Bacteriome Signature in SARS-CoV-2-Infected Patients Correlates with Increased Gut Permeability and Systemic Inflammatory Cytokines
Abstract
1. Introduction
2. Patients and Methods
2.1. Study Design, Ethical Aspects and Patients’ Enrollment
2.2. Bacteriome Characterization by 16S Sequencing
2.3. Zonulin and Secretory Immunoglobulin a Quantification by ELISA Assays
2.4. Cytokine Quantification by Cytometric Bead Array and ELISA Assay
2.5. Bioinformatic and Statistical Analyses
3. Results
3.1. Clinical and Demographic Characteristics of SARS-CoV-2 Infected Patients
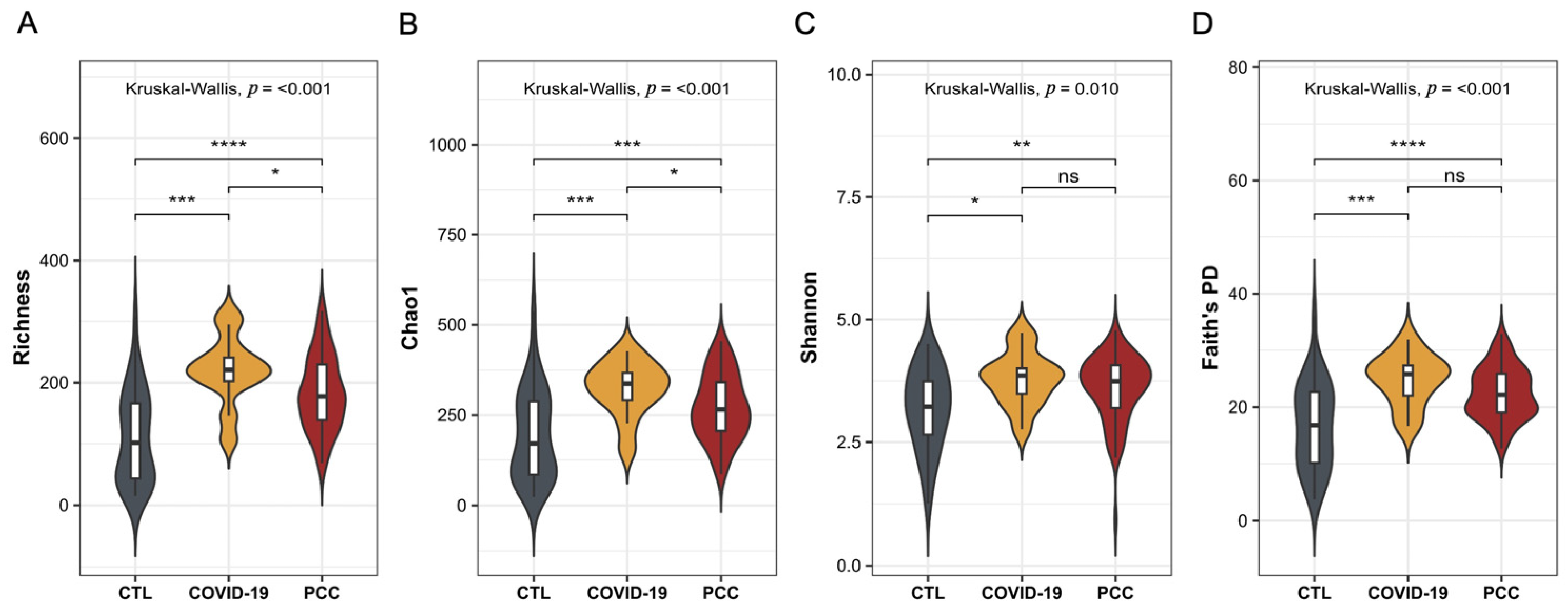
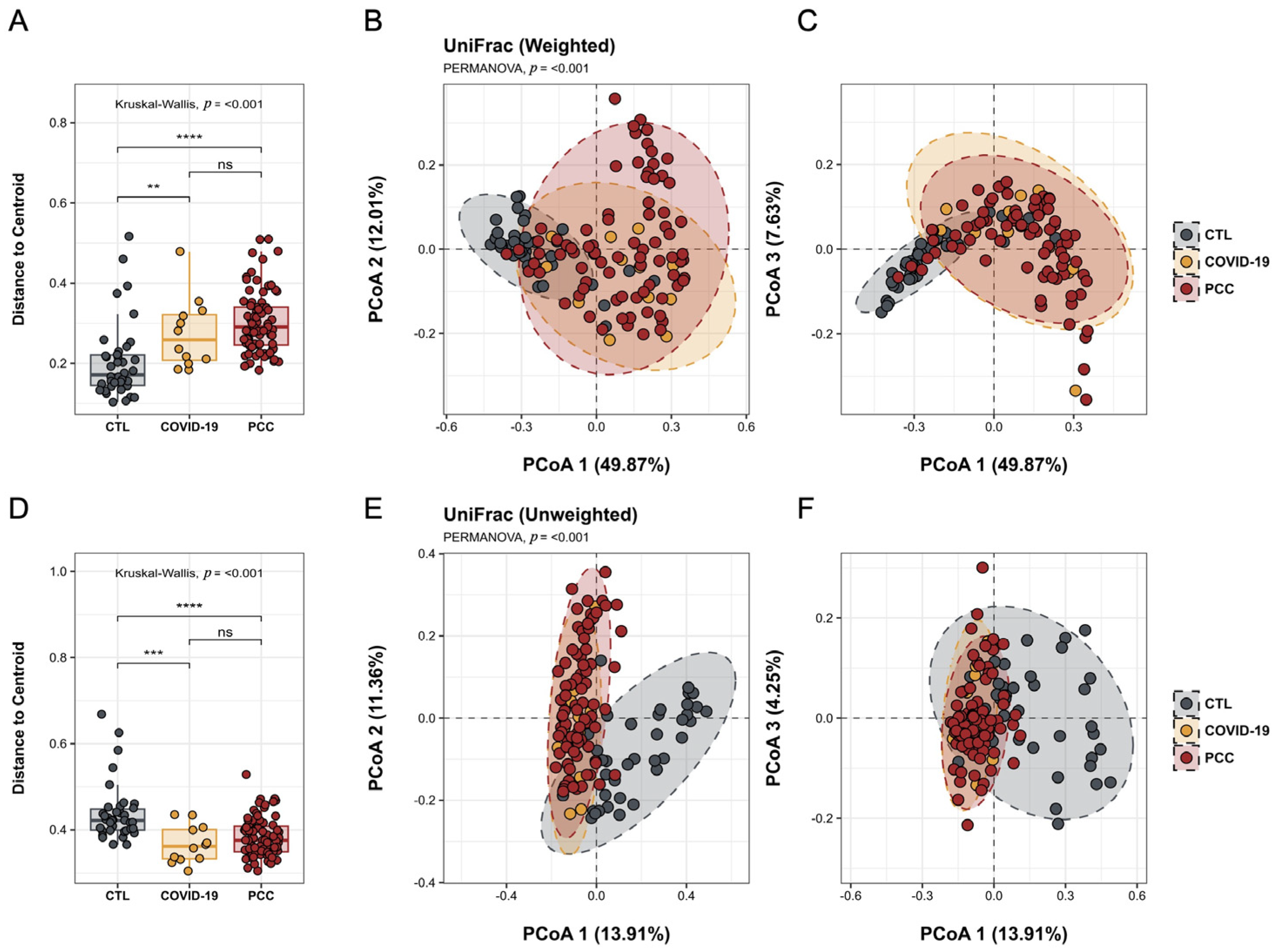
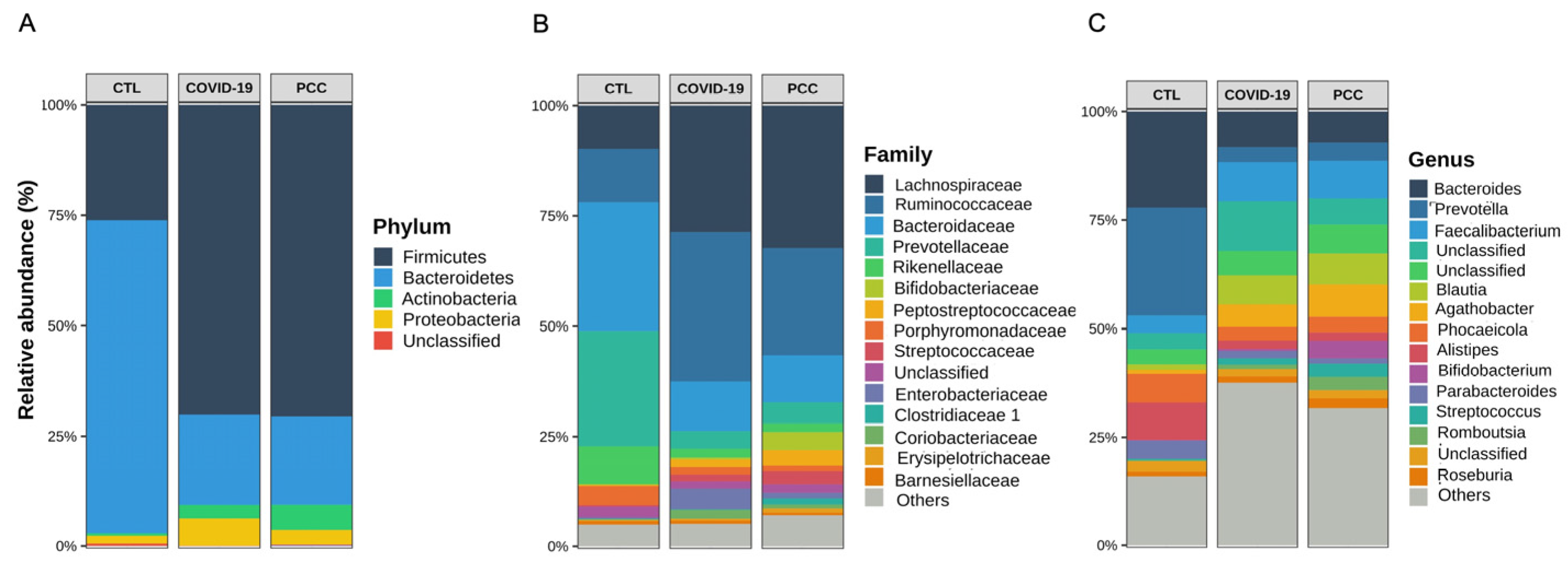
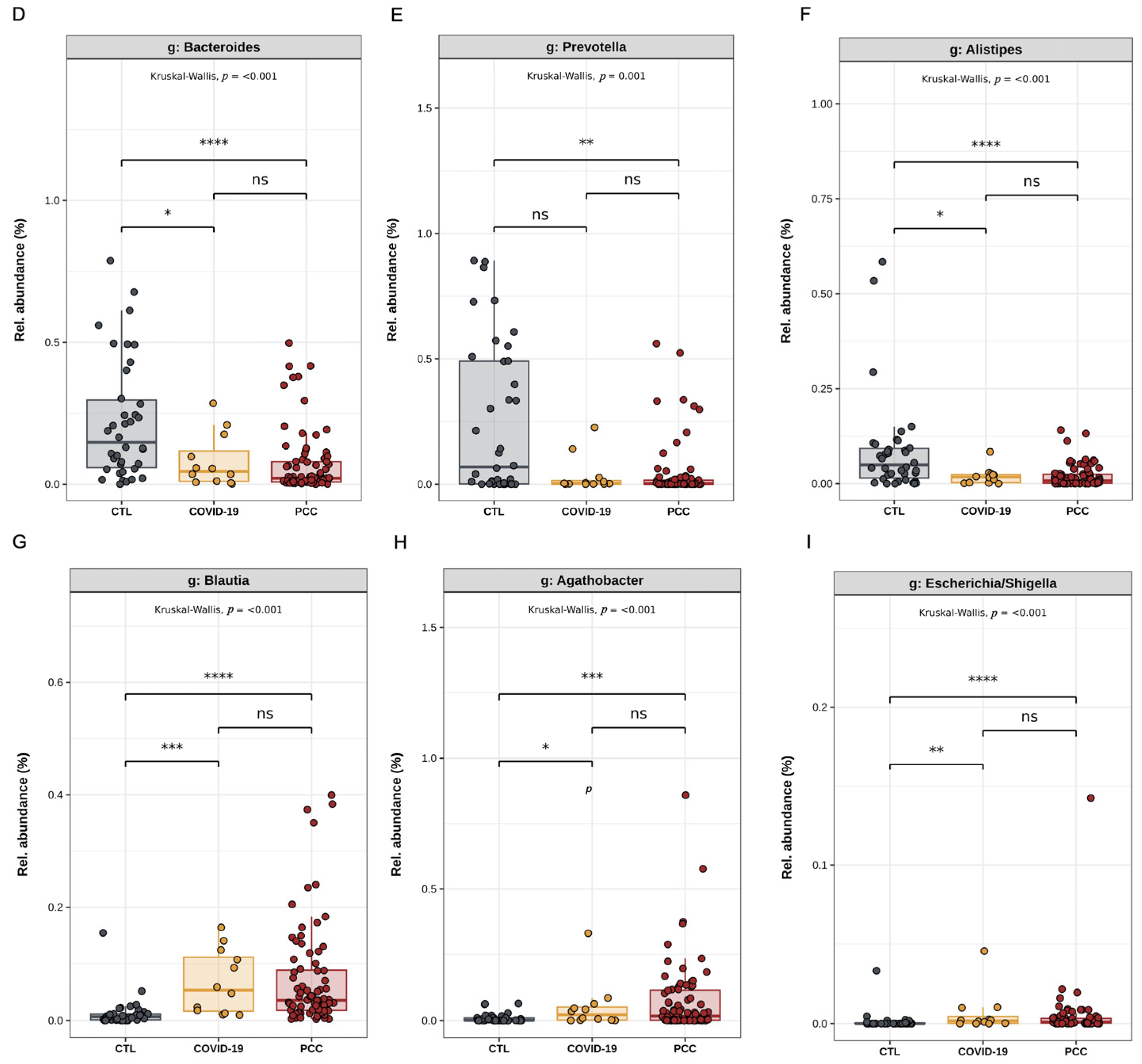
3.2. Bacteriome Signature in Patients Infected with SARS-CoV-2 Virus
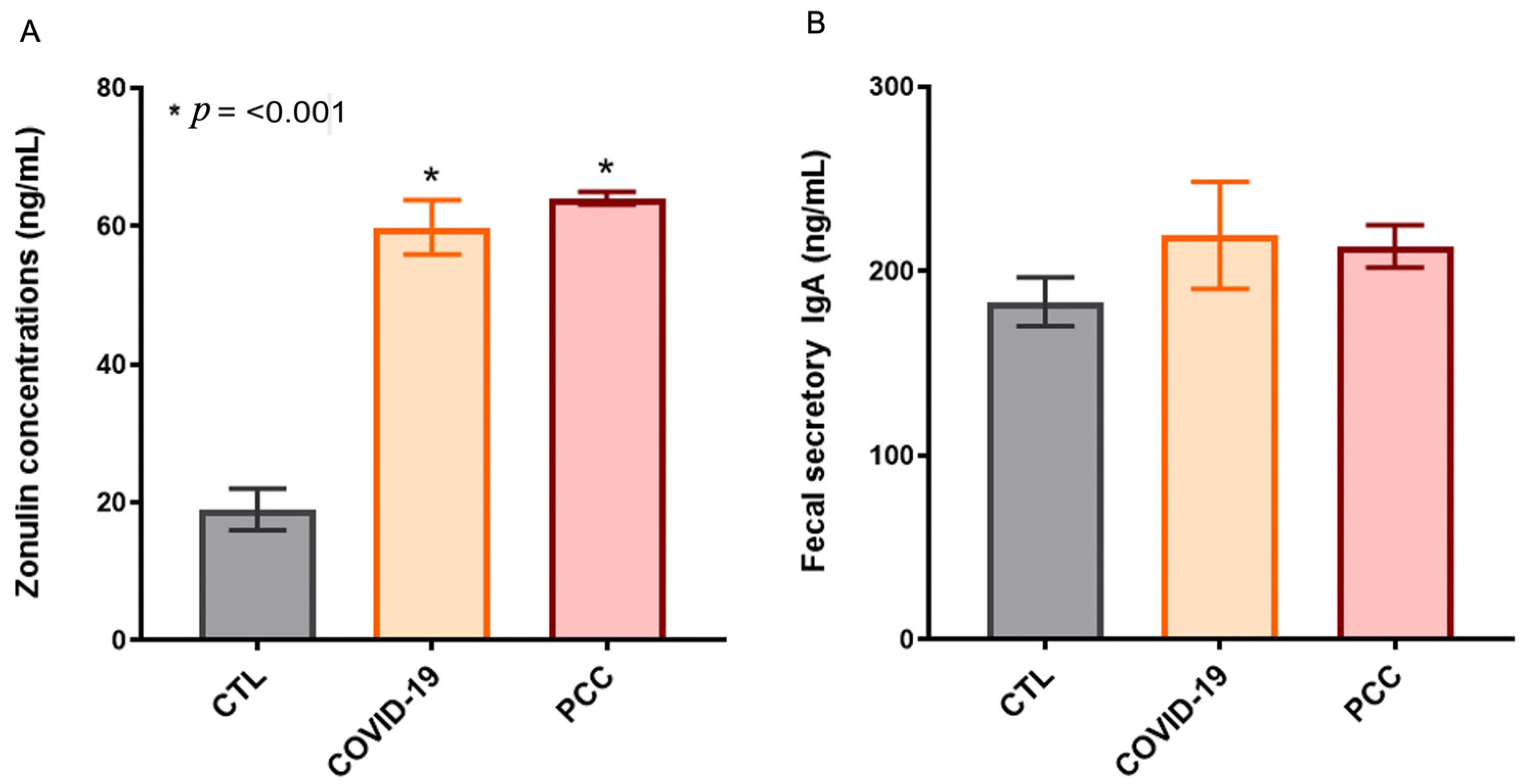
3.3. Increased Gut Permeability in SARS-CoV-2-Infected Patients
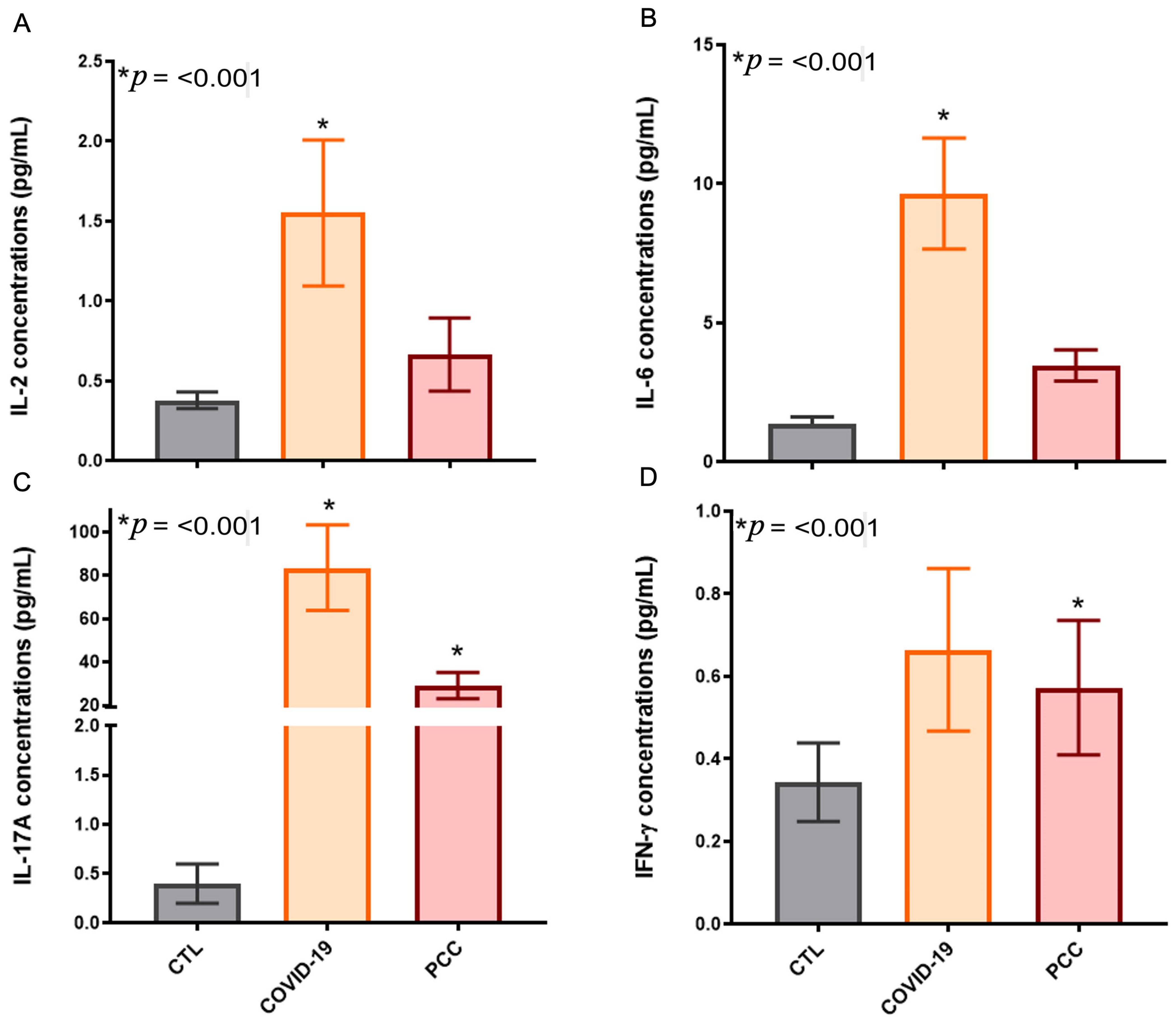
3.4. Increased Inflammatory Cytokines in SARS-CoV-2-Infected Patients
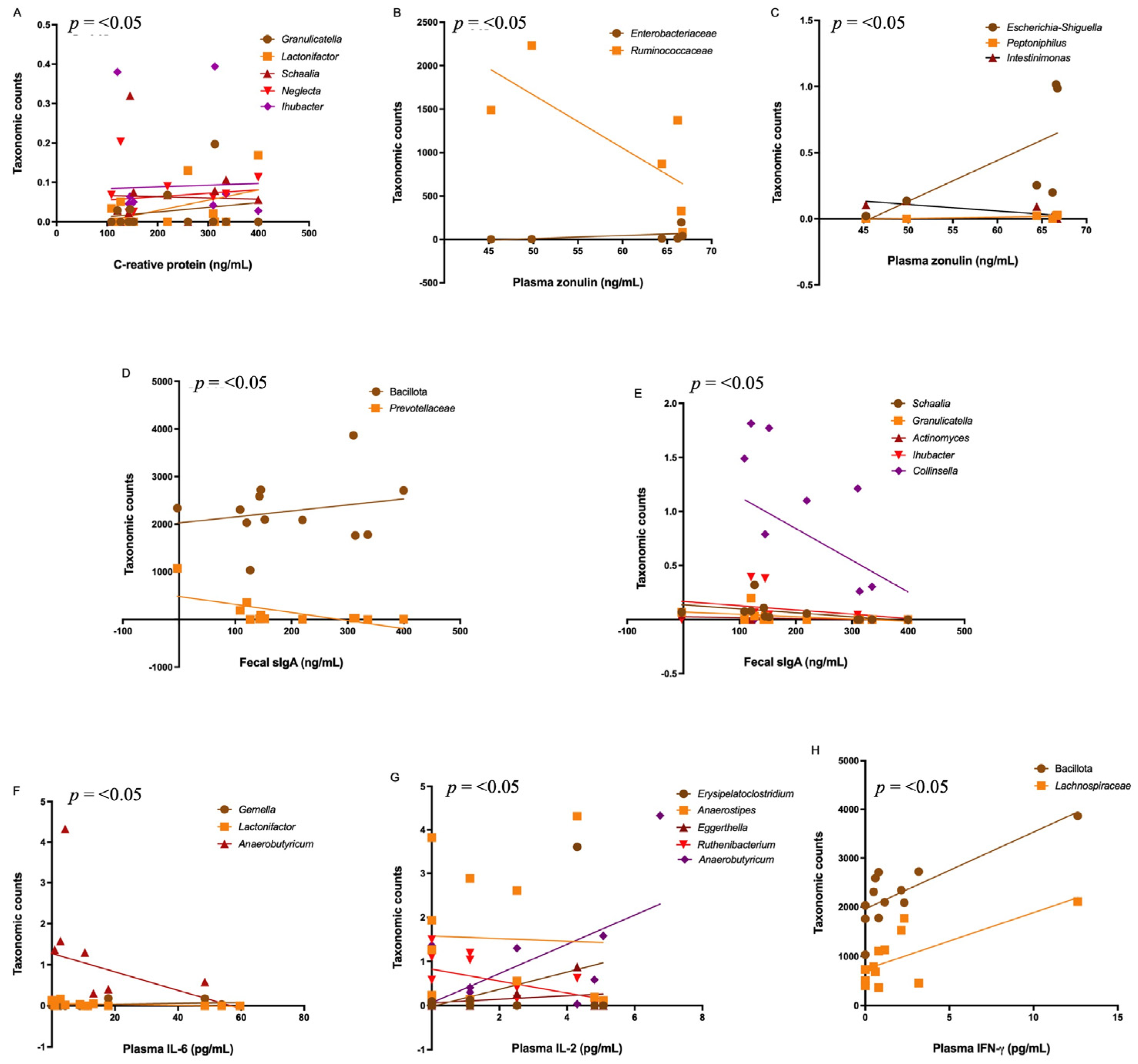
3.5. Correlations Among the Bacteriome, Clinical Data, Gut Permeability, and Cytokines
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Senevirathne, T.H.; Wekking, D.; Swain, J.W.R.; Solinas, C.; De Silva, P. COVID-19: From emerging variants to vaccination. Cytokine Growth Factor Rev. 2024, 76, 127–141. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO COVID-19 Dashboard. Available online: https://data.who.int/dashboards/covid19/cases?n=c (accessed on 10 June 2025).
- Brazilian Ministry of Health. Coronavirus Panel. Available online: https://covid.saude.gov.br/ (accessed on 10 June 2025).
- Wiersinga, W.J.; Rhodes, A.; Cheng, A.C.; Peacock, S.J.; Prescott, H.C. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA 2020, 324, 782–793. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, R.T.; Lynch, J.B.; Del Rio, C. Mild or Moderate Covid-19. N. Engl. J. Med. 2020, 383, 1757–1766. [Google Scholar] [CrossRef]
- Berlin, D.A.; Gulick, R.M.; Martinez, F.J. Severe Covid-19. N. Engl. J. Med. 2020, 383, 2451–2460. [Google Scholar] [CrossRef]
- Brodin, P. Immune determinants of COVID-19 disease presentation and severity. Nat. Med. 2021, 27, 28–33. [Google Scholar] [CrossRef]
- Gheorghita, R.; Soldanescu, I.; Lobiuc, A.; Caliman Sturdza, O.A.; Filip, R.; Constantinescu-Bercu, A.; Dimian, M.; Mangul, S.; Covasa, M. The knowns and unknowns of long COVID-19: From mechanisms to therapeutical approaches. Front. Immunol. 2024, 15, 1344086. [Google Scholar] [CrossRef]
- Hurme, A.; Viinanen, A.; Teräsjärvi, J.; Jalkanen, P.; Feuth, T.; Löyttyniemi, E.; Vuorinen, T.; Kantele, A.; Oksi, J.; He, Q.; et al. Post-COVID-19 condition in prospective inpatient and outpatient cohorts. Sci. Rep. 2025, 15, 6925. [Google Scholar] [CrossRef]
- Nalbandian, A.; Sehgal, K.; Gupta, A.; Madhavan, M.V.; McGroder, C.; Stevens, J.S.; Cook, J.R.; Nordvig, A.S.; Shalev, D.; Sehrawat, T.S.; et al. Post-acute COVID-19 syndrome. Nat. Med. 2021, 27, 601–615. [Google Scholar] [CrossRef]
- Parotto, M.; Gyöngyösi, M.; Howe, K.; Myatra, S.N.; Ranzani, O.; Shankar-Hari, M.; Herridge, M.S. Post-acute sequelae of COVID-19: Understanding and addressing the burden of multisystem manifestations. Lancet Respir. Med. 2023, 11, 739–754. [Google Scholar] [CrossRef]
- Santos, M.; Dorna, M.; Franco, E.; Geronutti, J.; Brizola, L.; Ishimoto, L.; Barros, Y.; Costa, A.; Breda, C.; Marin, C.; et al. Clinical and Physiological Variables in Patients with Post-COVID-19 Condition and Persistent Fatigue. J. Clin. Med. 2024, 13, 3876. [Google Scholar] [CrossRef] [PubMed]
- Davis, H.E.; McCorkell, L.; Vogel, J.M.; Topol, E.J. Long COVID: Major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 2023, 21, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Oelsner, E.C.; Sun, Y.; Balte, P.P.; Allen, N.B.; Andrews, H.; Carson, A.; Cole, S.A.; Coresh, J.; Couper, D.; Cushman, M.; et al. Epidemiologic Features of Recovery From SARS-CoV-2 Infection. JAMA Netw. Open 2024, 7, e2417440. [Google Scholar] [CrossRef] [PubMed]
- Soriano, J.B.; Murthy, S.; Marshall, J.C.; Relan, P.; Diaz, J.V. WHO Clinical Case Definition Working Group on Post-COVID-19 Condition. A clinical case definition of post-COVID-19 condition by a Delphi consensus. Lancet Infect. Dis. 2022, 22, e102–e107. [Google Scholar] [CrossRef]
- Lamers, M.M.; Haagmans, B.L. SARS-CoV-2 pathogenesis. Nat. Rev. Microbiol. 2022, 20, 270–284. [Google Scholar] [CrossRef]
- Steiner, S.; Kratzel, A.; Barut, G.T.; Lang, R.M.; Aguiar Moreira, E.; Thomann, L.; Kelly, J.N.; Thiel, V. SARS-CoV-2 biology and host interactions. Nat. Rev. Microbiol. 2024, 22, 206–225. [Google Scholar] [CrossRef]
- Pozdnyakova, V.; Weber, B.; Cheng, S.; Ebinger, J.E. Review of Immunologic Manifestations of COVID-19 Infection and Vaccination. Rheum. Dis. Clin. N. Am. 2025, 51, 111–121. [Google Scholar] [CrossRef]
- Karki, R.; Sharma, B.R.; Tuladhar, S.; Williams, E.P.; Zalduondo, L.; Samir, P.; Zheng, M.; Sundaram, B.; Banoth, B.; Malireddi, R.K.S.; et al. Synergism of TNF-α and IFN-γ Triggers Inflammatory Cell Death, Tissue Damage, and Mortality in SARS-CoV-2 Infection and Cytokine Shock Syndromes. Cell 2021, 184, 149–168.e17. [Google Scholar] [CrossRef]
- Diamond, M.S.; Kanneganti, T.D. Innate immunity: The first line of defense against SARS-CoV-2. Nat. Immunol. 2022, 23, 165–176. [Google Scholar] [CrossRef]
- Zheng, M.; Karki, R.; Williams, E.P.; Yang, D.; Fitzpatrick, E.; Vogel, P.; Jonsson, C.B.; Kanneganti, T.-D. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat. Immunol. 2021, 22, 829–838. [Google Scholar] [CrossRef]
- Sette, A.; Crotty, S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 2021, 184, 861–880. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Liu, B.; Wang, X.; Zhang, L. The humoral response and antibodies against SARS-CoV-2 infection. Nat. Immunol. 2022, 23, 1008–1020. [Google Scholar] [CrossRef]
- Phetsouphanh, C.; Darley, D.R.; Wilson, D.B.; Howe, A.; Munier, C.M.L.; Patel, S.K.; Juno, J.A.; Burrell, L.M.; Kent, S.J.; Dore, G.J.; et al. Immunological dysfunction persists for 8 months following initial mild-to-moderate SARS-CoV-2 infection. Nat. Immunol. 2022, 23, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.; Peluso, M.J.; Luo, X.; Thomas, R.; Shin, M.G.; Neidleman, J.; Andrew, A.; Young, K.C.; Ma, T.; Hoh, R.; et al. Long COVID manifests with T cell dysregulation, inflammation and an uncoordinated adaptive immune response to SARS-CoV-2. Nat. Immunol. 2024, 25, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Phetsouphanh, C.; Jacka, B.; Ballouz, S.; Jackson, K.J.L.; Wilson, D.B.; Manandhar, B.; Klemm, V.; Tan, H.-X.; Wheatley, A.; Aggarwal, A.; et al. Improvement of immune dysregulation in individuals with long COVID at 24-months following SARS-CoV-2 infection. Nat. Commun. 2024, 15, 3315. [Google Scholar] [CrossRef]
- Oliveira, G.L.V.; Oliveira, C.N.S.; Pinzan, C.F.; de Salis, L.V.V.; Cardoso, C.R.B. Microbiota Modulation of the Gut-Lung Axis in COVID-19. Front. Immunol. 2021, 12, 635471. [Google Scholar] [CrossRef]
- Budden, K.F.; Gellatly, S.L.; Wood, D.L.; Cooper, M.A.; Morrison, M.; Hugenholtz, P.; Hansbro, P.M. Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 2017, 15, 55–63. [Google Scholar] [CrossRef]
- Grayson, M.H.; Camarda, L.E.; Hussain, S.A.; Zemple, S.J.; Hayward, M.; Lam, V.; Hunter, D.A.; Santoro, J.L.; Rohlfing, M.; Cheung, D.S.; et al. Intestinal Microbiota Disruption Reduces Regulatory T Cells and Increases Respiratory Viral Infection Mortality Through Increased IFNγ Production. Front. Immunol. 2018, 9, 1587. [Google Scholar] [CrossRef]
- Dang, A.T.; Marsland, B.J. Microbes, metabolites, and the gut-lung axis. Mucosal Immunol. 2019, 12, 843–850. [Google Scholar] [CrossRef]
- Enaud, R.; Prevel, R.; Ciarlo, E.; Beaufils, F.; Wieërs, G.; Guery, B.; Delhaes, L. The Gut-Lung Axis in Health and Respiratory Diseases: A Place for Inter-Organ and Inter-Kingdom Crosstalks. Front. Cell. Infect. Microbiol. 2020, 10, 9. [Google Scholar] [CrossRef]
- Zhang, D.; Li, S.; Wang, N.; Tan, H.Y.; Zhang, Z.; Feng, Y. The Cross-Talk Between Gut Microbiota and Lungs in Common Lung Diseases. Front. Microbiol. 2020, 11, 301. [Google Scholar] [CrossRef] [PubMed]
- Stefan, K.L.; Kim, M.V.; Iwasaki, A.; Kasper, D.L. Commensal Microbiota Modulation of Natural Resistance to Virus Infection. Cell 2020, 183, 1312–1324.e10. [Google Scholar] [CrossRef] [PubMed]
- Ngo, V.L.; Lieber, C.M.; Kang, H.J.; Sakamoto, K.; Kuczma, M.; Plemper, R.K.; Gewirtz, A.T. Intestinal microbiota programming of alveolar macrophages influences severity of respiratory viral infection. Cell Host Microbe 2024, 32, 335–348.e8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ma, Y.; Sun, W.; Zhou, X.; Wang, R.; Xie, P.; Dai, L.; Gao, Y.; Li, J. Exploring gut-lung axis crosstalk in SARS-CoV-2 infection: Insights from a hACE2 mouse model. J. Med. Virol. 2024, 96, e29336. [Google Scholar] [CrossRef]
- Giron, L.B.; Dweep, H.; Yin, X.; Wang, H.; Damra, M.; Goldman, A.R.; Gorman, N.; Palmer, C.S.; Tang, H.-Y.; Shaikh, M.W.; et al. Plasma Markers of Disrupted Gut Permeability in Severe COVID-19 Patients. Front. Immunol. 2021, 12, 686240. [Google Scholar]
- Prasad, R.; Patton, M.J.; Floyd, J.L.; Fortmann, S.; DuPont, M.; Harbour, A.; Wright, J.; Lamendella, R.; Stevens, B.R.; Oudit, G.Y.; et al. Plasma Microbiome in COVID-19 Subjects: An Indicator of Gut Barrier Defects and Dysbiosis. Int. J. Mol. Sci. 2022, 23, 9141. [Google Scholar] [CrossRef]
- Bernard-Raichon, L.; Venzon, M.; Klein, J.; Axelrad, J.E.; Zhang, C.; Sullivan, A.P.; Hussey, G.A.; Casanovas-Massana, A.; Noval, M.G.; Valero-Jimenez, A.M.; et al. Gut microbiome dysbiosis in antibiotic-treated COVID-19 patients is associated with microbial translocation and bacteremia. Nat. Commun. 2022, 13, 5926. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics—Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Cole, J.R.; Chai, B.; Farris, R.J.; Wang, Q.; Kulam, S.A.; McGarrell, D.M.; Garrity, G.M.; Tiedje, J.M. The Ribosomal Database Project (RDP-II): Sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005, 33, D294–D296. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Schliep, K.P. phangorn: Phylogenetic analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, V. metagMisc: Miscellaneous Functions for Metagenomic Analysis. R package version 1.0.0. 2022. 2023. Available online: https://github.com/vmikk/metagMisc (accessed on 20 December 2024).
- Andersen, K.S.; Kirkegaard, R.H.; Karst, S.M.; Albertsen, M. ampvis2: An R Package to Analyse and Visualise 16S rRNA Amplicon Data. bioRxiv 2018. [Google Scholar] [CrossRef]
- Lahti, L.; Shetty, S. Microbiome: An R Package for Microbiome Analysis. [R Package]. 2012. Available online: https://microbiome.github.io/microbiome/ (accessed on 20 December 2024).
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’HAra, R.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. vegan: Community Ecology Package; 2022. Available online: https://cran.r-project.org/package=vegan (accessed on 20 December 2024).
- Arbizu, P.M. pairwiseAdonis: Pairwise Multilevel Comparison Using Adonis; 2017. Available online: https://github.com/pmartinezarbizu/pairwiseAdonis (accessed on 20 December 2024).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Sencio, V.; Machado, M.G.; Trottein, F. The lung-gut axis during viral respiratory infections: The impact of gut dysbiosis on secondary disease outcomes. Mucosal Immunol. 2021, 14, 296–304. [Google Scholar] [CrossRef]
- Allali, I.; Bakri, Y.; Amzazi, S.; Ghazal, H. Gut-Lung Axis in COVID-19. Interdiscip. Perspect. Infect. Dis. 2021, 2021, 6655380. [Google Scholar] [CrossRef]
- Yang, Y.; Huang, W.; Fan, Y.; Chen, G.Q. Gastrointestinal Microenvironment and the Gut-Lung Axis in the Immune Responses of Severe COVID-19. Front. Mol. Biosci. 2021, 8, 647508. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, L.; Wang, Y.; Dai, T.; Qin, Z.; Zhou, F.; Zhang, L. Alterations in microbiota of patients with COVID-19: Potential mechanisms and therapeutic interventions. Signal Transduct. Target. Ther. 2022, 7, 143. [Google Scholar] [CrossRef]
- Zhang, F.; Lau, R.I.; Liu, Q.; Su, Q.; Chan, F.K.L.; Ng, S.C. Gut microbiota in COVID-19: Key microbial changes, potential mechanisms and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 323–337. [Google Scholar] [CrossRef]
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.C.K.; Cheung, C.P.; Chen, N.; et al. Alterations in Gut Microbiota of Patients With COVID-19 During Time of Hospitalization. Gastroenterology 2020, 159, 944–955.e8. [Google Scholar] [CrossRef]
- Gu, S.; Chen, Y.; Wu, Z.; Chen, Y.; Gao, H.; Lv, L.; Guo, F.; Zhang, X.; Luo, R.; Huang, C.; et al. Alterations of the Gut Microbiota in Patients With Coronavirus Disease 2019 or H1N1 Influenza. Clin. Infect. Dis. 2020, 71, 2669–2678. [Google Scholar] [CrossRef]
- Zuo, T.; Liu, Q.; Zhang, F.; Lui, G.C.; Tso, E.Y.; Yeoh, Y.K.; Chen, Z.; Boon, S.; Chan, F.K.L.; Chan, P.; et al. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut 2021, 70, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Yeoh, Y.K.; Zuo, T.; Lui, G.C.; Zhang, F.; Liu, Q.; Li, A.Y.; Chung, A.C.; Cheung, C.P.; Tso, E.Y.; Fung, K.S.; et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 2021, 70, 698–706. [Google Scholar] [CrossRef] [PubMed]
- Nobre, J.G.; Delgadinho, M.; Silva, C.; Mendes, J.; Mateus, V.; Ribeiro, E.; Costa, D.A.; Lopes, M.; Pedroso, A.I.; Trigueiros, F.; et al. Gut microbiota profile of COVID-19 patients: Prognosis and risk stratification (MicroCOVID-19 study). Front. Microbiol. 2022, 13, 1035422. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, K.; Hirata, H.; Tokuhira, N.; Motooka, D.; Nakamura, S.; Ueda, A.; Tachino, J.; Koide, M.; Uchiyama, A.; Ogura, H.; et al. Dysbiosis of gut microbiota in patients with severe COVID-19. Acute Med. Surg. 2024, 11, e923. [Google Scholar] [CrossRef]
- Yokoyama, Y.; Ichiki, T.; Yamakawa, T.; Tsuji, Y.; Kuronuma, K.; Takahashi, S.; Narimatsu, E.; Katanuma, A.; Nakase, H. Gut microbiota and metabolites in patients with COVID-19 are altered by the type of SARS-CoV-2 variant. Front. Microbiol. 2024, 15, 1358530. [Google Scholar] [CrossRef]
- Bucci, V.; Ward, D.V.; Bhattarai, S.; Rojas-Correa, M.; Purkayastha, A.; Holler, D.; Da Qu, M.; Mitchell, W.G.; Yang, J.; Fountain, S.; et al. The intestinal microbiota predicts COVID-19 severity and fatality regardless of hospital feeding method. mSystems 2023, 8, e0031023. [Google Scholar] [CrossRef]
- Zhong, J.; Guo, L.; Wang, Y.; Jiang, X.; Wang, C.; Xiao, Y.; Wang, Y.; Zhou, F.; Wu, C.; Chen, L.; et al. Gut Microbiota Improves Prognostic Prediction in Critically Ill COVID-19 Patients Alongside Immunological and Hematological Indicators. Research 2024, 7, 0389. [Google Scholar] [CrossRef]
- Nagata, N.; Takeuchi, T.; Masuoka, H.; Aoki, R.; Ishikane, M.; Iwamoto, N.; Sugiyama, M.; Suda, W.; Nakanishi, Y.; Terada-Hirashima, J.; et al. Human Gut Microbiota and Its Metabolites Impact Immune Responses in COVID-19 and Its Complications. Gastroenterology 2023, 164, 272–288. [Google Scholar] [CrossRef]
- An, Y.; He, L.; Xu, X.; Piao, M.; Wang, B.; Liu, T.; Cao, H. Gut microbiota in post-acute COVID-19 syndrome: Not the end of the story. Front. Microbiol. 2024, 15, 1500890. [Google Scholar] [CrossRef]
- Chen, Y.; Gu, S.; Chen, Y.; Lu, H.; Shi, D.; Guo, J.; Wu, W.-R.; Yang, Y.; Li, Y.; Xu, K.-J.; et al. Six-month follow-up of gut microbiota richness in patients with COVID-19. Gut 2022, 71, 222–225. [Google Scholar] [CrossRef]
- Liu, Q.; Mak, J.W.Y.; Su, Q.; Yeoh, Y.K.; Lui, G.C.; Ng, S.S.S.; Zhang, F.; Li, A.Y.L.; Lu, W.; Hui, D.S.-C.; et al. Gut microbiota dynamics in a prospective cohort of patients with post-acute COVID-19 syndrome. Gut 2022, 71, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Ferreira-Junior, A.S.; Borgonovi, T.F.; De Salis, L.V.V.; Leite, A.Z.; Dantas, A.S.; De Salis, G.V.V.; Cruz, G.N.F.; De Oliveira, L.F.V.; Gomes, E.; Penna, A.L.B.; et al. Detection of Intestinal Dysbiosis in Post-COVID-19 Patients One to Eight Months after Acute Disease Resolution. Int. J. Environ. Res. Public. Health 2022, 19, 10189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wan, Y.; Zuo, T.; Yeoh, Y.K.; Liu, Q.; Zhang, L.; Zhan, H.; Lu, W.; Xu, W.; Lui, G.C.; et al. Prolonged Impairment of Short-Chain Fatty Acid and L-Isoleucine Biosynthesis in Gut Microbiome in Patients With COVID-19. Gastroenterology 2022, 162, 548–561.e4. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.; Lau, R.I.; Liu, Q.; Li, M.K.T.; Yan Mak, J.W.; Lu, W.; Lau, I.S.; Lau, L.H.; Yeung, G.T.; Cheung, C.P.; et al. The gut microbiome associates with phenotypic manifestations of post-acute COVID-19 syndrome. Cell Host Microbe 2024, 32, 651–660.e4. [Google Scholar] [CrossRef]
- Blankestijn, J.M.; Baalbaki, N.; Beijers, R.J.H.C.G.; Cornelissen, M.E.B.; Wiersinga, W.J.; Abdel-Aziz, M.I.; der Zee, A.H.M.-V. P4O2 Consortium. Exploring Heterogeneity of Fecal Microbiome in Long COVID Patients at 3 to 6 Months After Infection. Int. J. Mol. Sci. 2025, 26, 1781. [Google Scholar] [CrossRef]
- Darif, D.; Hammi, I.; Kihel, A.; El Idrissi Saik, I.; Guessous, F.; Akarid, K. The pro-inflammatory cytokines in COVID-19 pathogenesis: What goes wrong? Microb. Pathog. 2021, 153, 104799. [Google Scholar] [CrossRef]
- Zanza, C.; Romenskaya, T.; Manetti, A.C.; Franceschi, F.; La Russa, R.; Bertozzi, G.; Maiese, A.; Savioli, G.; Volonnino, G.; Longhitano, Y. Cytokine Storm in COVID-19: Immunopathogenesis and Therapy. Medicina 2022, 58, 144. [Google Scholar] [CrossRef]
- Deng, X.; Tang, K.; Wang, Z.; He, S.; Luo, Z. Impacts of Inflammatory Cytokines Variants on Systemic Inflammatory Profile and COVID-19 Severity. J. Epidemiol. Glob. Health 2024, 14, 363–378. [Google Scholar] [CrossRef]
- Islam, F.; Habib, S.; Badruddza, K.; Rahman, M.; Islam, M.R.; Sultana, S.; Nessa, A. The Association of Cytokines IL-2, IL-6, TNF-α, IFN-γ, and IL-10 With the Disease Severity of COVID-19: A Study From Bangladesh. Cureus 2024, 16, e57610. [Google Scholar] [CrossRef]
- Safont, G.; Villar-Hernández, R.; Smalchuk, D.; Stojanovic, Z.; Marín, A.; Lacoma, A.; Pérez-Cano, C.; López-Martínez, A.; Molina-Moya, B.; Solis, A.J.; et al. Measurement of IFN-γ and IL-2 for the assessment of the cellular immunity against SARS-CoV-2. Sci. Rep. 2024, 14, 1137. [Google Scholar] [CrossRef]
- Qin, C.; Zhou, L.; Hu, Z.; Zhang, S.; Yang, S.; Tao, Y.; Xie, C.; Ma, K.; Shang, K.; Wang, W.; et al. Dysregulation of Immune Response in Patients With Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020, 71, 762–768. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Huang, S.; Yin, L. The cytokine storm and COVID-19. J. Med. Virol. 2021, 93, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Kervevan, J.; Staropoli, I.; Slama, D.; Jeger-Madiot, R.; Donnadieu, F.; Planas, D.; Pietri, M.-P.; Loghmari-Bouchneb, W.; Tanah, M.A.; Robinot, R.; et al. Divergent adaptive immune responses define two types of long COVID. Front. Immunol. 2023, 14, 1221961. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, A.; Maddumage, J.; Eriksson, E.M.; Annesley, S.J.; Lawson, V.A.; Bryant, V.L.; Gras, S. Beyond acute infection: Mechanisms underlying post-acute sequelae of COVID-19 (PASC). Med. J. Aust. 2024, 9, S40–S48. [Google Scholar] [CrossRef]
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.R.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B.; et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 2020, 584, 463–469. [Google Scholar] [CrossRef]
- Mahmood, S.B.Z.; Majid, H.; Arshad, A.; Zaib-Un-Nisa; Niazali, N.; Kazi, K.; Aslam, A.; Ahmed, S.; Jamil, B.; Jafri, L. Interleukin-6 (IL-6) as a Predictor of Clinical Outcomes in Patients with COVID-19. Clin. Lab. 2023, 69, 1126–1133. [Google Scholar] [CrossRef]
- Herold, T.; Jurinovic, V.; Arnreich, C.; Lipworth, B.J.; Hellmuth, J.C.; von Bergwelt-Baildon, M.; Klein, M.; Weinberger, T. Elevated levels of IL-6 and CRP predict the need for mechanical ventilation in COVID-19. J. Allergy Clin. Immunol. 2020, 146, 128–136.e4. [Google Scholar] [CrossRef]
- Chen, X.; Zhao, B.; Qu, Y.; Chen, Y.; Xiong, J.; Feng, Y.; Men, D.; Huang, Q.; Liu, Y.; Yang, B.; et al. Detectable Serum Severe Acute Respiratory Syndrome Coronavirus 2 Viral Load (RNAemia) Is Closely Correlated With Drastically Elevated Interleukin 6 Level in Critically Ill Patients With Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 71, 1937–1942. [Google Scholar] [CrossRef]
- Zhang, F.; Mears, J.R.; Shakib, L.; Beynor, J.I.; Shanaj, S.; Korsunsky, I.; Nathan, A.; Donlin, L.T.; Raychaudhuri, S. IFN-γ and TNF-α drive a CXCL10+ CCL2+ macrophage phenotype expanded in severe COVID-19 lungs and inflammatory diseases with tissue inflammation. Genome Med. 2021, 13, 64. [Google Scholar] [CrossRef]
- Mouchati, C.; Durieux, J.C.; Zisis, S.N.; Labbato, D.; Rodgers, M.A.; Ailstock, K.; Reinert, B.L.; Funderburg, N.T.; McComsey, G.A. Increase in gut permeability and oxidized ldl is associated with post-acute sequelae of SARS-CoV-2. Front. Immunol. 2023, 14, 1182544. [Google Scholar] [CrossRef]
- Gallo, A.; Murace, C.A.; Corbo, M.M.; Sarlo, F.; De Ninno, G.; Baroni, S.; Fancello, G.; Masucci, L.; Covino, M.; Tosato, M.; et al. Gemelli against COVID-19 Post-Acute Care Team. Intestinal Inflammation and Permeability in Patients Recovered from SARS-CoV-2 Infection. Dig. Dis. 2025, 43, 1–10. [Google Scholar] [PubMed]
- Oliva, A.; Miele, M.C.; Di Timoteo, F.; De Angelis, M.; Mauro, V.; Aronica, R.; Al Ismail, D.; Ceccarelli, G.; Pinacchio, C.; d’Ettorre, G.; et al. Persistent Systemic Microbial Translocation and Intestinal Damage During Coronavirus Disease-19. Front. Immunol. 2021, 12, 708149. [Google Scholar] [CrossRef] [PubMed]
- Oliva, A.; Cammisotto, V.; Cangemi, R.; Ferro, D.; Miele, M.C.; De Angelis, M.; Cancelli, F.; Pignatelli, P.; Venditti, M.; Pugliese, F.; et al. Low-Grade Endotoxemia and Thrombosis in COVID-19. Clin. Transl. Gastroenterol. 2021, 12, e00348. [Google Scholar] [CrossRef] [PubMed]
- Palomino-Kobayashi, L.A.; Ymaña, B.; Ruiz, J.; Mayanga-Herrera, A.; Ugarte-Gil, M.F.; Pons, M.J. Zonulin, a marker of gut permeability, is associated with mortality in a cohort of hospitalised peruvian COVID-19 patients. Front. Cell. Infect. Microbiol. 2022, 12, 1000291. [Google Scholar]
| COVID-19 (n = 79) | PCC (n = 141) | CTL (n = 97) | |
|---|---|---|---|
| Biological sex Female/Male | 46 F/33 M | 88 F/53 M | 83 F/14 M |
| Age (Years) Mean ± SD | 51.8 ± 16.3 | 40.8 ± 13.9 | 43.8 ± 13.6 |
| BMI (Kg/m2) Mean ± SD | 28.6 ± 6.2 | 29.1 ± 5.4 | 25.2 ± 4.8 |
| CRP (mg/dL) Mean ± SD | 78.6 ± 67.0 | 8.4 ± 9.6 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Souza, L.S.; Ferreira-Junior, A.S.; Estella, P.C.; Noda, R.K.; Sousa, L.F.; Murata, M.T.Y.; Carvalho, L.A.L.; Brisotti, J.L.; Pinheiro, D.G.; Rodrigues, J.; et al. Bacteriome Signature in SARS-CoV-2-Infected Patients Correlates with Increased Gut Permeability and Systemic Inflammatory Cytokines. Microorganisms 2025, 13, 1407. https://doi.org/10.3390/microorganisms13061407
Souza LS, Ferreira-Junior AS, Estella PC, Noda RK, Sousa LF, Murata MTY, Carvalho LAL, Brisotti JL, Pinheiro DG, Rodrigues J, et al. Bacteriome Signature in SARS-CoV-2-Infected Patients Correlates with Increased Gut Permeability and Systemic Inflammatory Cytokines. Microorganisms. 2025; 13(6):1407. https://doi.org/10.3390/microorganisms13061407
Chicago/Turabian StyleSouza, Larissa S., Alexandre S. Ferreira-Junior, Pedro C. Estella, Ricardo K. Noda, Lhorena F. Sousa, Miguel T. Y. Murata, Lucas A. L. Carvalho, João L. Brisotti, Daniel G. Pinheiro, Josias Rodrigues, and et al. 2025. "Bacteriome Signature in SARS-CoV-2-Infected Patients Correlates with Increased Gut Permeability and Systemic Inflammatory Cytokines" Microorganisms 13, no. 6: 1407. https://doi.org/10.3390/microorganisms13061407
APA StyleSouza, L. S., Ferreira-Junior, A. S., Estella, P. C., Noda, R. K., Sousa, L. F., Murata, M. T. Y., Carvalho, L. A. L., Brisotti, J. L., Pinheiro, D. G., Rodrigues, J., Fortaleza, C. M. C. B., & de Oliveira, G. L. V. (2025). Bacteriome Signature in SARS-CoV-2-Infected Patients Correlates with Increased Gut Permeability and Systemic Inflammatory Cytokines. Microorganisms, 13(6), 1407. https://doi.org/10.3390/microorganisms13061407








