Ni2+ and Cd2+ Biosorption Capacity and Redox-Mediated Toxicity Reduction in Bacterial Strains from Highly Contaminated Soils of Uzbekistan
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteria Strains
2.2. Preparation of Selected Bacteria Biomass
2.3. Determination of Bacterial Biosorption Capacity Under Variable Environmental Conditions
2.4. Study of Redox Process Reducing the Toxicity of Heavy Metal Cations
2.5. Observation of Oxidation–Reduction of Ni2+ and Cd2+ by Bacteria
3. Results
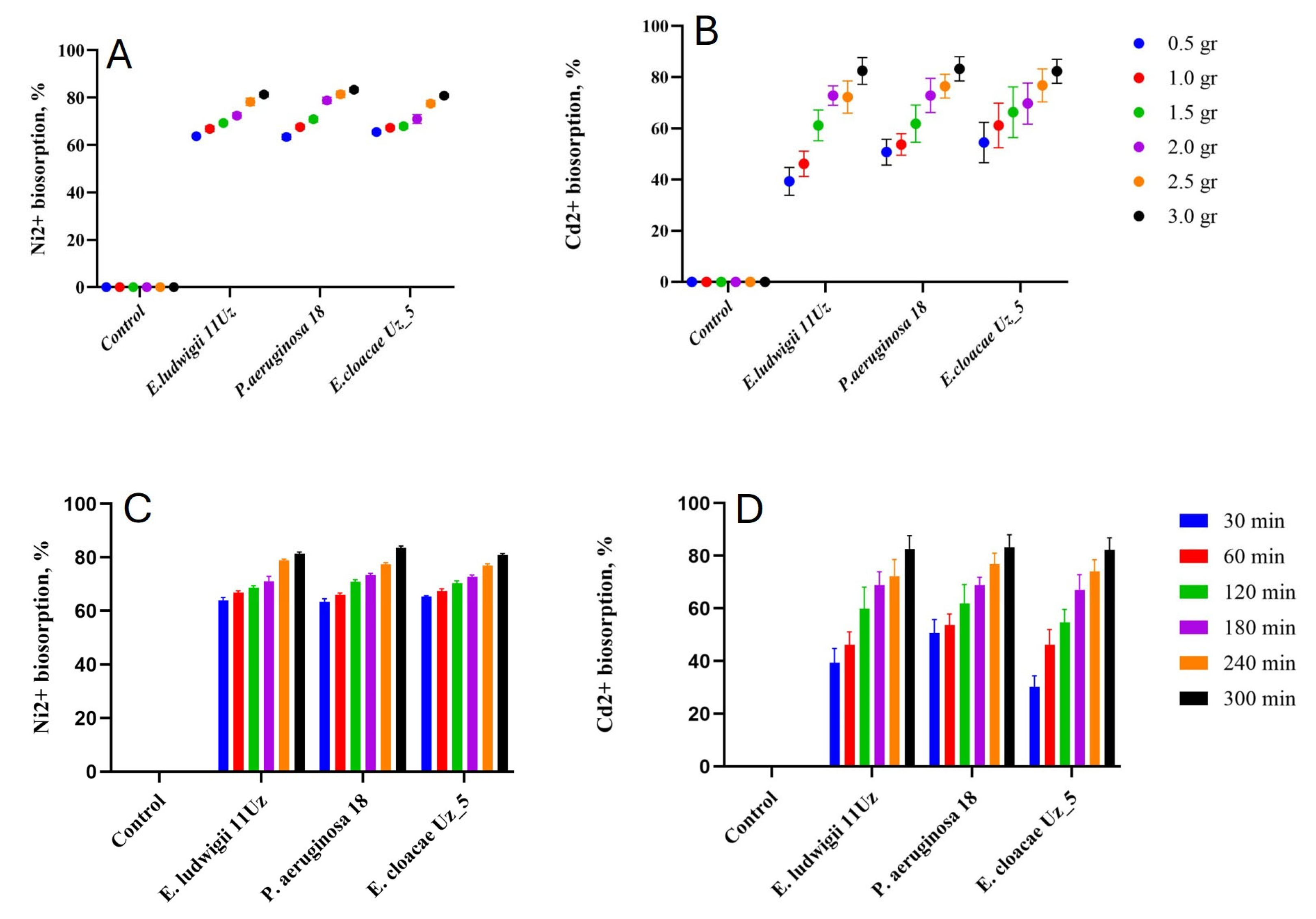
3.1. Effect of Growing and Environmental Conditions on the Biosorption Capacity of the Bacterial Strains
3.2. Identification of Bacterial Functional Groups Responsible for Heavy Metal Binding
3.3. Monitoring the Redox Process That Reduces the Toxicity of Heavy Metal Cations
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Karimov, H.N.; Uzakov, Z.Z.; Khushmurodov, J.P.; Usmonova, D.A.; Mallaeva, D.A. Pollution of irrigated soils and their biological treatment. Sci. Rev. Biol. Sci. 2021, 2, 34–40. [Google Scholar]
- Zhabbarov, Z.A.; Atoeva, G.R.; Sayitov, S.S. Pollution of soils with heavy metals around the landfill of municipal solid waste in Tashkent. Sci. Rev. Biol. Sci. 2021, 2, 17–23. [Google Scholar]
- Berdieva, D.S. Soil contamination with heavy metals in the sh. Rashidovsky district of Jizzakh region and methods of their decrease from the soil composition. E3S Web Conf 2021, 265, 03007. [Google Scholar] [CrossRef]
- Karimov, K.N.; Kadirova, G.K.; Riskiev, R.R.; Usmanova, D.A.; Mallaeva, D.A. Pollution of agricultural soils with mobile metals. J. Agric. Biol. Sci. 2024, 5, 76–84. [Google Scholar]
- Wang, Y.; Luo, Y.; Zeng, G.; Wu, X.; Wu, B.; Li, X.; Xu, H. Characteristics and in situ remediation effects of heavy metal immobilizing bacteria on cadmium and nickel co-contaminated soil. Ecotoxicol. Environ. Saf. 2020, 192, 110294. [Google Scholar] [CrossRef]
- Rahman, Z.; Singh, V.P. Bioremediation of toxic heavy metals (THMs) contaminated sites: Concepts, applications and challenges. Environ. Sci. Pollut. Res. 2020, 27, 27563–27581. [Google Scholar] [CrossRef]
- Kang, C.-H.; Kwon, Y.-J.; So, J.-S. Bioremediation of heavy metals by using bacterial mixtures. Ecol. Eng. 2016, 89, 64–69. [Google Scholar] [CrossRef]
- Hassan, M.U.; Chattha, M.U.; Khan, I.; Chattha, M.B.; Aamer, M.; Nawaz, M.; Ali, A.; Khan, M.A.U.; Khan, T.A. Nickel toxicity in plants: Reasons, toxic effects, tolerance mechanisms, and remediation possibilities—A review. Environ. Sci. Pollut. Res. 2019, 26, 12673–12688. [Google Scholar] [CrossRef]
- Nnaji, N.D.; Onyeaka, H.; Miri, T.; Ugwa, C. Bioaccumulation for heavy metal removal: A review. SN Appl. Sci. 2023, 5, 125. [Google Scholar] [CrossRef]
- Pathak, A.; Agarwal, M.; Rathore, R.S.; Chauhan, A. Gene Determinants for Mercury Bioremediation as Revealed by Draft Genome Sequence Analysis of Stenotrophomonas sp. Strain MA5. Microbiol. Resour. Announc. 2019, 8, e00130-19. [Google Scholar] [CrossRef]
- Tan, H.; Wang, C.; Zeng, G.; Luo, Y.; Li, H.; Xu, H. Bioreduction and biosorption of Cr(VI) by a novel Bacillus sp. CRB-B1 strain. J. Hazard. Mater. 2020, 15, 121628. [Google Scholar] [CrossRef] [PubMed]
- Pham, V.H.T.; Kim, J.; Chang, S.; Chung, W. Bacterial biosorbents, an efcient heavy metals green clean-up strategy: Prospects, challenges, and opportunities. Microorganisms 2022, 10, 610. [Google Scholar] [CrossRef]
- Parihar, J.K.; Parihar, P.K.; Pakade, Y.B.; Katnoria, J.K. Bioaccumulation potential of indigenous plants for heavy metal phytoremediation in rural areas of Shaheed Bhagat Singh Nagar, Punjab (India). Environ. Sci. Pollut. Res. Int. 2021, 28, 2426–2442. [Google Scholar] [CrossRef]
- Wood, B.W. Nickel deficiency symptoms are influenced by foliar Zn:Ni or Cu:Ni concentration ratio. Acta Hortic. 2010, 868, 163–170. [Google Scholar] [CrossRef]
- Maňkovská, B.; Godzik, B.; Badea, O.; Shparyk, Y.; Moravčík, P. Chemical and morphological characteristics of key tree species of the Carpathian Mountains. Environ. Pollut. 2004, 130, 41–54. [Google Scholar] [CrossRef]
- Usmonkulova, A.; Shonakhunov, T.; Kadirova, G. Activity of nitrogen-fixing cyanobacteria under salinity and heavy metals stress. J. Pharm. Negat. Results 2022, 13, 355–363. [Google Scholar] [CrossRef]
- Kang, S.M.; Shahzad, R.; Bilal, S.; Khan, A.L.; Park, Y.G.; Lee, K.E.; Lee, I.J. Indole3-acetic-acid and ACC deaminase producing Leclercia adecarboxylata MO1 improves Solanum lycopersicum L. growth and salinity stress tolerance by endogenous secondary metabolites regulation. BMC Microbiol. 2019, 19, 80. [Google Scholar] [CrossRef]
- Enshaei, M.; Khanafari, A.; Akhavan, S.A. Metallothionein induction in two species of Pseudomonas exposed to cadmium and copper contamination. Iran. J. Environ. Health Sci. Eng. 2010, 7, 287–298. [Google Scholar]
- Meng, D.; Li, J.; Liu, T.; Liu, Y.; Yan, M.; Hu, J.; Li, X.; Liu, X.; Liang, Y.; Liu, H.; et al. Effects of redox potential on soil cadmium solubility: Insight into microbial community. J. Environ. Sci. 2019, 75, 224–232. [Google Scholar] [CrossRef]
- Essa, A.M.M.; Al Abboud, M.A.; Khatib, S.I. Metal transformation as a strategy for bacterial detoxification of heavy metals. J. Basic Microbiol. 2018, 58, 17–29. [Google Scholar] [CrossRef]
- Liu, M.; Hu, Z.; Fan, Y.; Hua, B.; Yang, W.; Pang, S.; Mao, R.; Zhang, Y.; Bai, K.; Fadda, C.; et al. Effects of leguminous green manure–crop rotation on soil enzyme activity and stoichiometry. J. Plant Ecol. 2024, 17, rtae065. [Google Scholar] [CrossRef]
- Patil, A.; Chakraborty, S.; Yadav, Y.; Sharma, B.; Singh, S.; Arya, M. Bioremediation strategies and mechanisms of bacteria for resistance against heavy metals: A review. Bioremediat. J. 2024, 1–33. [Google Scholar] [CrossRef]
- Pagnucco, G.; Overfield, D.; Chamlee, Y.; Shuler, C.; Kassem, A.; Opara, S.; Najaf, H.; Abbas, L.; Coutinho, O.; Fortuna, A. Metal tolerance and biosorption capacities of bacterial strains isolated from an urban watershed. Front. Microbiol. 2023, 14, 1278886. [Google Scholar] [CrossRef] [PubMed]
- Mahle, R.; Kumbhakar, P.; Pramanik, A.; Kumbhakar, P.; Sahoo, S.; Mukherjee, R.; Tiwary, C.S.; Banerjee, R. Probing the bacterial detoxification of cadmium to form cadmium sulfide quantum dots and the underlying mechanism. Mater. Adv. 2020, 1, 1168–1175. [Google Scholar] [CrossRef]
- Amonov, M.R.; Shirinova, Q.G. Analytical chemistry in diagrams and tables [Analitik kimyo sxema va jadvallarda]. In Reference Book; BuxDU: Bukhara, Uzbekistan, 2019; p. 285. [Google Scholar]
- Agarwal, M.; Rathore, R.S.; Chauhan, A. A Rapid and High Throughput MIC Determination Method to Screen Uranium Resistant Microorganisms. Methods Protoc. 2020, 3, 21. [Google Scholar] [CrossRef]
- Govarthanan, M.; Park, S.-H.; Park, Y.-J.; Myung, H.; Krishnamurthy, R.R.; Lee, S.-H.; Lovanh, N.; Kamala-Kannan, S.; Oh, B.-T. Lead biotransformation potential of allochthonous Bacillus sp. SKK11 with sesame oil cake extract in mine soil. RSC Adv. 2015, 5, 54564–54570. [Google Scholar] [CrossRef]
- Mejias Carpio, I.E.; Ansari, A.; Rodrigues, D.F. Relationship of biodiversity with heavy metal tolerance and sorption capacity: A meta-analysis approach. Environ. Sci. Technol. 2018, 52, 184–194. [Google Scholar] [CrossRef]
- Green-Ruiz, C.; Rodriguez-Tirado, V.; Gomez-Gil, B. Cadmium and zinc removal from aqueous solutions by Bacillus Jeotgali: pH, salinity and temperature effects. Bioresour. Technol. 2008, 99, 3864–3870. [Google Scholar] [CrossRef]
- Agarwal, M.; Pathak, A.; Rathore, R.S.; Prakash, O.; Singh, R.; Jaswal, R.; Seaman, J.; Chauhan, A. Proteogenomic Analysis of Burkholderia Species Strains 25 and 46 Isolated from Uraniferous Soils Reveals Multiple Mechanisms to Cope with Uranium Stress. Cells 2018, 7, 269. [Google Scholar] [CrossRef]
- Andy, A.K.; Masih, S.A.; Gour, V.S. Isolation, screening and characterization of plant growth promoting rhizobacteria from rhizospheric soils of selected pulses. Biocatal. Agric. Biotechnol. 2020, 27, 101685. [Google Scholar] [CrossRef]
- Gupta, K.; Chatterjee, C.; Gupta, B. Isolation and characterization of heavy metal tolerant Gram-positive bacteria with bioremedial properties from municipal waste rich soil of Kestopur canal (Kolkata), West Bengal, India. Biologia 2012, 67, 827–836. [Google Scholar] [CrossRef]
- Edulamudi, P.; Antony Masilamani, A.J.; Vanga, U.R.; Divi, V.R.S.G.; Konada, V.M. Nickel tolerance and biosorption potential of rhizobia associated with horse gram [Macrotyloma uniflorum (Lam.) Verdc.]. Int. J. Phytoremediation 2021, 23, 1184–1190. [Google Scholar] [CrossRef] [PubMed]
- Kara, A.K.; Fakıoğlu, Ö.; Kotan, R.; Atamanalp, M.; Alak, G. The investigation of bioremediation potential of Bacillus subtilis and B. thuringiensis isolates under controlled conditions in freshwater. Arch. Microbiol. 2021, 203, 2075–2085. [Google Scholar] [CrossRef]
- Halim, M.A.; Rahman, M.M.; Megharaj, M.; Naidu, R. Cadmium Immobilization in the Rhizosphere and Plant Cellular Detoxification: Role of Plant-Growth-Promoting Rhizobacteria as a Sustainable Solution. J. Agric. Food Chem. 2020, 68, 13497–13529. [Google Scholar] [CrossRef] [PubMed]
- Rocco, D.H.E.; Freire, B.M.; Oliveira, T.J.; Alves, P.L.M.; Júnior, J.M.d.O.; Batista, B.L.; Grotto, D.; Jozala, A.F. Bacillus subtilis as an efective tool for bioremediation of lead, copper and cadmium in water. Discov. Appl. Sci. 2024, 6, 430. [Google Scholar] [CrossRef]
- Ayangbenro, A.S.; Babalola, O.O. A New Strategy for Heavy Metal Polluted Environments: A Review of Microbial Biosorbents. Int. J. Environ. Res. Public Health 2017, 14, 94. [Google Scholar] [CrossRef]
- Kassem, A.; Abbas, L.; Coutinho, O.; Opara, S.; Najaf, H.; Kasperek, D.; Pokhrel, K.; Li, X.; Tiquia-Arashiro, S. Applications of Fourier Transform-Infrared spectroscopy in microbial cell biology and environmental microbiology: Advances, challenges, and future perspectives. Front. Microbiol. 2023, 21, 1304081. [Google Scholar] [CrossRef]
- Ma, Y.; Bantec, T.N.; Oliveira, R.S.; Coutinho, A.; Zhang, C.; Freitas, H. The role of bacteria in metal bioaccumulation and biosorption In Advances in Microbe-Assisted Phytoremediation of Polluted Sites; Bauddh, K., Ma, Y., Eds.; Elsevier: New York, NY, USA, 2022; pp. 103–112. [Google Scholar]
- Camacho-Chab, J.C.; Castañeda-Chávez, M.D.R.; Chan-Bacab, M.J.; Aguila-Ramírez, R.N.; Galaviz-Villa, I.; Bartolo-Pérez, P.; Lango-Reynoso, F.; Tabasco-Novelo, C.; Gaylarde, C.; Ortega-Morales, B.O. Biosorption of Cadmium by Non-Toxic Extracellular Polymeric Substances (EPS) Synthesized by Bacteria from Marine Intertidal Biofilms. Int. J. Environ. Res. Public Health 2018, 15, 314. [Google Scholar] [CrossRef]
- Tunali, S.; Çabuk, A.; Akar, T. Removal of lead and copper ions from aqueous solutions by bacterial strain isolated from soil. Chem. Eng. J. 2006, 115, 203–211. [Google Scholar] [CrossRef]
- Nascimento, T.L.S.; Oliveira, K.F.S.; Junior, J.O.D.; Pimenta, A.S.; Melo, D.M.A.; Melo, M.A.F.; Braga, R.M. Biosorption of nickel and cadmium using Pachira aquatica Aubl. peel biochar. Sci. Rep. 2024, 14, 5086. [Google Scholar] [CrossRef]
- Nascimento, F.X.; Rossi, M.J.; Glick, B.R. Role of ACC deaminase in stress control of leguminous plants. In Plant Growth Promoting Actinobacteria; Subramanian, G., Ed.; Springer Science: Berlin/Heidelberg, Germany, 2016; pp. 179–192. [Google Scholar]
- Pande, V.; Pandey, S.C.; Sati, D.; Bhatt, P.; Samant, M. Microbial Interventions in Bioremediation of Heavy Metal Contaminants in Agroecosystem. Front. Microbiol. 2022, 6, 824084. [Google Scholar] [CrossRef] [PubMed]
- Afzal, A.M.; Rasool, M.H.; Waseem, M.; Aslam, B. Assessment of heavy metal tolerance and biosorptive potential of Klebsiella variicola isolated from industrial effluents. AMB Express 2017, 7, 184. [Google Scholar] [CrossRef] [PubMed]
- Salbitani, G.; Maresca, V.; Cianciullo, P.; Bossa, R.; Carfagna, S.; Basile, A. Non-Protein Thiol Compounds and Antioxidant Responses Involved in Bryophyte Heavy-Metal Tolerance. Int. J. Mol. Sci. 2023, 10, 5302. [Google Scholar] [CrossRef] [PubMed]
- Fein, J.B.; Yu, Q.; Nam, J.; Yee, N. Bacterial cell envelope and extracellular sulfhydryl binding sites: Their roles in metal binding and bioavailability. Chem. Geol. 2019, 521, 28–38. [Google Scholar] [CrossRef]
- Usmonkulova, A.A.; Kadirova, G.K.H.; Khusanov, T.S.; Shonakhunov, T.E.; Shukurov, N. Determination of local bacteria synthesizing ACC deaminase on plant growth indicators under nickel and cadmium stress conditions. SABRAO J. Breed. Genet. 2024, 56, 2033–2044. [Google Scholar] [CrossRef]

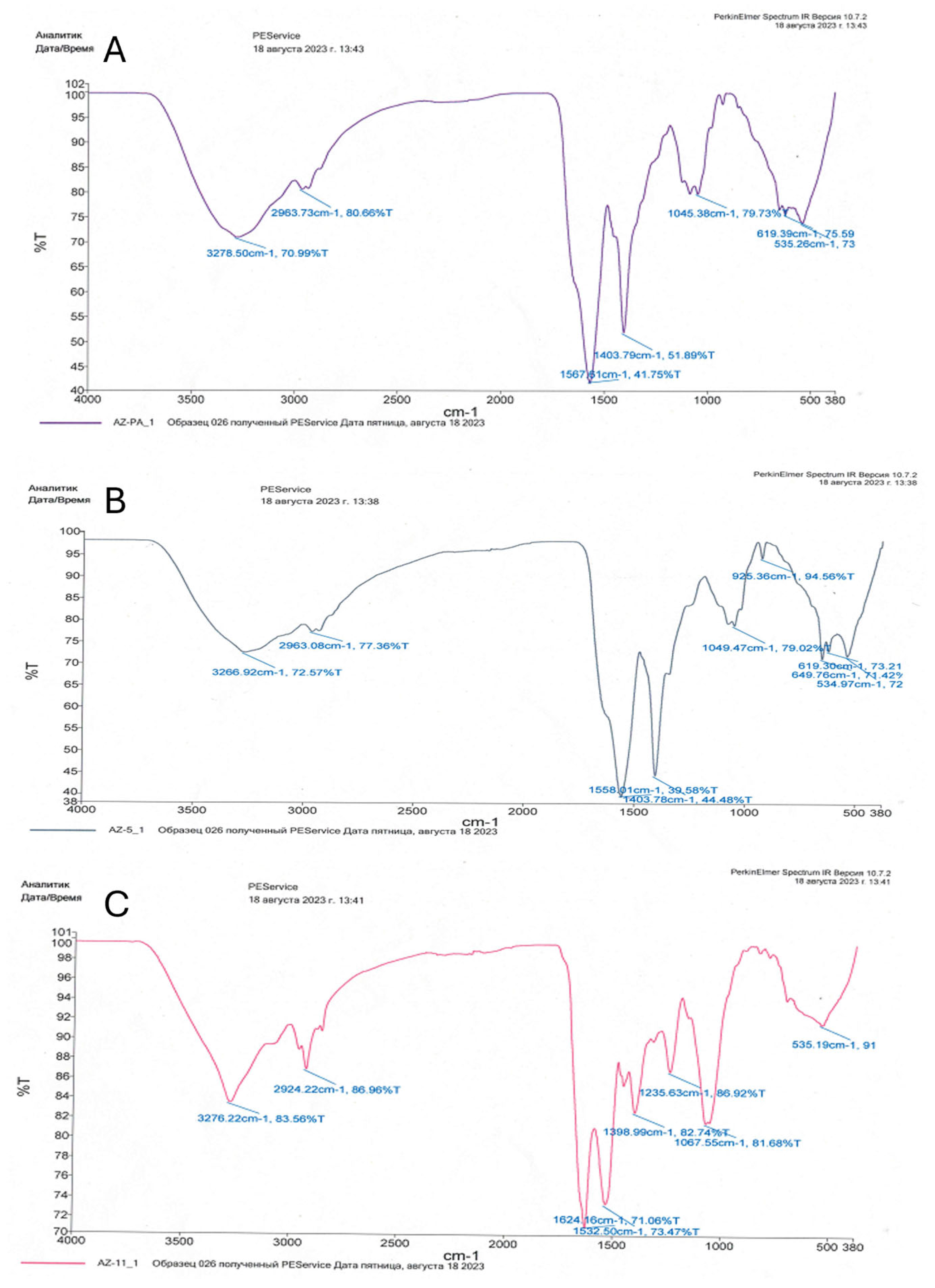
| Functional Group | Wavelength (cm−1) | ||
|---|---|---|---|
| Pseudomonas aeruginosa 18 | Enterobacter ludwigii 11Uz | Enterobacter cloacae Uz_5 | |
| NH2 (amino) | 535.26 619.39 | 535.19 | 534.97 649.76 619.30 |
| S=O (sulfoxide) | 1045.38 | 1067.55 | 1049.47 |
| COOH (carboxyl) | 1403.79 | - | 1403.78 |
| N-H (amid) | 1567.61 | 1532.50 1624.16 | 1558.01 |
| CH (aromatic) | 2963.73 | - | 2963.08 |
| C=O (ketonic) | 3278.50 | 2924.22 | 3266.92 |
| C-OH (alcoholic) | 3278 | 3276.22 | 3266.92 |
| P-O (phosphoryl) | - | 1235.63 | - |
| C-N (aromatic) | - | 1398.99 | - |
| C-H (alkenes) | - | - | 925.36 |
| Strain | Amount of Ni2+ and Cd2+ Cations Added to the Growth Medium | Indicator Used | Cysteine Formation |
|---|---|---|---|
| Pseudomonas aeruginosa 18 | Ni2+ 2 mM | Na2HPO4 | − |
| Na2CO3 | − | ||
| NaOH | − | ||
| Cd2+ 3 mM | Na2HPO4 | + | |
| Na2CO3 | + | ||
| NaOH | + | ||
| Enterobacter ludwigii 11Uz | Ni2+ 3 mM | Na2HPO4 | − |
| Na2CO3 | − | ||
| NaOH | − | ||
| Cd2+ 1 mM | Na2HPO4 | − | |
| Na2CO3 | − | ||
| NaOH | − | ||
| Enterobacter Cloacae Uz_5 | Ni2+ 2 mM | Na2HPO4 | − |
| Na2CO3 | − | ||
| NaOH | − | ||
| Cd2+ 1 mM | Na2HPO4 | + | |
| Na2CO3 | + | ||
| NaOH | + | ||
| Control | Ni2+ 2 mM | Na2HPO4 | + |
| Na2CO3 | + | ||
| NaOH | + | ||
| Cd2+ 3 mM | Na2HPO4 | + | |
| Na2CO3 | + | ||
| NaOH | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Usmonkulova, A.; Malusa, E.; Kadirova, G.; Khalilov, I.; Canfora, L.; Abdulmyanova, L. Ni2+ and Cd2+ Biosorption Capacity and Redox-Mediated Toxicity Reduction in Bacterial Strains from Highly Contaminated Soils of Uzbekistan. Microorganisms 2025, 13, 1485. https://doi.org/10.3390/microorganisms13071485
Usmonkulova A, Malusa E, Kadirova G, Khalilov I, Canfora L, Abdulmyanova L. Ni2+ and Cd2+ Biosorption Capacity and Redox-Mediated Toxicity Reduction in Bacterial Strains from Highly Contaminated Soils of Uzbekistan. Microorganisms. 2025; 13(7):1485. https://doi.org/10.3390/microorganisms13071485
Chicago/Turabian StyleUsmonkulova, Aziza, Eligio Malusa, Gulchekhra Kadirova, Ilkhom Khalilov, Loredana Canfora, and Liliya Abdulmyanova. 2025. "Ni2+ and Cd2+ Biosorption Capacity and Redox-Mediated Toxicity Reduction in Bacterial Strains from Highly Contaminated Soils of Uzbekistan" Microorganisms 13, no. 7: 1485. https://doi.org/10.3390/microorganisms13071485
APA StyleUsmonkulova, A., Malusa, E., Kadirova, G., Khalilov, I., Canfora, L., & Abdulmyanova, L. (2025). Ni2+ and Cd2+ Biosorption Capacity and Redox-Mediated Toxicity Reduction in Bacterial Strains from Highly Contaminated Soils of Uzbekistan. Microorganisms, 13(7), 1485. https://doi.org/10.3390/microorganisms13071485







