Heterologous Expression and Antimicrobial Mechanism of a Cysteine-Rich Peptide from Barnacle Pollicipes pollicipes
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Prediction and Identification of Cysteine-Rich AMPs
2.3. Expression and Purification of Recombinant PpRcys1 (rPpRcys1)
2.4. Identification of PpRcys1 by Liquid Chromatography–Mass Spectrometry (LC-MS)
2.5. Antimicrobial Activity Assay of rPpRcys1
2.6. Molecular Dynamics (MD) Simulations
2.7. Microorganism-Binding Assay
2.8. Binding Assay for Pathogen-Associated Molecular Patterns
2.9. Electron Microscopy
2.10. Membrane Permeability Assay
2.11. Evaluation of Respiratory Chain Dehydrogenase Activity
2.12. Protease Inhibition Assay
2.13. DNA Binding Assay
2.14. Hemolytic Activity Assay
2.15. Statistical Analysis
3. Results
3.1. Sequence and Structure Characterization of PpRcys1
3.2. Recombinant Expression and Purification
3.3. Results of LC-MS Identification
3.4. Antimicrobial Activity of rPpRcys1
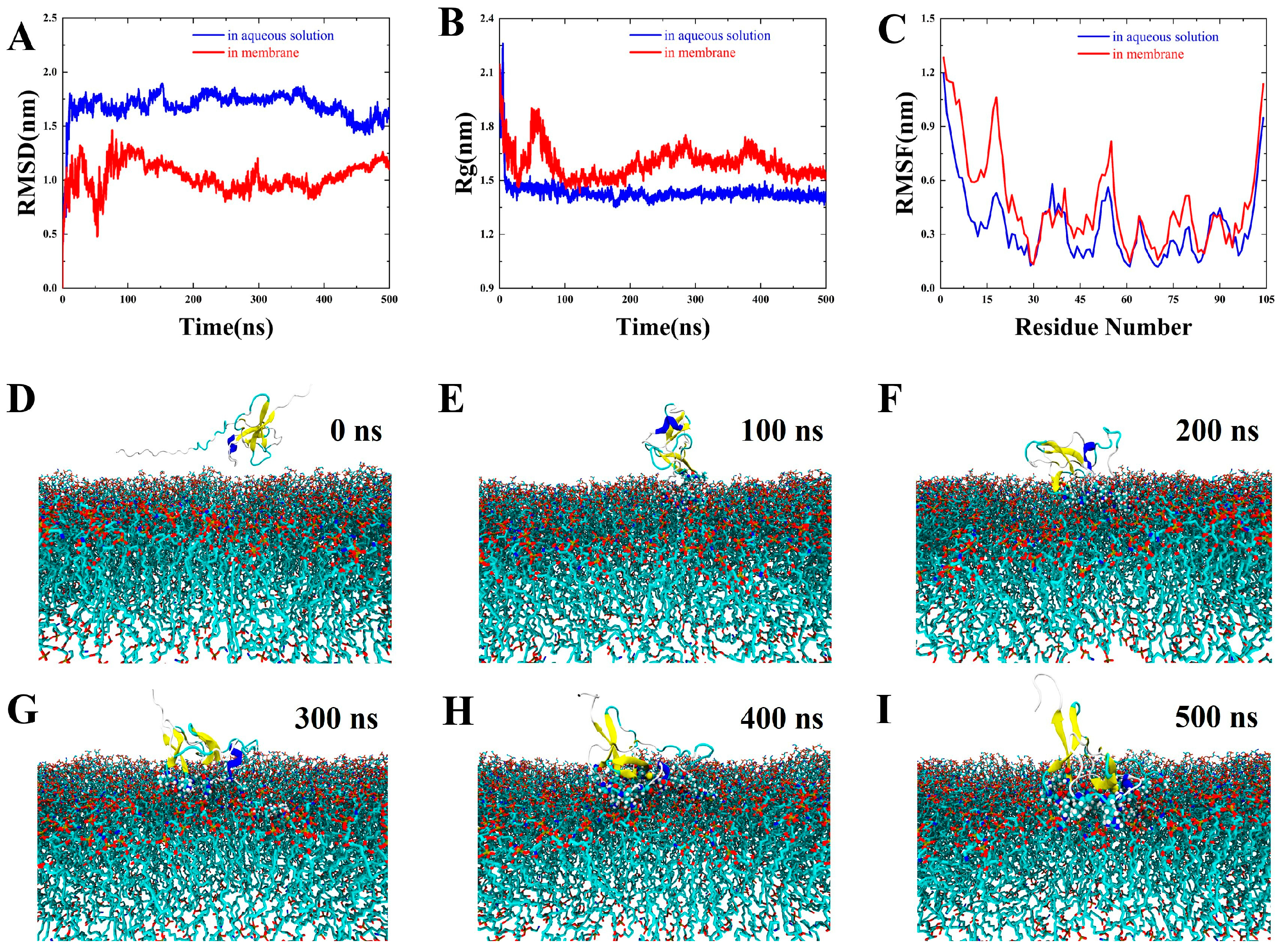
3.5. Results of Molecular Dynamics (MD) Simulations
3.6. Microorganism-Binding Activity of rPpRcys1
3.7. Pathogen-Associated Molecular Patterns Binding (PAMP) Activity of rPpRcys1
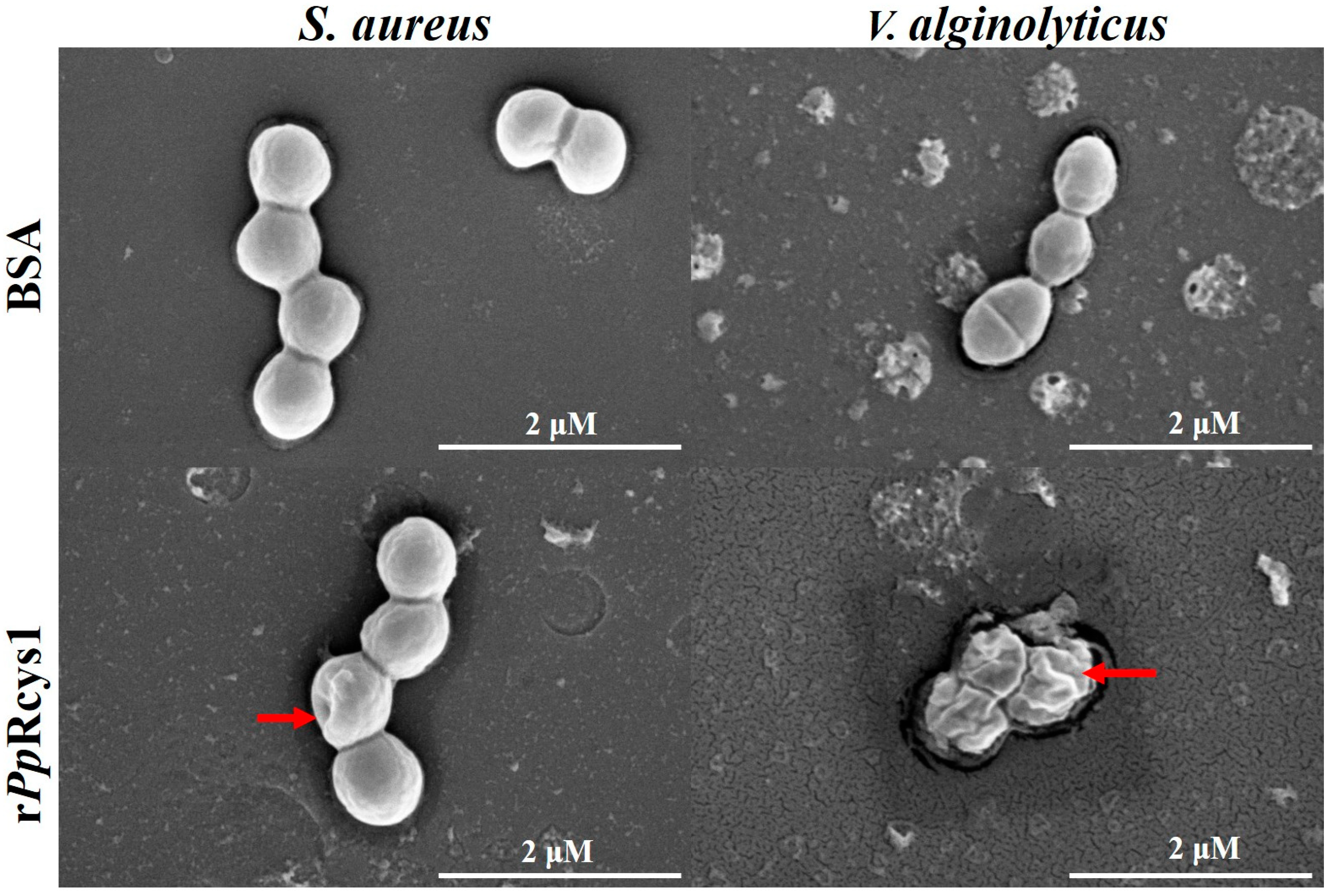
3.8. Effects of rPpRcys1 on Bacterial Morphology
3.9. Effects of rPpRcys1 on Membrane Permeability and the Respiratory Chain
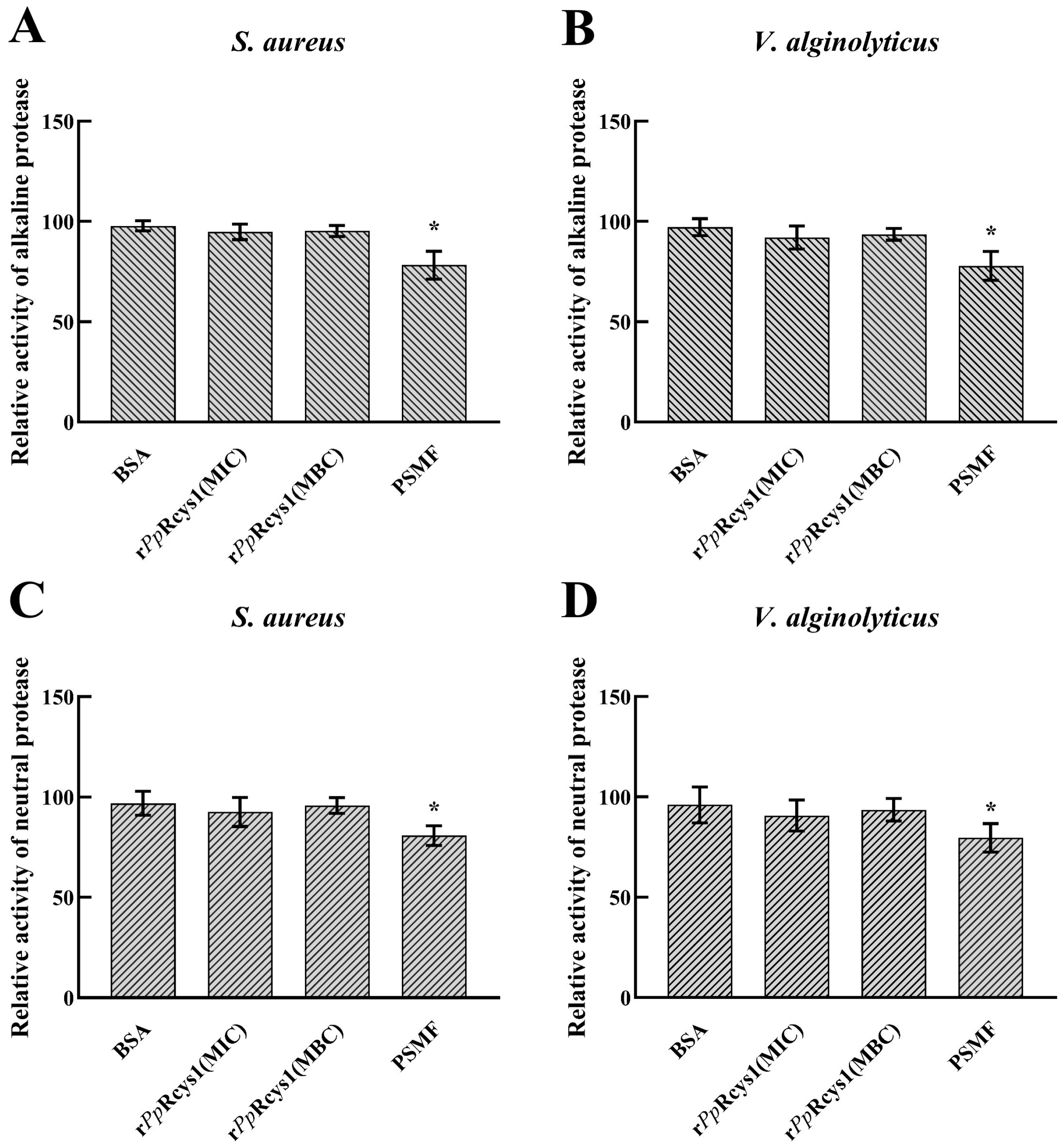
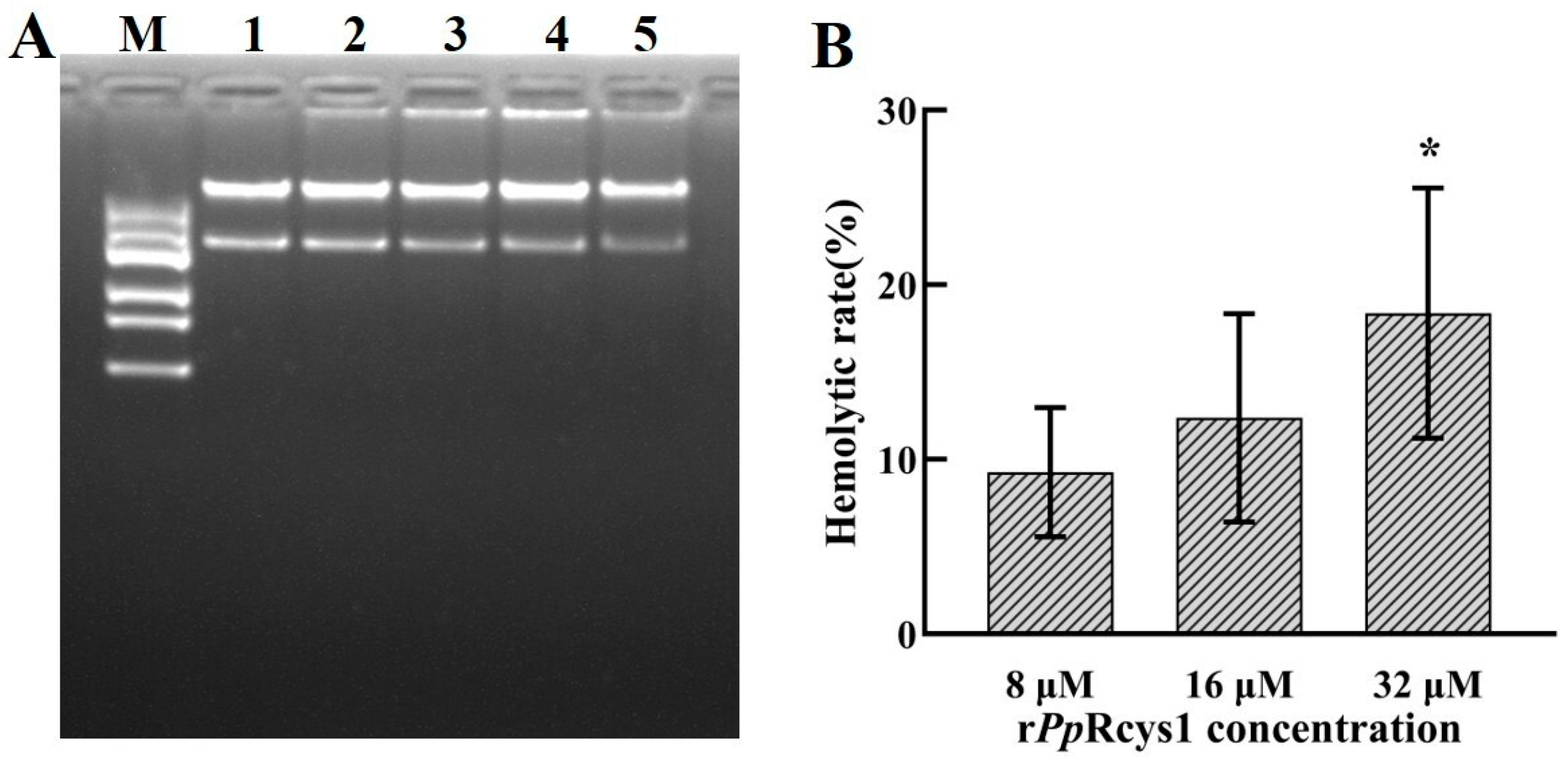
3.10. Effects of rPpRcys1 Protease Activity and DNA Migration
3.11. Hemolytic Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMP | antimicrobial peptide |
| LTA | lipoteichoic acid |
| PGN | peptidoglycan |
| LPS | lipopolysaccharide |
| Rg | gyration |
| RMSD | root mean square distances |
| RMSF | root mean square fluctuations |
| POPE | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine |
| POPG | phosphatidylglycerol |
References
- Bondad-Reantaso, M.G.; MacKinnon, B.; Karunasagar, I.; Fridman, S.; Alday-Sanz, V.; Brun, E.; Le Groumellec, M.; Li, A.; Surachetpong, W.; Karunasagar, I. Review of Alternatives to Antibiotic Use in Aquaculture. Rev. Aquac. 2023, 15, 1421–1451. [Google Scholar] [CrossRef]
- Mo, W.Y.; Chen, Z.; Leung, H.M.; Leung, A.O.W. Application of Veterinary Antibiotics in China’s Aquaculture Industry and Their Potential Human Health Risks. Environ. Sci. Pollut. Res. 2017, 24, 8978–8989. [Google Scholar] [CrossRef] [PubMed]
- Salem, M.; Keshavarzi Arshadi, A.; Yuan, J.S. AMPDeep: Hemolytic Activity Prediction of Antimicrobial Peptides Using Transfer Learning. BMC Bioinform. 2022, 23, 389. [Google Scholar] [CrossRef]
- Hu, J.; Li, S.; Miao, M.; Li, F. Characterization of the Antibacterial and Opsonic Functions of the Antimicrobial Peptide LvCrustinVI from Litopenaeus Vannamei. Dev. Comp. Immunol. 2024, 154, 105146. [Google Scholar] [CrossRef]
- Wang, G.; Mishra, B. The Importance of Amino Acid Composition in Natural AMPs: An Evolutional, Structural, and Functional Perspective. Front. Immunol. 2012, 3, 31946. [Google Scholar]
- Hollmann, A.; Martínez, M.; Noguera, M.E.; Augusto, M.T.; Disalvo, A.; Santos, N.C.; Semorile, L.; Maffía, P.C. Role of Amphipathicity and Hydrophobicity in the Balance between Hemolysis and Peptide–Membrane Interactions of Three Related Antimicrobial Peptides. Colloids Surf. B Biointerfaces 2016, 141, 528–536. [Google Scholar] [CrossRef] [PubMed]
- Schnapp, D.; Kemp, G.D.; Smith, V.J. Purification and Characterization of a Proline-rich Antibacterial Peptide, with Sequence Similarity to Bactenecin-7, from the Haemocytes of the Shore Crab, Carcinus Maenas. Eur. J. Biochem. 1996, 240, 532–539. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD3: The Antimicrobial Peptide Database as a Tool for Research and Education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef]
- Su, T.; Han, M.; Cao, D.; Xu, M. Molecular and Biological Properties of Snakins: The Foremost Cysteine-Rich Plant Host Defense Peptides. J. Fungi 2020, 6, 220. [Google Scholar] [CrossRef]
- Dimarcq, J.; Bulet, P.; Hetru, C.; Hoffmann, J. Cysteine-rich Antimicrobial Peptides in Invertebrates. Pept. Sci. 1998, 47, 465–477. [Google Scholar] [CrossRef]
- Reddy, K.V.R.; Yedery, R.D.; Aranha, C. Antimicrobial Peptides: Premises and Promises. Int. J. Antimicrob. Agents 2004, 24, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Dashora, K.; Ameta, K.L.; Singh, N.P.; El-Enshasy, H.A.; Pagano, M.C.; Hesham, A.E.; Sharma, G.D.; Sharma, M.; Bhargava, A. Cysteine-rich Antimicrobial Peptides from Plants: The Future of Antimicrobial Therapy. Phytother. Res. 2021, 35, 256–277. [Google Scholar] [CrossRef] [PubMed]
- Smith, V.J.; Fernandes, J.M.O.; Kemp, G.D.; Hauton, C. Crustins: Enigmatic WAP Domain-Containing Antibacterial Proteins from Crustaceans. Dev. Comp. Immunol. 2008, 32, 758–772. [Google Scholar] [CrossRef]
- Ma, H.; Feng, Y.; Cao, Q.; Jia, J.; Ali, M.; Shah, D.; Meyers, B.C.; He, H.; Zhang, Y. Evolution of Antimicrobial Cysteine-Rich Peptides in Plants. Plant Cell Rep. 2023, 42, 1517–1527. [Google Scholar] [CrossRef]
- Holzknecht, J.; Marx, F. Navigating the Fungal Battlefield: Cysteine-Rich Antifungal Proteins and Peptides from Eurotiales. Front. Fungal Biol. 2024, 5, 1451455. [Google Scholar] [CrossRef] [PubMed]
- Bernot, J.P.; Avdeyev, P.; Zamyatin, A.; Dreyer, N.; Alexeev, N.; Pérez-Losada, M.; Crandall, K.A. Chromosome-Level Genome Assembly, Annotation, and Phylogenomics of the Gooseneck Barnacle Pollicipes Pollicipes. Gigascience 2022, 11, giac021. [Google Scholar] [CrossRef]
- Clements, K.; Giménez, L.; Jones, D.L.; Wilson, J.; Malham, S.K. Epizoic Barnacles Act as Pathogen Reservoirs on Shellfish Beds. J. Shellfish. Res. 2013, 32, 533–538. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, X.; Zhang, J.; Ye, T.; Zhou, Q.; Xu, Y.; Li, W.; Hu, Z.; Shang, C. Discovery and Characterization of a New Crustin Antimicrobial Peptide from Amphibalanus Amphitrite. Pharmaceutics 2022, 14, 413. [Google Scholar] [CrossRef]
- Cruz, T.; Jacinto, D.; Sousa, A.; Penteado, N.; Pereira, D.; Fernandes, J.N.; Silva, T.; Castro, J.J. The State of the Fishery, Conservation and Management of the Stalked Barnacle Pollicipes Pollicipes in Portugal. Mar. Environ. Res. 2015, 112, 73–80. [Google Scholar] [CrossRef]
- Liu, X.; Jin, H.; Xu, G.; Lai, R.; Wang, A. Bioactive Peptides from Barnacles and Their Potential for Antifouling Development. Mar. Drugs 2023, 21, 480. [Google Scholar] [CrossRef]
- Yang, S.; Cai, X. Genome-Wide Screening of the Classical Cadherin Gene Family and Cadherin-1 Expression Response Infected with Streptococcus agalactiae in Nile Tilapia (Oreochromis niloticus). Aquac. Rep. 2020, 17, 100393. [Google Scholar] [CrossRef]
- Li, G.; Chen, J.; Li, J.; Shang, C.; Wang, C. Structural Characteristics, Prokaryotic Expression and Activity Analysis of Antimicrobial Peptide ALFPm10 from Penaeus Monodon. Int. J. Pept. Res. Ther. 2021, 28, 25. [Google Scholar] [CrossRef]
- Nagy, K.; Végh, A.G.; Kereszt, A.; Kondorosi, É.; Váró, G.; Szegletes, Z. Interaction of Cysteine-Rich Cationic Antimicrobial Peptides with Intact Bacteria and Model Membranes. Gen. Physiol. Biophys. 2015, 34, 135–144. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ho, T.N.T.; Turner, A.; Pham, S.H.; Nguyen, H.T.; Nguyen, L.T.T.; Nguyen, L.T.; Dang, T.T. Cysteine-Rich Peptides: From Bioactivity to Bioinsecticide Applications. Toxicon 2023, 230, 107173. [Google Scholar] [CrossRef]
- Baindara, P.; Kapoor, A.; Korpole, S.; Grover, V. Cysteine-Rich Low Molecular Weight Antimicrobial Peptides from Brevibacillus and Related Genera for Biotechnological Applications. World J. Microbiol. Biotechnol. 2017, 33, 124. [Google Scholar] [CrossRef]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 Predicts All Five Types of Signal Peptides Using Protein Language Models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Nakai, K. Protein Subcellular Localization Prediction with WoLF PSORT. In Proceedings of the 4th Asia-Pacific Bioinformatics Conference, Taipei, Taiwan, 13–16 February 2006; World Scientific: Singapore, 2006; pp. 39–48. [Google Scholar]
- Cramer, P. AlphaFold2 and the Future of Structural Biology. Nat. Struct. Mol. Biol. 2021, 28, 704–705. [Google Scholar] [CrossRef] [PubMed]
- Forstinus, N.O.; Arnon, C.; Pawarisa, T.; Chinwe, N.L.; Komwit, S.; Sittiruk, R.; Piyawan, V.S.; Sarunyou, C. Synergistic Effects of Polymyxin and Vancomycin Combinations on Carbapenem- and Polymyxin-Resistant Klebsiella Pneumoniae and Their Molecular Characteristics. Microbiol. Spectr. 2023, 11, e01199-23. [Google Scholar] [CrossRef]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell Jr, A.D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone ϕ, ψ and Side-Chain Χ1 and Χ2 Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef]
- Murzyn, K.; Róg, T.; Pasenkiewicz-Gierula, M. Phosphatidylethanolamine-Phosphatidylglycerol Bilayer as a Model of the Inner Bacterial Membrane. Biophys. J. 2005, 88, 1091–1103. [Google Scholar] [CrossRef]
- Balatti, G.E.; Martini, M.F.; Pickholz, M. A Coarse-Grained Approach to Studying the Interactions of the Antimicrobial Peptides Aurein 1.2 and Maculatin 1.1 with POPG/POPE Lipid Mixtures. J. Mol. Model. 2018, 24, 208. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.C.; van Postma, J.P.M.; Van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Darden, T.; York, D. An N⋅ Log (N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Bernetti, M.; Bussi, G. Pressure Control Using Stochastic Cell Rescaling. J. Chem. Phys. 2020, 153, 114107. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Zhang, W.; Wei, L.; Chen, P.; Ning, B.; Wang, J.; He, P.; Shang, C.; Yu, D. Discovery and Characterization of an Atypical Crustin Antimicrobial Peptide from Pollicipes Pollicipes. Mar. Drugs 2024, 22, 526. [Google Scholar] [CrossRef]
- Chen, Y.-C.; Yang, Y.; Zhang, C.; Chen, H.-Y.; Chen, F.; Wang, K.-J. A Novel Antimicrobial Peptide Sparamosin26–54 From the Mud Crab Scylla Paramamosain Showing Potent Antifungal Activity Against Cryptococcus Neoformans. Front. Microbiol. 2021, 12, 746006. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Tian, Z.; Zhou, L.; Zhu, L.; Sun, C.; Huang, M.; Peng, J.; Guo, G. In Vitro Antifungal Activity of a Novel Antimicrobial Peptide AMP-17 Against Planktonic Cells and Biofilms of Cryptococcus Neoformans. Infect. Drug Resist. 2022, 15, 233–248. [Google Scholar] [CrossRef]
- Yang, Z.; He, S.; Wei, Y.; Li, X.; Shan, A.; Wang, J. Antimicrobial Peptides in Combination with Citronellal Efficiently Kills Multidrug Resistance Bacteria. Phytomedicine 2023, 120, 155070. [Google Scholar] [CrossRef]
- Seyfi, R.; Kahaki, F.A.; Ebrahimi, T.; Montazersaheb, S.; Eyvazi, S.; Babaeipour, V.; Tarhriz, V. Antimicrobial Peptides (AMPs): Roles, Functions and Mechanism of Action. Int. J. Pept. Res. Ther. 2020, 26, 1451–1463. [Google Scholar] [CrossRef]
- Peng, C.; Shi, C.; Cao, X.; Li, Y.; Liu, F.; Lu, F. Factors Influencing Recombinant Protein Secretion Efficiency in Gram-Positive Bacteria: Signal Peptide and Beyond. Front. Bioeng. Biotechnol. 2019, 7, 139. [Google Scholar] [CrossRef]
- Zhang, L.; Li, X.; Wei, D.; Wang, J.; Shan, A.; Li, Z. Expression of Plectasin in Bacillus Subtilis Using SUMO Technology by a Maltose-Inducible Vector. J. Ind. Microbiol. Biotechnol. 2015, 42, 1369–1376. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Feng, J.; Kang, Y.; Pan, P.; Ge, J.; Wang, Y.; Wang, M.; Wu, Z.; Zhang, X.; Yu, J. Discovery of Antimicrobial Peptides with Notable Antibacterial Potency by an LLM-Based Foundation Model. Sci. Adv. 2025, 11, eads8932. [Google Scholar] [CrossRef]
- Luo, Y.; Song, Y. Mechanism of Antimicrobial Peptides: Antimicrobial, Anti-Inflammatory and Antibiofilm Activities. Int. J. Mol. Sci. 2021, 22, 11401. [Google Scholar] [CrossRef] [PubMed]
- Semreen, M.H.; El-Gamal, M.I.; Abdin, S.; Alkhazraji, H.; Kamal, L.; Hammad, S.; El-Awady, F.; Waleed, D.; Kourbaj, L. Recent Updates of Marine Antimicrobial Peptides. Saudi Pharm. J. 2018, 26, 396–409. [Google Scholar] [CrossRef]
- Wu, R.; Patocka, J.; Nepovimova, E.; Oleksak, P.; Valis, M.; Wu, W.; Kuca, K. Marine Invertebrate Peptides: Antimicrobial Peptides. Front. Microbiol. 2021, 12, 785085. [Google Scholar] [CrossRef]
- Thomas, A.M.; Antony, S.P. Marine Antimicrobial Peptides: An Emerging Nightmare to the Life-Threatening Pathogens. Probiotics Antimicrob. Proteins 2023, 16, 552–578. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Ding, J.; Liao, C.; Xu, J.; Liu, X.; Lu, W. Defensins: The Natural Peptide Antibiotic. Adv. Drug Deliv. Rev. 2021, 179, 114008. [Google Scholar] [CrossRef] [PubMed]
- Brady, D.; Grapputo, A.; Romoli, O.; Sandrelli, F. Insect Cecropins, Antimicrobial Peptides with Potential Therapeutic Applications. Int. J. Mol. Sci. 2019, 20, 5862. [Google Scholar] [CrossRef]
- Wan, F.; Wong, F.; Collins, J.J.; de la Fuente-Nunez, C. Machine Learning for Antimicrobial Peptide Identification and Design. Nat. Rev. Bioeng. 2024, 2, 392–407. [Google Scholar] [CrossRef]
- Torres, M.D.T.; Cao, J.; Franco, O.L.; Lu, T.K.; de la Fuente-Nunez, C. Synthetic Biology and Computer-Based Frameworks for Antimicrobial Peptide Discovery. ACS Nano 2021, 15, 2143–2164. [Google Scholar] [CrossRef]
- Waghu, F.H.; Barai, R.S.; Gurung, P.; Idicula-Thomas, S. CAMPR3: A Database on Sequences, Structures and Signatures of Antimicrobial Peptides. Nucleic Acids Res. 2016, 44, D1094–D1097. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Dong, M.; Wang, Q.; Sun, Y.; Hang, B.; Zhang, H.; Hu, J.; Zhang, G. Soluble Expression of Antimicrobial Peptide BSN-37 from Escherichia Coli by SUMO Fusion Technology. Protein J. 2023, 42, 563–574. [Google Scholar] [CrossRef]
- Park, A.R.; Kim, S.W.; Kim, S.Y.; Kwon, K.-C. Expression of Antimicrobial Peptide (AMP), Cecropin B, in a Fused Form to SUMO Tag with or without Three-Glycine Linker in Escherichia Coli and Evaluation of Bacteriolytic Activity of the Purified AMP. Probiotics Antimicrob. Proteins 2021, 13, 1780–1789. [Google Scholar] [CrossRef]
- Wang, X.; van Beekveld, R.A.M.; Xu, Y.; Parmar, A.; Das, S.; Singh, I.; Breukink, E. Analyzing Mechanisms of Action of Antimicrobial Peptides on Bacterial Membranes Requires Multiple Complimentary Assays and Different Bacterial Strains. Biochim. Biophys. Acta (BBA)-Biomembr. 2023, 1865, 184160. [Google Scholar] [CrossRef]
- Matsuzaki, K. Membrane Permeabilization Mechanisms. In Antimicrobial Peptides: Basics for Clinical Application; Spring: Berlin/Heidelberg, Germany, 2019; pp. 9–16. [Google Scholar]
- Shai, Y.; Oren, Z. From “Carpet” Mechanism to de-Novo Designed Diastereomeric Cell-Selective Antimicrobial Peptides. Peptides 2001, 22, 1629–1641. [Google Scholar] [CrossRef]
- Ciulla, M.G.; Gelain, F. Structure–Activity Relationships of Antibacterial Peptides. Microb. Biotechnol. 2023, 16, 757–777. [Google Scholar] [CrossRef]
- Powers, J.-P.S.; Hancock, R.E.W. The Relationship between Peptide Structure and Antibacterial Activity. Peptides 2003, 24, 1681–1691. [Google Scholar] [CrossRef]
- Koprivnjak, T.; Weidenmaier, C.; Peschel, A.; Weiss, J.P. Wall Teichoic Acid Deficiency in Staphylococcus Aureus Confers Selective Resistance to Mammalian Group IIA Phospholipase A2 and Human β-Defensin 3. Infect. Immun. 2008, 76, 2169–2176. [Google Scholar] [CrossRef]
- Malanovic, N.; Lohner, K. Antimicrobial Peptides Targeting Gram-Positive Bacteria. Pharmaceuticals 2016, 9, 59. [Google Scholar] [CrossRef]
- Zheng, S.; Tu, Y.; Li, B.; Qu, G.; Li, A.; Peng, X.; Li, S.; Shao, C. Antimicrobial Peptide Biological Activity, Delivery Systems and Clinical Translation Status and Challenges. J. Transl. Med. 2025, 23, 292. [Google Scholar] [CrossRef]
- Chen, P.; Zhang, T.; Li, C.; Praveen, P.; Parisi, K.; Beh, C.; Ding, S.; Wade, J.D.; Hong, Y.; Li, S. Aggregation-Prone Antimicrobial Peptides Target Gram-Negative Bacterial Nucleic Acids and Protein Synthesis. Acta Biomater. 2025, 192, 446–460. [Google Scholar] [CrossRef]
- Necula, G.; Bacalum, M.; Radu, M. Interaction of Tryptophan-and Arginine-Rich Antimicrobial Peptide with E. Coli Outer Membrane—A Molecular Simulation Approach. Int. J. Mol. Sci. 2023, 24, 2005. [Google Scholar] [CrossRef]
- Khunsri, I.; Prombutara, P.; Htoo, H.H.; Wanvimonsuk, S.; Samernate, T.; Pornsing, C.; Tharntada, S.; Jaree, P.; Chaikeeratisak, V.; Somboonwiwat, K. Roles of QseC Mutation in Bacterial Resistance against Anti-Lipopolysaccharide Factor Isoform 3 (ALFPm3). PLoS ONE 2023, 18, e0286764. [Google Scholar] [CrossRef]
- Zhou, L.; Li, G.; Jiao, Y.; Huang, D.; Li, A.; Chen, H.; Liu, Y.; Li, S.; Li, H.; Wang, C. Molecular and Antimicrobial Characterization of a Group G Anti-Lipopolysaccharide Factor (ALF) from Penaeus Monodon. Fish. Shellfish. Immunol. 2019, 94, 149–156. [Google Scholar] [CrossRef]
- Zhuang, H.; Ou, Y.; Chen, R.; Huang, D.; Wang, C. Comparing the Ability of Secretory Signal Peptides for Heterologous Expression of Anti-Lipopolysaccharide Factor 3 in Chlamydomonas Reinhardtii. Mar. Drugs 2023, 21, 346. [Google Scholar] [CrossRef]
- Yang, H.; Li, S.; Li, F.; Xiang, J. Structure and Bioactivity of a Modified Peptide Derived from the LPS-Binding Domain of an Anti-Lipopolysaccharide Factor (ALF) of Shrimp. Mar. Drugs 2016, 14, 96. [Google Scholar] [CrossRef] [PubMed]
- Capecchi, A.; Cai, X.; Personne, H.; Köhler, T.; van Delden, C.; Reymond, J.-L. Machine Learning Designs Non-Hemolytic Antimicrobial Peptides. Chem. Sci. 2021, 12, 9221–9232. [Google Scholar] [CrossRef] [PubMed]
- Han, F.-F.; Liu, Y.-F.; Xie, Y.-G.; Gao, Y.-H.; Luan, C.; Wang, Y.-Z. Antimicrobial Peptides Derived from Different Animals: Comparative Studies of Antimicrobial Properties, Cytotoxicity and Mechanism of Action. World J. Microbiol. Biotechnol. 2011, 27, 1847–1857. [Google Scholar] [CrossRef]
- Chen, R.; Khormaee, S.; Eccleston, M.E.; Slater, N.K.H. The Role of Hydrophobic Amino Acid Grafts in the Enhancement of Membrane-Disruptive Activity of PH-Responsive Pseudo-Peptides. Biomaterials 2009, 30, 1954–1961. [Google Scholar] [CrossRef]




| Microorganism | Minimal Inhibitory Concentrations (μM) | Minimum Bactericidal Concentration (μM) | |||
|---|---|---|---|---|---|
| rPpRcys1 | Ampicillin | rPpRcys1 | Ampicillin | ||
| Gram+ | S. aureus | 8 | 2 | 16 | 4 |
| Bacillus sp. T2 | 8 | - | 32 | ||
| S. agalactiae | 16 | 4 | 32 | 16 | |
| Gram− | A. hydrophila | 32 | - | 64 | - |
| Acinetobacter sp. L32 | 32 | - | 64 | - | |
| E. coli | 16 | 64 | 32 | 256 | |
| V. alginolyticus | 16 | 64 | 32 | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Z.; Fei, Z.; Shi, H.; Huang, M.; Wei, L.; Wang, J.; He, P.; Zhang, W. Heterologous Expression and Antimicrobial Mechanism of a Cysteine-Rich Peptide from Barnacle Pollicipes pollicipes. Microorganisms 2025, 13, 1381. https://doi.org/10.3390/microorganisms13061381
He Z, Fei Z, Shi H, Huang M, Wei L, Wang J, He P, Zhang W. Heterologous Expression and Antimicrobial Mechanism of a Cysteine-Rich Peptide from Barnacle Pollicipes pollicipes. Microorganisms. 2025; 13(6):1381. https://doi.org/10.3390/microorganisms13061381
Chicago/Turabian StyleHe, Zhicheng, Zixun Fei, Huishao Shi, Meichuan Huang, Liumi Wei, Junjian Wang, Peng He, and Wei Zhang. 2025. "Heterologous Expression and Antimicrobial Mechanism of a Cysteine-Rich Peptide from Barnacle Pollicipes pollicipes" Microorganisms 13, no. 6: 1381. https://doi.org/10.3390/microorganisms13061381
APA StyleHe, Z., Fei, Z., Shi, H., Huang, M., Wei, L., Wang, J., He, P., & Zhang, W. (2025). Heterologous Expression and Antimicrobial Mechanism of a Cysteine-Rich Peptide from Barnacle Pollicipes pollicipes. Microorganisms, 13(6), 1381. https://doi.org/10.3390/microorganisms13061381






