] [CrossRef] [PubMed]
- Ricci, V.; Attah, V.; Overton, T.; Grainger, D.C.; Piddock, L.J.V. CsrA Maximizes Expression of the AcrAB Multidrug Resistance Transporter. Nucleic Acids Res. 2017, 45, 12798–12807. [Google Scholar] [CrossRef]
- Alexander, D.C.; Valvano, M.A. Role of the Rfe Gene in the Biosynthesis of the Escherichia coli O7-Specific Lipopolysaccharide and Other O-Specific Polysaccharides Containing N-Acetylglucosamine. J. Bacteriol. 1994, 176, 7079–7084. [Google Scholar] [CrossRef]
- Barua, S.; Yamashino, T.; Hasegawa, T.; Yokoyama, K.; Torii, K.; Ohta, M. Involvement of Surface Polysaccharides in the Organic Acid Resistance of Shiga Toxin-Producing Escherichia coli O157:H7. Mol. Microbiol. 2002, 43, 629–640. [Google Scholar] [CrossRef]
- Roney, I.J.; Rudner, D.Z. Two Broadly Conserved Families of Polyprenyl-Phosphate Transporters. Nature 2023, 613, 729–734. [Google Scholar] [CrossRef]
- Tiwari, V.; Sharma, A.; Braga, R.; Garcia, E.; Appiah, R.; Fleeman, R.; Abuaita, B.H.; Patrauchan, M.; Doerrler, W.T. Klebsiella Pneumoniae DedA Family Proteins Have Redundant Roles in Divalent Cation Homeostasis and Resistance to Phagocytosis. Microbiol. Spectr. 2024, 12, e0380723. [Google Scholar] [CrossRef]
- Kumar, S.; Doerrler, W.T. Members of the Conserved DedA Family Are Likely Membrane Transporters and Are Required for Drug Resistance in Escherichia coli. Antimicrob. Agents Chemother. 2014, 58, 923–930. [Google Scholar] [CrossRef]
- Thompkins, K.; Chattopadhyay, B.; Xiao, Y.; Henk, M.C.; Doerrler, W.T. Temperature Sensitivity and Cell Division Defects in an Escherichia coli Strain with Mutations in yghB and yqjA, Encoding Related and Conserved Inner Membrane Proteins. J. Bacteriol. 2008, 190, 4489–4500. [Google Scholar] [CrossRef]
- Wahl, A.; My, L.; Dumoulin, R.; Sturgis, J.N.; Bouveret, E. Antagonistic Regulation of dgkA and plsB Genes of Phospholipid Synthesis by Multiple Stress Responses in Escherichia coli. Mol. Microbiol. 2011, 80, 1260–1275. [Google Scholar] [CrossRef]
- Van Horn, W.D.; Sanders, C.R. Prokaryotic Diacylglycerol Kinase and Undecaprenol Kinase. Annu. Rev. Biophys. 2012, 41, 81–101. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.L.; Sugiman-Marangos, S.; Zhang, K.; Valvano, M.A.; Wright, G.D.; Junop, M.S. Structural and Kinetic Characterization of the LPS Biosynthetic Enzyme D-Alpha,Beta-D-Heptose-1,7-Bisphosphate Phosphatase (GmhB) from Escherichia coli. Biochemistry 2010, 49, 1033–1041. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; She, Y.; Dong, N.; Li, P.; He, H.; Borio, A.; Wu, Q.; Lu, S.; Ding, X.; Cao, Y.; et al. Alpha-Kinase 1 Is a Cytosolic Innate Immune Receptor for Bacterial ADP-Heptose. Nature 2018, 561, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Holmes, C.L.; Smith, S.N.; Gurczynski, S.J.; Severin, G.B.; Unverdorben, L.V.; Vornhagen, J.; Mobley, H.L.T.; Bachman, M.A. The ADP-Heptose Biosynthesis Enzyme GmhB Is a Conserved Gram-Negative Bacteremia Fitness Factor. Infect. Immun. 2022, 90, e0022422. [Google Scholar] [CrossRef]
- Corti, S.; Chevalier, J.; Cremieux, A. Intracellular Accumulation of Norfloxacin in Mycobacterium Smegmatis. Antimicrob. Agents Chemother. 1995, 39, 2466–2471. [Google Scholar] [CrossRef][Green Version]
- Piddock, L.J.; Williams, K.J.; Ricci, V. Accumulation of Rifampicin by Mycobacterium Aurum, Mycobacterium Smegmatis and Mycobacterium Tuberculosis. J. Antimicrob. Chemother. 2000, 45, 159–165. [Google Scholar] [CrossRef]
- Goldman, R.C. Why Are Membrane Targets Discovered by Phenotypic Screens and Genome Sequencing in Mycobacterium Tuberculosis? Tuberculosis 2013, 93, 569–588. [Google Scholar] [CrossRef]
- Tahlan, K.; Wilson, R.; Kastrinsky, D.B.; Arora, K.; Nair, V.; Fischer, E.; Barnes, S.W.; Walker, J.R.; Alland, D.; Barry, C.E.; et al. SQ109 Targets MmpL3, a Membrane Transporter of Trehalose Monomycolate Involved in Mycolic Acid Donation to the Cell Wall Core of Mycobacterium Tuberculosis. Antimicrob. Agents Chemother. 2012, 56, 1797–1809. [Google Scholar] [CrossRef] [PubMed]
- Grzegorzewicz, A.E.; Pham, H.; Gundi, V.A.K.B.; Scherman, M.S.; North, E.J.; Hess, T.; Jones, V.; Gruppo, V.; Born, S.E.M.; Korduláková, J.; et al. Inhibition of Mycolic Acid Transport across the Mycobacterium Tuberculosis Plasma Membrane. Nat. Chem. Biol. 2012, 8, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Moorey, A.R.; Cabanillas, A.; Batt, S.M.; Ghidelli-Disse, S.; Urones, B.; Sanz, O.; Lelievre, J.; Bantscheff, M.; Cox, L.R.; Besra, G.S. The Multi-Target Aspect of an MmpL3 Inhibitor: The BM212 Series of Compounds Bind EthR2, a Transcriptional Regulator of Ethionamide Activation. Cell Surf. 2021, 7, 100068. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.T.; Haiderer, E.R.; Coulson, G.B.; Conner, K.N.; Ellsworth, E.; Chen, C.; Alvarez-Cabrera, N.; Li, W.; Jackson, M.; Dick, T.; et al. Identification of New MmpL3 Inhibitors by Untargeted and Targeted Mutant Screens Defines MmpL3 Domains with Differential Resistance. Antimicrob. Agents Chemother. 2019, 63, e00547-19. [Google Scholar] [CrossRef]
- Williams, J.T.; Giletto, M.; Haiderer, E.R.; Aleiwi, B.; Krieger-Burke, T.; Ellsworth, E.; Abramovitch, R.B. The Mycobacterium Tuberculosis MmpL3 Inhibitor MSU-43085 Is Active in a Mouse Model of Infection. Microbiol. Spectr. 2024, 12, e0367723. [Google Scholar] [CrossRef]
| Reference # | Strain Name | Species | Antibiotic | Source |
|---|---|---|---|---|
| ALR1247 | K–12 derivative BW25113 | E. coli | [31] | |
| JLD1285 | K–12 ΔtolC | E. coli | Kan | [32] |
| SCA1496 | K12 ΔtolC, ΔgmhB | E. coli | Kan | This Study |
| SCA1497 | K12 ΔtolC, Δ81gmhB | E. coli | Kan | This Study |
| SCA1544 | K12 ΔtolC, ΔgmhB-pBAD33-gmhB | E. coli | Kan, Cm | This Study |
| SCA1547 | K12 ΔtolC, Δ81gmhB-pBAD33-gmhB | E. coli | Kan, Cm | This Study |
| CSD001 | SL1344 | S. Typhimurium | Str | [33] |
| SCA1498 | SL1344 ΔgmhB | S. Typhimurium | Str, Kan | This Study |
| SCA1499 | SL1344 Δ81gmhB | S. Typhimurium | Str, Kan | This Study |
| SCA1546 | SL1344 ΔgmhB-pBAD33-gmhB | S. Typhimurium | Str, Kan, Cm | This Study |
| CSD1347 | 14028s; MZ0431 | S. Typhimurium | Str | [34] |
| CSD1351 | 14028s:CM; MZ2758 | S. Typhimurium | Str, Cm | [34] |
| CSD1352 | 14028s:KAN; MZ2770 | S. Typhimurium | Str, Kan | [34] |
| SCA1375 | 14028s STM14_2915; ΔdedA | S. Typhimurium | Kan | [34] |
| SCA1373 | 14028s STM14_4716; Δrfe | S. Typhimurium | Kan | [34] |
| SCA1369 | 14028s STM14_3566; ΔbarA | S. Typhimurium | Kan | [34] |
| SCA1371 | 14028s STM14_5093; ΔdgkA | S. Typhimurium | Kan | [34] |
| SCA1376 | 14028s STM14_2915; ΔdedA | S. Typhimurium | Cm | [34] |
| SCA1374 | 14028s STM14_4716; Δrfe | S. Typhimurium | Cm | [34] |
| SCA1370 | 14028s STM14_3566; ΔbarA | S. Typhimurium | Cm | [34] |
| SCA1372 | 14028s STM14_5093; ΔdgkA | S. Typhimurium | CM | [34] |
| # | Primer | Primer Sequence |
|---|---|---|
| 1 | gmhB LR FWD | 5′GTGTAGGCTGGAGCTGCTTCTCGAAACATGCGATACTAGCGTCACATGCCTTATTAAGGAGCTATAAAAG |
| 2 | gmhB LR Del REV | 5′AGGAAGACAAGCGGAAAAATGCATTTTTATTTCAACCGCTCATCTTTTAAATGGGAATTAGCCATGGTCC |
| 3 | gmhB LR Trunc REV | 5′CCCTGCGGATGATGCGGGCAATAATAGATACCATCCAGATCGACATCTCGATGGGAATTAGCCATGGTCC |
| 4 | pBAD33 amplification FWD | 5′GCAAAAACCGGCACAATGATCTGATTCGTTACCAATTATGACAACTTGACGGCTACAT |
| 5 | pBAD33 amplification REV | 5′CTCTTCGCATAAACCTGATTGGAAGATCGGGCTCGCC |
| 6 | gmhB gene amplification FWD | 5′AGTGGCGAGCCCGATCTTCCAATCAGGTTTATGCGAAGAGCACTT |
| 7 | gmhB gene amplification REV | 5′CATAATTGGTAACGAATCAGATCATTGTGCCGGTTTTTGCTG |
| Strain | Gene | Nt Position | Mutation | Effect |
|---|---|---|---|---|
| Mutant 1 | gmhB | 223,075 | TTC → ATT | Premature stop |
| Mutant 2 | gmhB | 223,174 | CAC → CGC | His to Arg |
| Mutant 3 | gmhB | 223,184 | CTTTT → CTTA | Frameshift |
| Mutant 4 | gmhB | 223,192 | GCA → GGA | Ala to Gly |
| Mutant 5 | gmhB | 223,200 | TAT → TTT | Tyr to Phe |
| Mutant 6 | ygeG | 2,991,548 | CT → CTT | Frameshift |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Allgood, S.C.; Ewing, C.A.; Chu, W.; Porwollik, S.; McClelland, M.; Detweiler, C.S. Mutations in Genes with a Role in Cell Envelope Biosynthesis Render Gram-Negative Bacteria Highly Susceptible to the Anti-Infective Small Molecule D66. Microorganisms 2025, 13, 1521. https://doi.org/10.3390/microorganisms13071521
Allgood SC, Ewing CA, Chu W, Porwollik S, McClelland M, Detweiler CS. Mutations in Genes with a Role in Cell Envelope Biosynthesis Render Gram-Negative Bacteria Highly Susceptible to the Anti-Infective Small Molecule D66. Microorganisms. 2025; 13(7):1521. https://doi.org/10.3390/microorganisms13071521
Chicago/Turabian StyleAllgood, Samual C., Calvin A. Ewing, Weiping Chu, Steffen Porwollik, Michael McClelland, and Corrella S. Detweiler. 2025. "Mutations in Genes with a Role in Cell Envelope Biosynthesis Render Gram-Negative Bacteria Highly Susceptible to the Anti-Infective Small Molecule D66" Microorganisms 13, no. 7: 1521. https://doi.org/10.3390/microorganisms13071521
APA StyleAllgood, S. C., Ewing, C. A., Chu, W., Porwollik, S., McClelland, M., & Detweiler, C. S. (2025). Mutations in Genes with a Role in Cell Envelope Biosynthesis Render Gram-Negative Bacteria Highly Susceptible to the Anti-Infective Small Molecule D66. Microorganisms, 13(7), 1521. https://doi.org/10.3390/microorganisms13071521






