Molecular Structure and Biosynthesis of Pyoverdines Produced by Pseudomonas fulva
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Culture
2.2. Genomic Analysis
2.3. Purification of PVDs
2.4. LC-MS Analysis
2.5. Amino Acid Analysis
2.6. Bioinformatic Analysis
3. Results and Discussion
3.1. Characteristics of the P. fulva Genome
3.2. Identification of Secondary Metabolite BGCs Using antiSMASH
3.3. Molecular Structures of the P. fulva PVDs
3.4. Amino Acid Compositions of the P. fulva PVDs
3.5. Molecular Mass Distribution of PVDs in Pseudomonas Strains
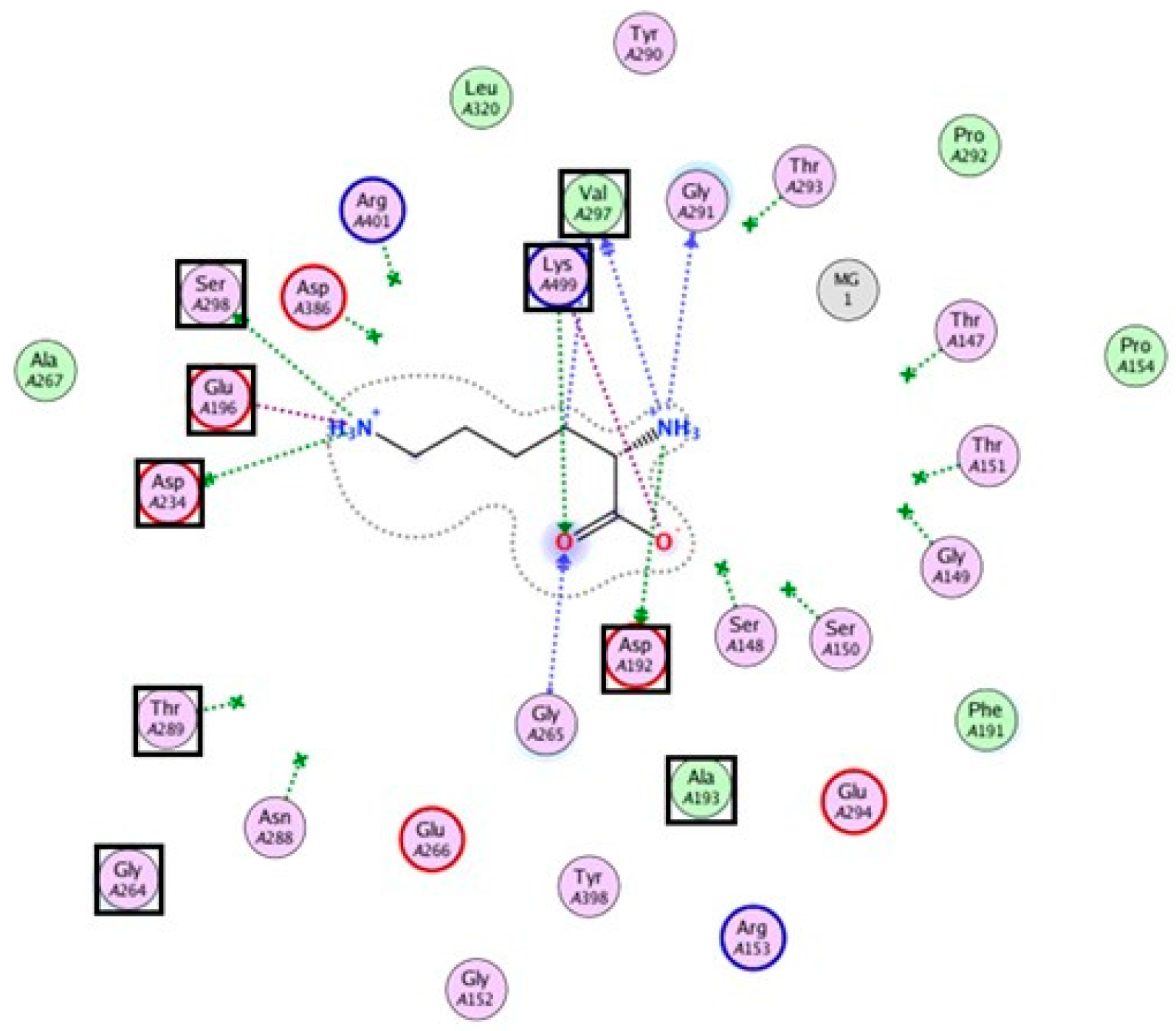
3.6. Bioinformatic Analysis of the Lysine-Incorporating Adenylation Domain of P. fulva PVDs
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PVD | pyoverdine |
| A-domain | adenylation domain |
| NRPS | non-ribosomal peptide synthetase |
| C-domain | condensation domain |
| PCP | peptidyl carrier protein |
| Pvd | pyoverdine biosynthesis protein |
| Pls-A | ε-poly-L-lysine synthetase |
| ORF | open reading flame |
| HPLC | high-performance liquid chromatography |
| MS | mass spectrometry |
| T-domain | thiolation domain |
Appendix A
| Fragment Type | Chemical Formula | Observed m/z (Theoretical) |
|---|---|---|
| A4 | C26H32N7O12 | 634.21 (634.210) |
| A5 | C33H44N9O15 | 806.29 (806.295) |
| B1 | C17H17N4O6 | 373.11 (373.114) |
| B2 | C21H22N5O9 | 488.14 (488.141) |
| B3 | C24H27N6O11 | 575.17 (575.173) |
| B4 | C27H32N7O13 | 662.20 (662.205) |
| B5 | C34H44N9O16 | 834.29 (834.290) |
| B6 | C40H56N11O17 | 962.39 (962.385) |
| B7 | C46H68N13O18 | 1090.48 (1090.480) |
| Y″2 | C11H23N4O3 | 259.18 (259.176) |
| Y″5 | C27H52N9O9 | 646.39 (646.388) |
| Y″6 | C30H57N10O11 | 733.42 (733.420) |
| Y″7 | C34H62N11O14 | 848.44 (848.447) |
| Fragment Type | Chemical Formula | Observed m/z (Theoretical) |
|---|---|---|
| A4 | C26H32N7O11 | 619.20 (619.215) |
| B1 | C17H17N4O5 | 357.12 (357.119) |
| B2 | C21H22N5O8 | 472.15 (472.146) |
| B3 | C24H27N6O10 | 560.16 (559.178) |
| B4 | C27H32N7O12 | 646.21 (646.210) |
| B5 | C34H44N9O15 | 818.30 (818.295) |
| B6 | C40H56N11O16 | 946.39 (946.390) |
| B7 | C46H68N13O17 | 1075.47 (1074.485) |
| Y″2 | C11H23N4O3 | 259.18 (259.176) |
| Y″6 | C30H57N10O11 | 733.42 (733.420) |
| Y″7 | C34H62N11O14 | 848.44 (848.447) |
| Fragment Type | Chemical Formula | Observed m/z (Theoretical) |
|---|---|---|
| A3 | C23H25N5O9 | 514.92 (514.165) |
| A4 | C26H30N6O11 | 601.19 (601.197) |
| B1 | C17H15N3O5 | 340.09 (340.101) |
| B2 | C21H20N4O8 | 455.12 (455.128) |
| B3 | C24H25N5O10 | 542.15 (542.160) |
| B4 | C27H30N6O12 | 629.18 (629.192) |
| B5 | C34H42N8O15 | 801.27 (801.276) |
| B6 | C40H54N10O16 | 929.36 (929.371) |
| B7 | C46H66N12O17 | 1057.46 (1057.466) |
| Y″2 | C11H23N4O3 | 259.18 (259.176) |
| Y″4 | C24H47N8O7 | 559.18 (559.356) |
| Y″5 | C27H52N9O9 | 646.21 (646.388) |
| Y″6 | C30H57N10O11 | 733.42 (733.420) |
| Y″7 | C34H62N11O14 | 848.45 (848.447) |
| Fragment Type | Chemical Formula | Observed m/z (Theoretical) |
|---|---|---|
| A4 | C23H26N6O10 | 545.16 (545.170) |
| B2 | C18H16N4O7 | 399.09 (399.101) |
| B3 | C21H21N5O9 | 486.13 (486.133) |
| B4 | C24H26N6O11 | 573.16 (573.165) |
| B5 | C31H38N8O14 | 746.25 (746.250) |
| B6 | C37H50N10O15 | 873.34 (873.345) |
| B7 | C43H62N12O16 | 1001.43 (1001.440) |
| Y″6 | C30H57N10O11 | 733.42 (733.420) |
Appendix B
References
- Neilands, J.B. Siderophores: Structure and function of microbial iron transport compounds. J. Biol. Chem. 1995, 270, 26723–26726. [Google Scholar] [CrossRef] [PubMed]
- Varma, A.; Chincholkar, S.B. Microbial Siderophores; Springer: Berlin, Germany, 2007. [Google Scholar]
- Hider, R.C.; Kong, X. Chemistry and biology of siderophores. Nat. Prod. Rep. 2010, 27, 637–657. [Google Scholar] [CrossRef] [PubMed]
- Visca, P.; Imperi, F.; Lamont, I.L. Pyoverdine siderophores: From biogenesis to biosignificance. Trends Microbiol. 2007, 15, 22–30. [Google Scholar] [CrossRef]
- Schalk, I.J.; Guillon, L. Pyoverdine biosynthesis and secretion in Pseudomonas aeruginosa: Implications for metal homeostasis. Environ. Microbiol. 2013, 15, 1661–1673. [Google Scholar] [CrossRef]
- Cézard, C.; Farvacques, N.; Sonnet, P. Chemistry and biology of pyoverdines, Pseudomonas primary siderophores. Curr. Med. Chem. 2015, 22, 165–186. [Google Scholar] [CrossRef]
- Ghssein, G.; Ezzeddine, Z. A review of Pseudomonas aeruginosa metallophores: Pyoverdine, pyochelin and pseudopaline. Biology 2022, 11, 1711. [Google Scholar] [CrossRef]
- Díaz-Pérez, S.P.; Solis, C.S.; López-Bucio, J.S.; Alarcón, J.J.V.; Villegas, J.; de la Cruz, H.R.; Campos-Garcia, J. Pathogenesis in Pseudomonas aeruginosa PAO1 biofilm-associated is dependent on the pyoverdine and pyocyanin siderophores by quorum sensing modulation. Microb. Ecol. 2023, 86, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Owen, J.G.; Ackerley, D.F. Characterization of pyoverdine and achromobactin in Pseudomonas syringae pv. phaseolicola 1448a. BMC Microbiol. 2011, 11, 218. [Google Scholar] [CrossRef]
- Moon, C.D.; Zhang, X.X.; Matthijs, S.; Schafer, M.; Budzikiewicz, H.; Rainey, P.B. Genomic, genetic and structural analysis of pyoverdine-mediated iron acquisition in the plant growth-promoting bacterium Pseudomonas fluorescens SBW25. BMC Microbiol. 2008, 8, 7. [Google Scholar] [CrossRef]
- Ye, L.; Ballet, S.; Hildebrand, F.; Laus, G.; Guillemyn, K.; Raes, J.; Matthijs, S.; Martins, J.; Cornelis, P. A combinatorial approach to the structure elucidation of a pyoverdine siderophore produced by a Pseudomonas putida isolate and the use of pyoverdine as a taxonomic marker for typing P. putida subspecies. Biometals 2013, 26, 561–575. [Google Scholar] [CrossRef]
- Parker, D.L.; Lee, S.W.; Geszvain, K.; Davis, R.E.; Gruffaz, C.; Meyer, J.M.; Torpey, J.W.; Tebo, B.M. Pyoverdine synthesis by the Mn(II)-oxidizing bacterium Pseudomonas putida GB-1. Front. Microbiol. 2014, 5, 202. [Google Scholar] [CrossRef]
- Sexton, D.J.; Glover, R.C.; Loper, J.E.; Schuster, M. Pseudomonas protegens Pf-5 favours self-produced siderophore over free-loading in interspecies competition for iron. Environ. Microbiol. 2017, 19, 3514–3525. [Google Scholar] [CrossRef]
- Patel, J.S.; Selvaraj, V.; More, P.; Bahmani, R.; Borza, T.; Prithiviraj, B. A plant biostimulant from ascophyllum nodosum potentiates plant growth promotion and stress protection activity of Pseudomonas protegens CHA0. Plants 2023, 12, 1208. [Google Scholar] [CrossRef]
- Dell’Anno, F.; Vitale, G.A.; Buonocore, C.; Vitale, L.; Palma Esposito, F.; Coppola, D.; Della Sala, G.; Tedesco, P.; de Pascale, D. Novel insights on pyoverdine: From biosynthesis to biotechnological application. Int. J. Mol. Sci. 2022, 23, 11507. [Google Scholar] [CrossRef]
- Budzikiewicz, H.; Schafer, M.; Fernandez, D.U.; Matthijs, S.; Cornelis, P. Characterization of the chromophores of pyoverdins and related siderophores by electrospray tandem mass spectrometry. Biometals 2007, 20, 135–144. [Google Scholar] [CrossRef]
- Sussmuth, R.D.; Mainz, A. Nonribosomal Peptide Synthesis-Principles and Prospects. Angew. Chem. Int. Ed. Engl. 2017, 56, 3770–3821. [Google Scholar] [CrossRef]
- Ringel, M.T.; Bruser, T. The biosynthesis of pyoverdines. Microb. Cell 2018, 5, 424–437. [Google Scholar] [CrossRef]
- Almuzara, M.N.; Vazquez, M.; Tanaka, N.; Turco, M.; Ramirez, M.S.; Lopez, E.L.; Pasteran, F.; Rapoport, M.; Procopio, A.; Vay, C.A. First case of human infection due to Pseudomonas fulva, an environmental bacterium isolated from cerebrospinal fluid. J. Clin. Microbiol. 2010, 48, 660–664. [Google Scholar] [CrossRef]
- Seok, Y.; Shin, H.; Lee, Y.; Cho, I.; Na, S.; Yong, D.; Jeong, S.H.; Lee, K. First report of bloodstream infection caused by Pseudomonas fulva. J. Clin. Microbiol. 2010, 48, 2656–2657. [Google Scholar] [CrossRef]
- Adeniji, A.A.; Ayangbenro, A.S.; Loots, D.T. Genome sequence resource of Pseudomonas fulva HARBPS9.1-candidate biocontrol agent. Phytopathology 2021, 111, 896–898. [Google Scholar] [CrossRef]
- Adeniji, A.A.; Babalola, O.O. Evaluation of Pseudomonas fulva PS9.1 and Bacillus velezensis NWUMFkBS10.5 as candidate plant growth promoters during maize-fusarium interaction. Plants 2022, 11, 324. [Google Scholar] [CrossRef]
- Thiem, D.; Złoch, M.; Gadzała-Kopciuch, R.; Szymańska, S.; Baum, C.; Hrynkiewicz, K. Cadmium-induced changes in the production of siderophores by a plant growth promoting strain of Pseudomonas fulva. J. Basic Microbiol. 2018, 58, 623–632. [Google Scholar] [CrossRef]
- Ye, L.; Hildebrand, F.; Dingemans, J.; Ballet, S.; Laus, G.; Matthijs, S.; Berendsen, R.; Cornelis, P. Draft genome sequence analysis of a Pseudomonas putida W15Oct28 strain with antagonistic activity to Gram-positive and Pseudomonas sp. pathogens. PLoS ONE 2014, 9, e110038. [Google Scholar] [CrossRef]
- Hesse, C.; Schulz, F.; Bull, C.T.; Shaffer, B.T.; Yan, Q.; Shapiro, N.; Hassan, K.A.; Varghese, N.; Elbourne, L.D.H.; Paulsen, I.T.; et al. Genome-based evolutionary history of Pseudomonas spp. Environ. Microbiol. 2018, 20, 2142–2159. [Google Scholar] [CrossRef]
- Uchino, M.; Shida, O.; Uchimura, T.; Komagata, K. Recharacterization of Pseudomonas fulva Iizuka and Komagata 1963, and proposals of Pseudomonas parafulva sp. nov. and Pseudomonas cremoricolorata sp. nov. J. Gen. Appl. Microbiol. 2001, 47, 247–261. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Rehm, K.; Vollenweider, V.; Kümmerli, R.; Bigler, L. Rapid identification of pyoverdines of fluorescent Pseudomonas spp. by UHPLC-IM-MS. Biometals 2023, 36, 19–34. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Okamoto, T.; Yamanaka, K.; Hamano, Y.; Nagano, S.; Hino, T. Crystal structure of the adenylation domain from an epsilon-poly-l-lysine synthetase provides molecular mechanism for substrate specificity. Biochem. Biophys. Res. Commun. 2022, 596, 43–48. [Google Scholar] [CrossRef]
- Pena, A.; Busquets, A.; Gomila, M.; Mulet, M.; Gomila, R.M.; Reddy, T.B.; Huntemann, M.; Pati, A.; Ivanova, N.; Markowitz, V.; et al. High quality draft genome sequences of Pseudomonas fulva DSM 17717(T), Pseudomonas parafulva DSM 17004(T) and Pseudomonas cremoricolorata DSM 17059(T) type strains. Stand. Genomic Sci. 2016, 11, 55. [Google Scholar] [CrossRef]
- Conti, E.; Stachelhaus, T.; Marahiel, M.A.; Brick, P. Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J. 1997, 16, 4174–4183. [Google Scholar] [CrossRef]
- Stachelhaus, T.; Mootz, H.D.; Marahiel, M.A. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 1999, 6, 493–505. [Google Scholar] [CrossRef]
- Vandenende, C.S.; Vlasschaert, M.; Seah, S.Y. Functional characterization of an aminotransferase required for pyoverdine siderophore biosynthesis in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2004, 186, 5596–5602. [Google Scholar] [CrossRef]
- Lamont, I.L.; Martin, L.W.; Sims, T.; Scott, A.; Wallace, M. Characterization of a gene encoding an acetylase required for pyoverdine synthesis in Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 3149–3152. [Google Scholar] [CrossRef]
- Rehm, K.; Vollenweider, V.; Kümmerli, R.; Bigler, L. A comprehensive method to elucidate pyoverdines produced by fluorescent Pseudomonas spp. by UHPLC-HR-MS/MS. Anal. Bioanal. Chem. 2022, 414, 2671–2685. [Google Scholar] [CrossRef]
- Braud, A.; Hoegy, F.; Jezequel, K.; Lebeau, T.; Schalk, I.J. New insights into the metal specificity of the Pseudomonas aeruginosa pyoverdine-iron uptake pathway. Environ. Microbiol. 2009, 11, 1079–1091. [Google Scholar] [CrossRef]
- Wichard, T.; Bellenger, J.P.; Morel, F.M.; Kraepiel, A.M. Role of the siderophore azotobactin in the bacterial acquisition of nitrogenase metal cofactors. Environ. Sci. Technol. 2009, 43, 7218–7224. [Google Scholar] [CrossRef]
- Sultana, R.; Siddiqui, B.S.; Taraz, K.; Budzikiewicz, H.; Meyer, J.M. An isopyoverdin from Pseudomonas putida CFML 90-44. Z. für Naturforschung C J. Biosci. 2001, 56, 303–307. [Google Scholar] [CrossRef]
- Budzikiewicz, H.; Kilz, S.; Taraz, K.; Meyer, J.-M. Identical pyoverdines from Pseudomonas fluorescens 9AW and from Pseudomonas putida 9BW. Zeitschrift für Naturforschung C 1997, 52, 721–728. [Google Scholar] [CrossRef][Green Version]
- Sultana, R.; Siddiqui, B.S.; Taraz, K.; Budzikiewicz, H.; Meyer, J.M. A pyoverdine from Pseudomonas putida CFML 90-51 with a Lys epsilon-amino link in the peptide chain. Biometals 2000, 13, 147–152. [Google Scholar] [CrossRef]
- Smith, E.E.; Sims, E.H.; Spencer, D.H.; Kaul, R.; Olson, M.V. Evidence for diversifying selection at the pyoverdine locus of Pseudomonas aeruginosa. J. Bacteriol. 2005, 187, 2138–2147. [Google Scholar] [CrossRef]
- Greenwald, J.; Nader, M.; Celia, H.; Gruffaz, C.; Geoffroy, V.; Meyer, J.M.; Schalk, I.J.; Pattus, F. FpvA bound to non-cognate pyoverdines: Molecular basis of siderophore recognition by an iron transporter. Mol. Microbiol. 2009, 72, 1246–1259. [Google Scholar] [CrossRef]
- Weiner, J., 3rd; Bornberg-Bauer, E. Evolution of circular permutations in multidomain proteins. Mol. Biol. Evol. 2006, 23, 734–743. [Google Scholar] [CrossRef]


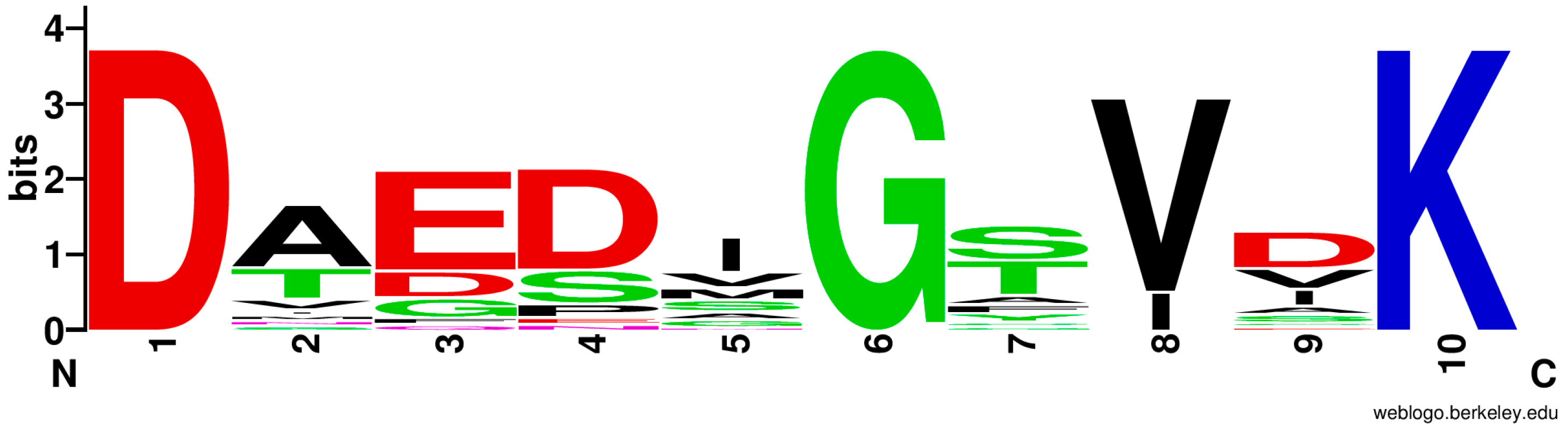

| Base | Count | Percentage |
|---|---|---|
| A | 939,138 | 19.10% |
| T | 1,499,495 | 30.50% |
| G | 1,528,630 | 31.10% |
| C | 950,693 | 19.30% |
| GC | 3,028,125 | 61.60% |
| All | 4,917,956 | 100.0% |
| Coding regions predicted by RAST | ||
| Coding sequences (protein-coding regions) | 4400 | |
| Misc RNA (other RNA-coding regions) | 68 | |
| rRNA (ribosomal RNA-coding regions) | 68 | |
| tmRNA (transfer-messenger RNA) | 1 | |
| tRNA (transfer RNA-coding regions) | 76 | |
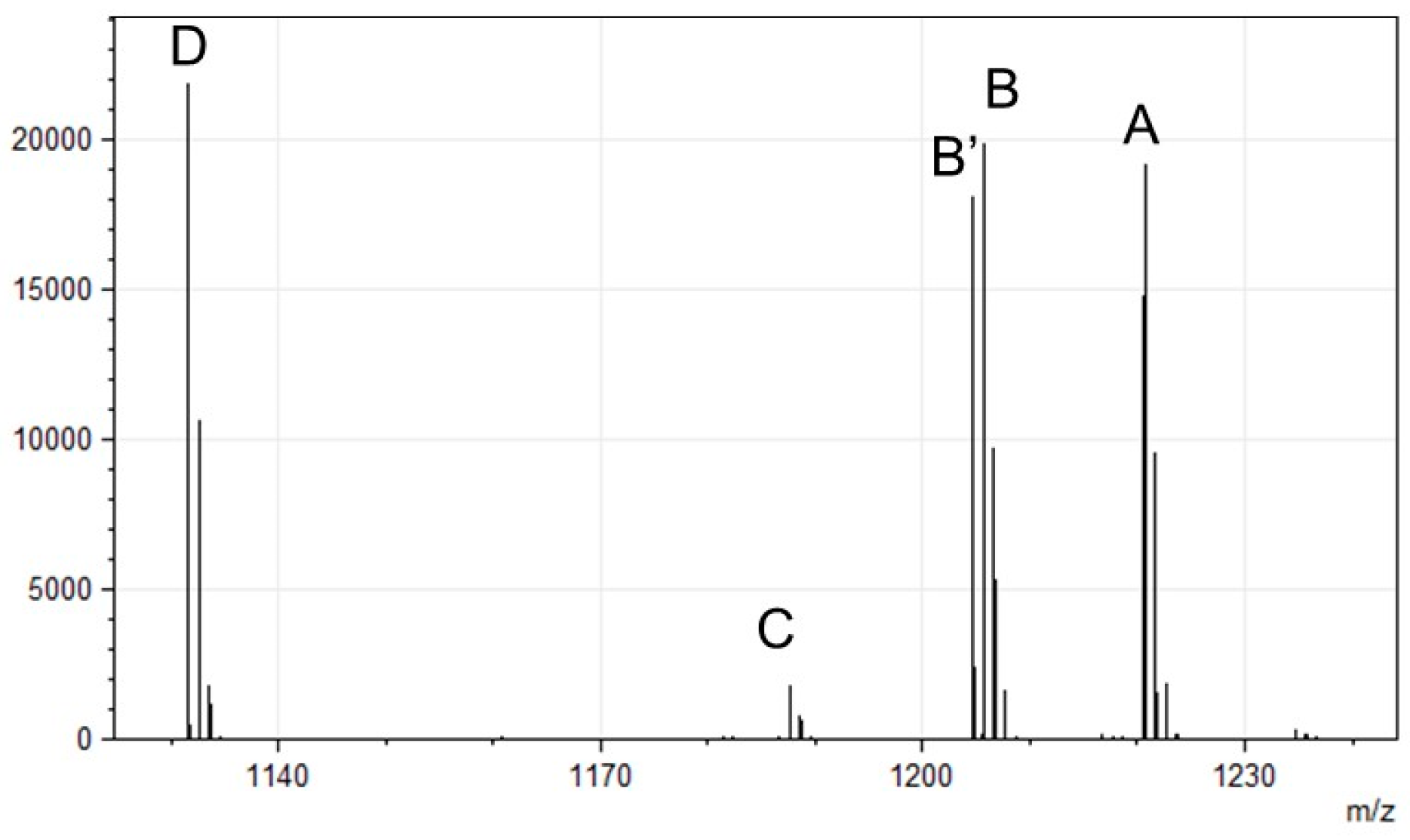
| Peak | [M+H]+ |
|---|---|
| A | 1220.56 |
| B | 1205.54 |
| B’ | 1204.57 |
| C | 1187.52 |
| D | 1131.50 |
| Amino Acid | Amount (nmol) | Ratio |
|---|---|---|
| Asp | 2.94 | 1.01 |
| Ser | 5.84 | 2 |
| Orn | 3.28 | 1.12 |
| Lys | 6.22 | 2.13 |
| Strain | Observed Mass | Sequence | Ref. |
|---|---|---|---|
| P. fulva (NBRC:16637) | 1115.47 1187.53 1204.56 1220.55 1238.56 | Asp-Ser-Ser-AcOHOrn-Lys-Lys-cOHOrn | This study |
| P. fulva (NBRC:16639) | 1131.50 1187.53 1204.56 1220.55 | Asp-Ser-Ser-AcOHOrn-Lys-Lys-cOHOrn | This study |
| P. parafulva (NBRC:16635) | 1190.48 1191.49 1206.50 1207.48 | Asp-Orn-(OHAsp-Dab)-Gly-Ser-cOHOrn | [11] |
| P. parafulva (NBRC:16636) | 1188.45 1206.47 | Not assigned | |
| P. putida (NBRC:3778) | 1368.51 1388.57 1404.57 | Not assigned | [43] |
| P. putida (NBRC:12996) | 1089.41 1091.42 1107.42 1122.43 | Asp-εLys-OHAsp-Ser-Ala-Ser-cOHOrn | [44] |
| P. putida (NBRC:14164) | 1319.50 1336.52 1337.51 1352.52 | Ser-Gln-Dab-Asp-Thr-Gly-Asp-Thr-Thr-Gly | [11] |
| P. putida (NBRC:14671) | 1342.58 1358.58 1359.58 1374.59 | Not assigned | |
| P. putida (NBRC:14796) | 1012.47 1150.42 1166.42 1184.43 | Asp-Orn-Dab-Gly-Ser-Ser-OHAsp-Thr | [43] |
| P. putida (NBRC:15366) | 1305.52 1321.51 1323.53 1338.54 1339.52 1352.55 1354.53 1368.55 1369.53 | Asp-Lys-OHAsp-Ser-Ala-Thr-Thr-Thr-cOHOrn | [11] |
| P. putida (NBRC:100650) | 1056.40 1073.42 1074.41 1092.42 | Asp-Orn-(OHAsp-Dab)-Gly-Ser-cOHOrn | [11] |
| P. putida (NBRC:109349) | 1187.49 1199.49 1217.50 1235.51 1251.52 | Asp-εLys-OHAsp-Ser-Gly-aThr-Lys-cOHOrn | [45] |
| P. putida (NBRC:110474) | 1323.53 1338.54 1339.52 1354.53 1356.55 1372.58 | Ser-Gln-Dab-Asp-Thr-Gly-Asp-Thr-Thr-Gly | [11] |
| Species | Domain | Signature |
|---|---|---|
| Pseudomonas fulva | PvdL(ORF3)A2 | D A E D H G T V T K |
| Pseudomonas fulva | PvdL(ORF3)A3 | D A E D H G T V T K |
| Pseudomonas protegens Pf-5 | PvdD A3 | D A E D N G T V S K |
| Pseudomonas tolaasii | APU91751 A6 | D A E S V G T I I K |
| Streptomyces albulus NBRC14147 | Pls-A | D A E S I G T V V K |
| Xenorhabdus miraniensis | Xmir_01407 A6 | D A E S I G T V I K |
| Planktothrix agardhii NIVA-CYA 126/8 | apnA A2 | D A E D I G S V V K |
| Nostoc punctiforme PCC 73102 | Npun_F2460 A2 | D A E D I G S V I K |
| Stigmatella aurantiaca Sg a15 | mxcG A1 | D A E D I G T V V K |
| Anabaena sp. 90 | aptA1 A2 | D T E D I G S V I K |
| Anabaena sp. 90 | aptA2 A2 | D T E D I G S V V K |
| Mycobacterium tuberculosis H37Rv | Rv0101 A1 | D I E D V G S V V K |
| Bacillus licheniformis | bacB A1 | D A E S I G S V C K |
| Paenibacillus larvae subsp. larvae DSM 25430 | ERIC2_c18070 A1 | D M E D V G S V D K |
| Paenibacillus sp. OSY-SE | pbtB A2 | D V G D V G S I D K |
| Streptomyces fradiae | lptC A1 | D A D D A G T V D K |
| Streptomyces sp. NRRL F-4415 | gobR A2 | D A D D G G F V D K |
| Uncultured bacterium | mlcL A1 | D T D D M G Y V D K |
| Streptomyces albus subsp. chlorinus | FM076_21195 A1 | D T E D M G Y V D K |
| Streptomyces viridosporus ATCC 14672 | cip22 A5 | D T D D M G F I D K |
| Burkholderia sp. B8(2020) | necA A1 | D T E N I G T I S K |
| Mycobacterium tuberculosis H37Rv | MbtF A1 | D A Q D A G C V E K |
| Myxococcus virescens | benD A1 | D N E S G G T V A K |
| Brevibacillus laterosporus | BogB A2 | D S G P S G A V D K |
| Brevibacillus laterosporus | BogC A4 | D A G P S G A V D K |
| Streptomyces filamentosus NRRL 15998 | SSGG_00679 A2 | D V F E S G G V A K |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ochiai, E.; Kawabe, T.; Shionyu, M.; Hasegawa, M. Molecular Structure and Biosynthesis of Pyoverdines Produced by Pseudomonas fulva. Microorganisms 2025, 13, 1409. https://doi.org/10.3390/microorganisms13061409
Ochiai E, Kawabe T, Shionyu M, Hasegawa M. Molecular Structure and Biosynthesis of Pyoverdines Produced by Pseudomonas fulva. Microorganisms. 2025; 13(6):1409. https://doi.org/10.3390/microorganisms13061409
Chicago/Turabian StyleOchiai, Eri, Takeru Kawabe, Masafumi Shionyu, and Makoto Hasegawa. 2025. "Molecular Structure and Biosynthesis of Pyoverdines Produced by Pseudomonas fulva" Microorganisms 13, no. 6: 1409. https://doi.org/10.3390/microorganisms13061409
APA StyleOchiai, E., Kawabe, T., Shionyu, M., & Hasegawa, M. (2025). Molecular Structure and Biosynthesis of Pyoverdines Produced by Pseudomonas fulva. Microorganisms, 13(6), 1409. https://doi.org/10.3390/microorganisms13061409






