The Impact of Jujube Witches’ Broom Phytoplasma on the Community Structure of Endophytes in Jujube
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction and Sequencing
2.3. Sequence Analysis
2.4. Statistical Analyses
3. Results
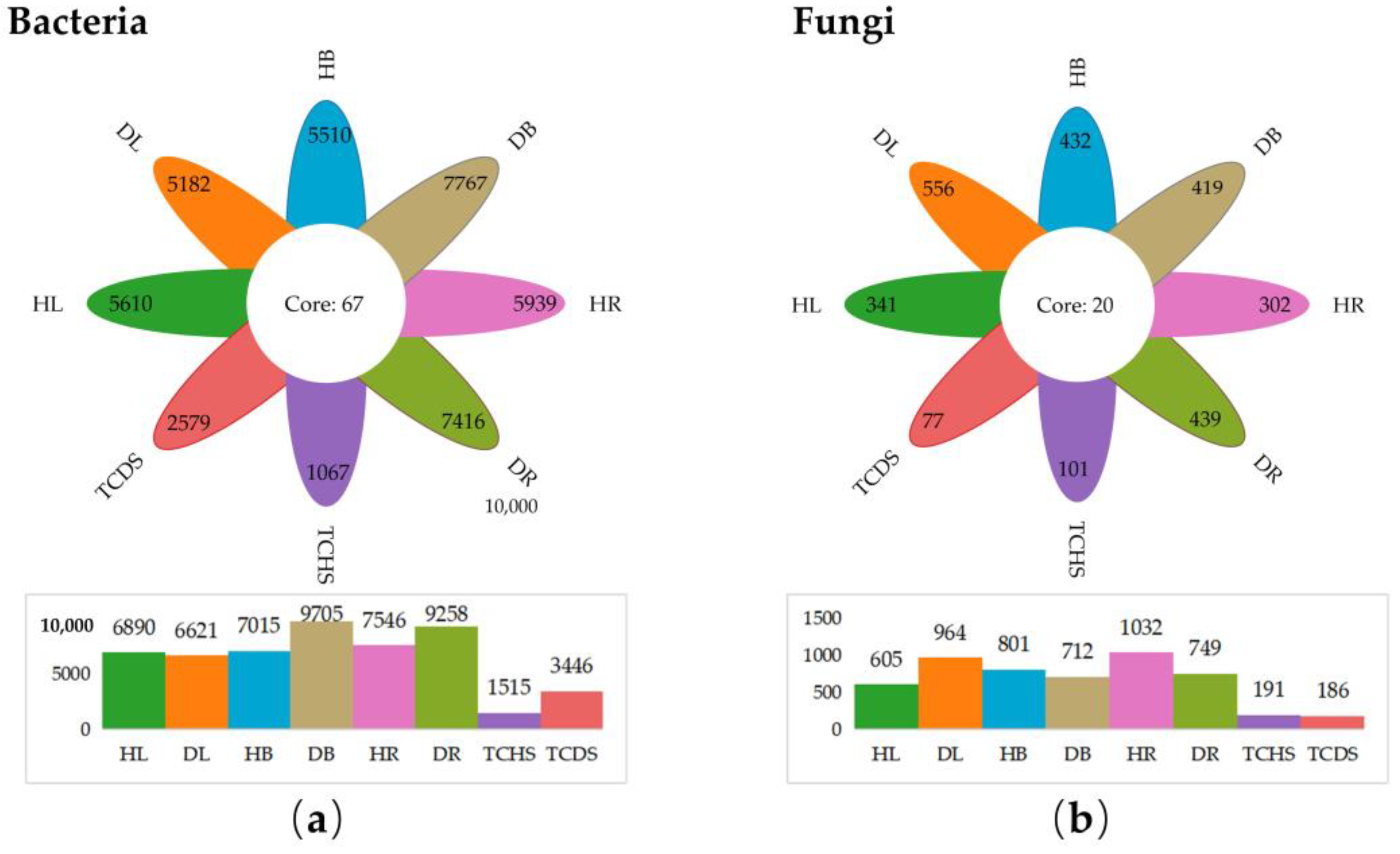
3.1. HiSeq Sequencing Output and Amplicon Sequence Variant Analysis
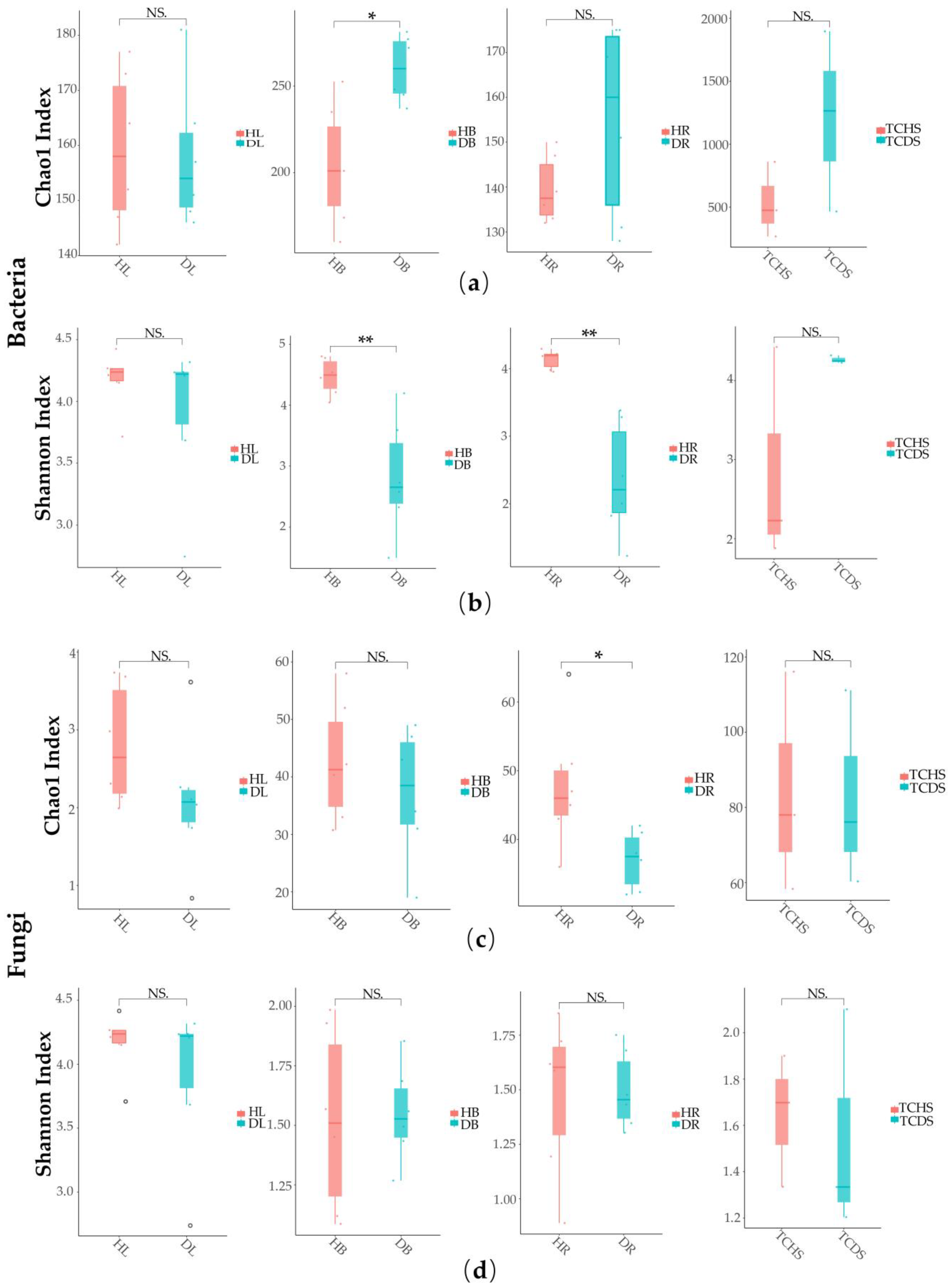
3.2. Effect of Jujube Witches’ Broom on Microbial Community Structure
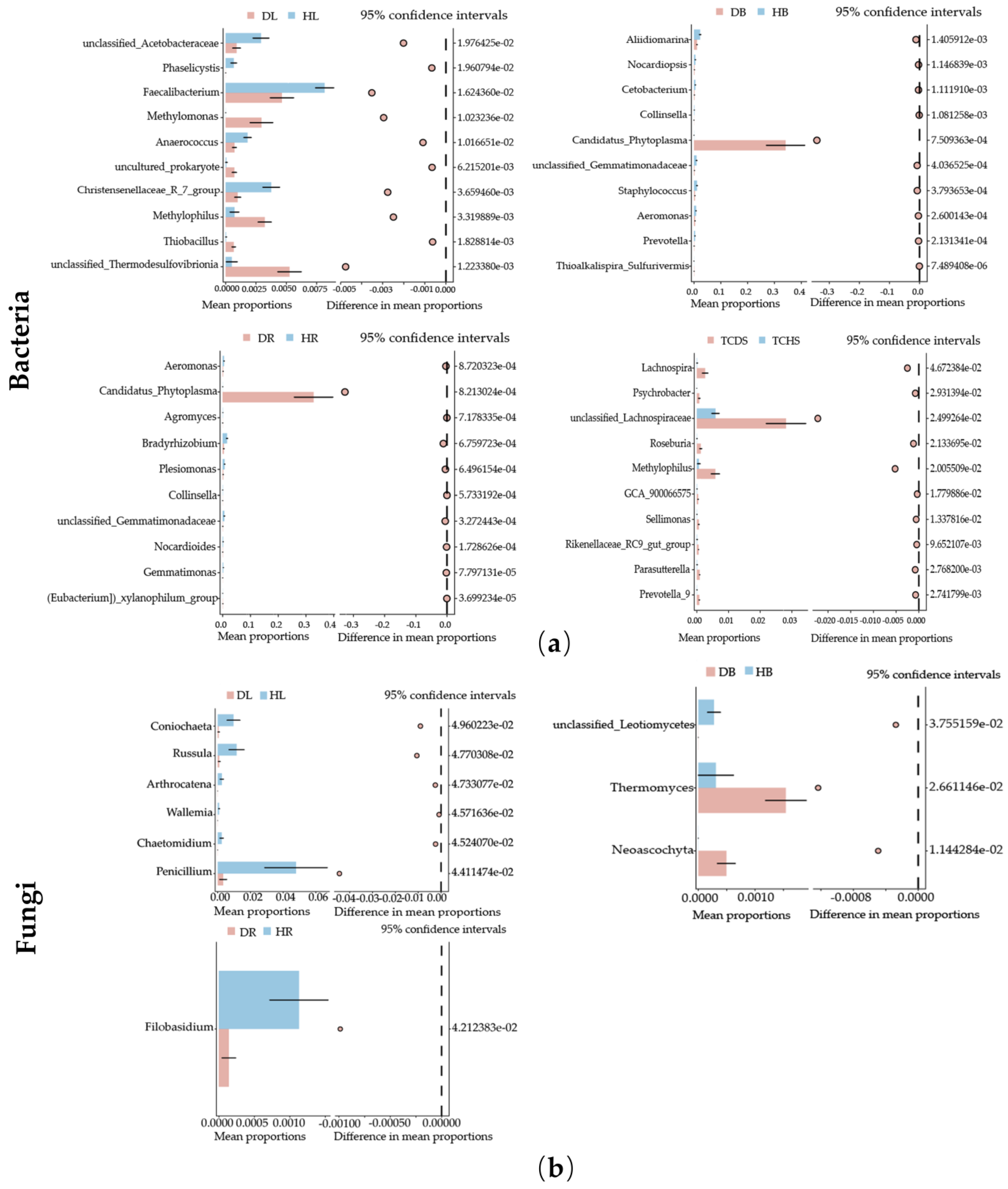
3.3. Effect of Jujube Witches’ Broom on Endophytic Community Composition
3.4. The Impact of Jujube Witches’ Broom on the Functions of the Endophytic Microbial Community
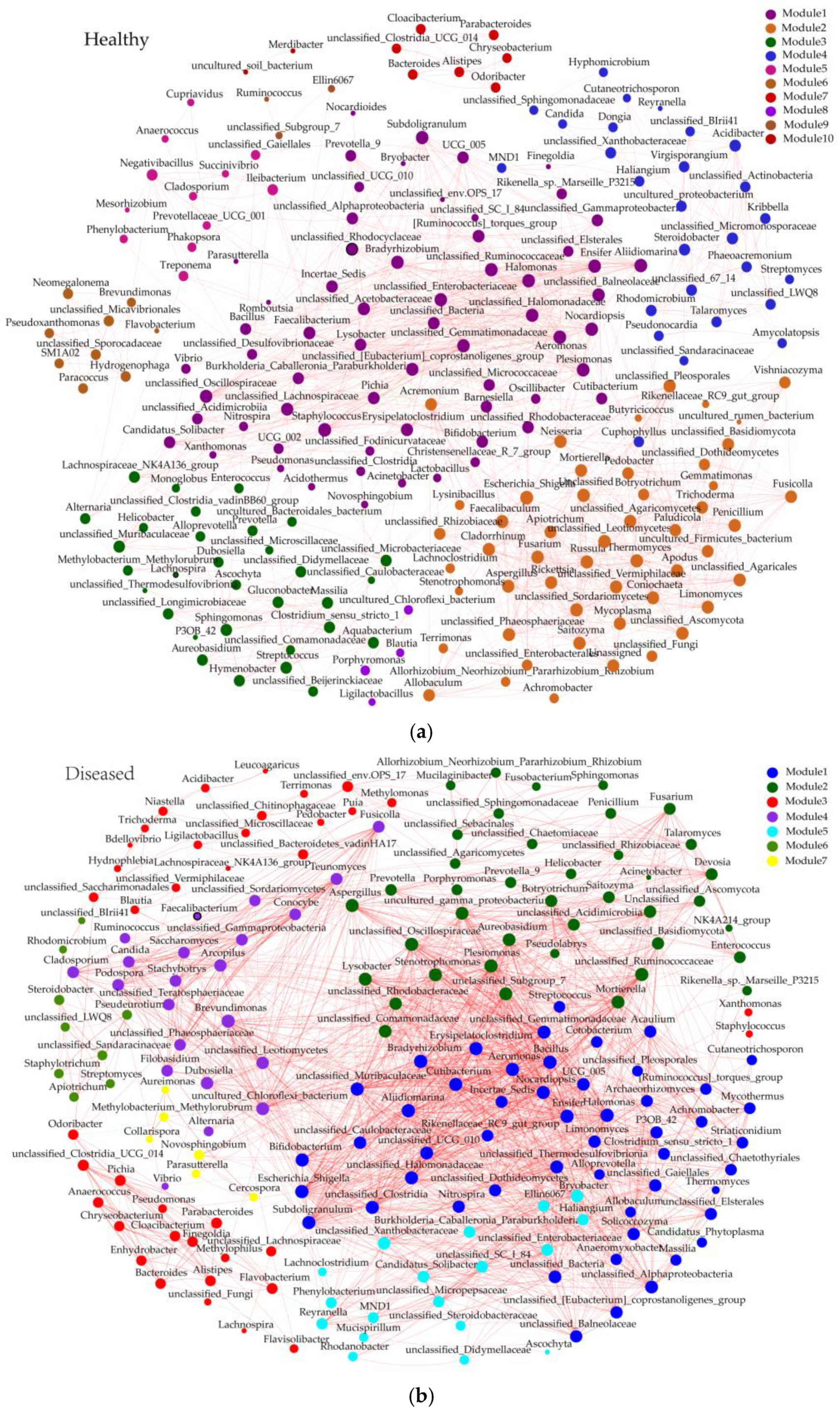
3.5. The Impact of Jujube Witches’ Broom on the Co-Network of Endophytic Bacteria–Fungi in Jujube
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cui, J.; Nie, F.; Zhao, Y.; Zhang, D.; Zhou, D.; Wu, J.; Qu, L.; Xiao, L.; Liu, L. A review on plant endophytes in response to abiotic stress. Environ. Pollut. Bioavailab. 2024, 36, 2323123. [Google Scholar] [CrossRef]
- Ganie, S.A.; Bhat, J.A.; Devoto, A. The influence of endophytes on rice fitness under environmental stresses. Plant Mol. Biol. 2022, 109, 447–467. [Google Scholar] [CrossRef] [PubMed]
- Qiang, M.; Xu, Y.; Gao, R. Research progress in stress resistance of plants mediated by endophytic bacteria. Bot. Res. 2020, 9, 226–239. [Google Scholar] [CrossRef]
- Schmidt, C.S.; Mrnka, L.; Lovecká, P.; Frantík, T.; Fenclová, M.; Demnerová, K.; Vosátka, M. Bacterial and fungal endophyte communities in healthy and diseased oilseed rape and their potential for biocontrol of Sclerotinia and Phoma disease. Sci. Rep. 2021, 11, 3810. [Google Scholar] [CrossRef] [PubMed]
- Esmaeili, M.; Shahryari, F.; Sarikhani, S. Unveiling the potential of endophytic bacteria in combatting walnut bacterial canker disease. J. Plant Pathol. 2025, 107, 551–563. [Google Scholar] [CrossRef]
- Tang, J.; Xiao, Y.; Xu, X.; Tang, M.; Zhang, X.; Yi, Y. Root microbiota alters response to root rot in Rhododendron delavayi Franch. Front. Microbiol. 2023, 14, 1236110. [Google Scholar] [CrossRef]
- Shi, W.; Li, J.; Xie, S.; Wang, X.; Zhang, Y.; Yao, H.; Chen, M.; Li, J.; Deng, Z. Selection of the dominant endophytes based on Illumina sequencing analysis for controlling bacterial wilt of patchouli caused by Ralstonia solanacearum. Plant Dis. 2024, 108, 996–1004. [Google Scholar] [CrossRef]
- Lahlali, R.; Ezrari, S.; Radouane, N.; Kenfaoui, J.; Esmaeel, Q.; El Hamss, H.; Belabess, Z.; Barka, E.A. Biological control of plant pathogens: A global perspective. Microorganisms 2022, 10, 596. [Google Scholar] [CrossRef]
- Hogenhout, S.A.; Oshima, K.; Ammar, E.-D.; Kakizawa, S.; Kingdom, H.N.; Namba, S. Phytoplasmas: Bacteria that manipulate plants and insects. Mol. Plant Pathol. 2008, 9, 403–423. [Google Scholar] [CrossRef]
- Garnier, M.; Foissac, X.; Gaurivaud, P.; Laigret, F.; Renaudin, J.; Saillard, C.; Bové, J.M. Mycoplasmas, plants, insect vectors: A matrimonial triangle. Comptes Rendus De L’academie Des Sci. Ser. III 2001, 324, 923–928. [Google Scholar] [CrossRef]
- Li, Q.C.; Chen, P.; Yang, Q.Q.; Chen, L.C.; Zhang, Y.; Li, J.D.; Feng, J.C. First report of ‘Candidatus Phytoplasma ziziphi’ in sweet potato in China. Plant Dis. 2022, 106, 1515. [Google Scholar] [CrossRef] [PubMed]
- Bulgari, D.; Bozkurt, A.I.; Casati, P.; Cağlayan, K.; Quaglino, F.; Bianco, P.A. Endophytic bacterial community living in roots of healthy and ‘Candidatus Phytoplasma mali’-infected apple (Malus domestica, Borkh.) trees. Antonie Van Leeuwenhoek 2012, 102, 677–687. [Google Scholar] [CrossRef]
- Liu, M.; Wang, J.; Wang, L.; Liu, P.; Zhao, J.; Zhao, Z.; Yao, S.; Stănică, F.; Liu, Z.; Wang, L.; et al. The historical and current research progress on jujube–a superfruit for the future. Hortic. Res. 2020, 7, 119. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Gu, L.; Zhang, Y.; Wu, Y.; Tan, B.; Zheng, X.; Ye, X.; Cheng, J.; Wang, W.; Bi, S.; et al. Jujube witches’ broom (‘Zaofeng’) disease: Bacteria that drive the plants crazy. Fruit Res. 2023, 3, 35. [Google Scholar] [CrossRef]
- Crawford, R.; Shan, F.; McCarthy, A. Chinese jujube: A developing industry in Australia. Acta Hortic. 2013, 993, 29–36. [Google Scholar] [CrossRef]
- Chen, P.; Chen, L.; Ye, X.; Tan, B.; Zheng, X.; Cheng, J.; Wang, W.; Yang, Q.; Zhang, Y.; Li, J.; et al. Phytoplasma effector Zaofeng6 induces shoot proliferation by decreasing the expression of ZjTCP7 in Ziziphus jujuba. Hortic. Res. 2022, 9, uhab032. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, Z.; Liu, M. The resistance of jujube trees to jujube witches’ broom disease in China. In Sustainable Management of Phytoplasma Diseases in Crops Grown in the Tropical Belt: Biology and Detection; Olivier, C.Y., Dumonceaux, T.J., Pérez-López, E., Eds.; Springer Nature Switzerland: Cham, Switzerland, 2019; pp. 219–232. [Google Scholar]
- Wang, H.; Ye, X.; Li, J.; Tan, B.; Chen, P.; Jiang, Y.; Cheng, J.; Wang, W.; Zheng, X.; Feng, J. Combination of iTRAQ proteomics and RNA-seq transcriptomics reveals jasmonate-related-metabolisms central regulation during the process of Jujube witches’ broom recovery by tetracycline treatment. Sci. Hortic. 2019, 243, 197–206. [Google Scholar] [CrossRef]
- Jung, H.Y.; Sawayanagi, T.; Kakizawa, S.; Nishigawa, H.; Wei, W.; Oshima, K.; Miyata, S.I.; Ugaki, M.; Hibi, T.; Namba, S. ‘Candidatus Phytoplasma ziziphi’, a novel phytoplasma taxon associated with jujube witches’-broom disease. Int. J. Syst. Evol. Microbiol. 2003, 53, 1037–1041. [Google Scholar] [CrossRef]
- Hamaila, H.; Han, G.; Li, X. Effects of actinomycete Act12 on soil microbial community structure and plant traits of Ziziphus jujuba Mill. Agronomy 2024, 14, 1411. [Google Scholar] [CrossRef]
- Lang, J.F.; Tian, X.L.; Shi, M.W.; Ran, L.X. Identification of endophytes with biocontrol potential from Ziziphus jujuba and its inhibition effects on Alternaria alternata, the pathogen of jujube shrunken-fruit disease. PLoS ONE 2018, 13, e0199466. [Google Scholar] [CrossRef]
- Liu, M. Chinese jujube: Botany and horticulture. Hortic. Rev. 2006, 32, 229–298. [Google Scholar] [CrossRef]
- Lorenz, K.H.; Schneider, B.; Ahrens, U.; Seemuller, E. Detection of the apple proliferation and pear decline phytoplasmas by PCR amplification of ribosomal and nonribosomal DNA. Phytopathology 1995, 85, 771–776. [Google Scholar] [CrossRef]
- Beckers, B.; Op De Beeck, M.; Weyens, N.; Boerjan, W.; Vangronsveld, J. Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees. Microbiome 2017, 5, 25. [Google Scholar] [CrossRef]
- Wang, X.; Xiao, C.; Ji, C.; Li, H.; Song, X.; Liu, Y.; Li, C.; Yan, D.; Li, H.; Qin, Y.; et al. Isolation and characterisation of endophytic bacteria for controlling root rot disease of Chinese jujube. J. Appl. Microbiol. 2021, 130, 926–936. [Google Scholar] [CrossRef]
- Miya, M.; Sato, Y.; Fukunaga, T.; Sado, T.; Poulsen, J.Y.; Sato, K.; Minamoto, T.; Yamamoto, S.; Yamanaka, H.; Araki, H.; et al. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: Detection of more than 230 subtropical marine species. R. Soc. Open Sci. 2015, 2, 150088. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Modi, A.; Vai, S.; Caramelli, D.; Lari, M. The Illumina sequencing protocol and the NovaSeq 6000 system. Methods Mol. Biol. 2021, 2242, 15–42. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Martin, M. CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef] [PubMed]
- Zgadzaj, R.; Garrido-Oter, R.; Jensen, D.B.; Koprivova, A.; Schulze-Lefert, P.; Radutoiu, S. Root nodule symbiosis in Lotus japonicus drives the establishment of distinctive rhizosphere, root, and nodule bacterial communities. Proc. Natl. Acad. Sci. USA 2016, 113, E7996–E8005. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.; O’Hara, B.; Simpson, G.; Solymos, P.; Stevens, H.; Wagner, H. Vegan: Community Ecology Package, R Package Version 2.2-1. 2015, Volume 2, pp. 1–2. Available online: https://cran.r-project.org/src/contrib/Archive/vegan/ (accessed on 18 October 2024).
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Wu, C.; Wang, F.; Ge, A.; Zhang, H.; Chen, G.; Deng, Y.; Yang, J.; Chen, J.; Ge, T. Enrichment of microbial taxa after the onset of wheat yellow mosaic disease. Agric. Ecosyst. Environ. 2021, 322, 107651. [Google Scholar] [CrossRef]
- Li, D.; Yu, Y.; Tian, C.; Lu, S.; Jiang, S. The impact of pine wilt disease on the endophytic microbial communities structure of Pinus koraiensis. Front. Microbiol. 2024, 15, 1493808. [Google Scholar] [CrossRef]
- Cao, R.; Dong, X.; Zhao, Y.; Yin, J. Effects of blister blight disease on endophytic microbial diversity and community structure in tea (Camellia sinensis) leaves. 3 Biotech 2023, 13, 421. [Google Scholar] [CrossRef]
- Bulgari, D.; Casati, P.; Quaglino, F.; Bianco, P.A. Endophytic bacterial community of grapevine leaves influenced by sampling date and phytoplasma infection process. BMC Microbiol. 2014, 14, 198. [Google Scholar] [CrossRef]
- Trivedi, P.; Duan, Y.; Wang, N. Huanglongbing, a systemic disease, restructures the bacterial community associated with citrus roots. Appl. Environ. Microbiol. 2010, 76, 3427–3436. [Google Scholar] [CrossRef] [PubMed]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- Bertaccini, A. Plants and Phytoplasmas: When bacteria modify plants. Plants 2022, 11, 1425. [Google Scholar] [CrossRef]
- Bertaccini, A.; Paltrinieri, S.; Martini, M.; Tedeschi, M.; Contaldo, N. Micropropagation and maintenance of phytoplasmas in tissue culture. Methods Mol. Biol. 2013, 938, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Orlikowska, T.; Nowak, K.; Reed, B.M. Bacteria in the plant tissue culture environment. Plant Cell Tissue Organ Cult. 2017, 128, 487–508. [Google Scholar] [CrossRef]
- Zakurin, A.; Li, J.; Barbieri, C.; Pacini, F.; Bertaccini, A. ‘Candidatus Phytoplasma ziziphi’ co-cultivation from micro-propagated jujube witches’ broom infected shoots. Phytopathogenic Mollicutes 2024, 14, 74–80. [Google Scholar] [CrossRef]
- Hong, C.; Si, Y.; Xing, Y.; Li, Y. Illumina MiSeq sequencing investigation on the contrasting soil bacterial community structures in different iron mining areas. Environ. Sci. Pollut. Res. 2015, 22, 10788–10799. [Google Scholar] [CrossRef]
- Orgiazzi, A.; Lumini, E.; Nilsson, R.H.; Girlanda, M.; Vizzini, A.; Bonfante, P.; Bianciotto, V. Unravelling soil fungal communities from different Mediterranean land-use backgrounds. PloS ONE 2012, 7, e34847. [Google Scholar] [CrossRef]



| Name | Raw Reads | Clean Reads | Sequence Length | Amplicon Sequence Variants (ASVs) |
|---|---|---|---|---|
| Bacteria | 3,280,457 | 2,127,470 | Less than 450 bp | 44,768 |
| Fungi | 3,361,862 | 3,050,660 | 190 bp to 460 bp | 3767 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, N.; Wang, M.; Jiang, Z.; Zhang, W.; You, Z.; Zhao, X.; Yao, J.; Gong, C.; Bertaccini, A.; Li, J. The Impact of Jujube Witches’ Broom Phytoplasma on the Community Structure of Endophytes in Jujube. Microorganisms 2025, 13, 1371. https://doi.org/10.3390/microorganisms13061371
Wang N, Wang M, Jiang Z, Zhang W, You Z, Zhao X, Yao J, Gong C, Bertaccini A, Li J. The Impact of Jujube Witches’ Broom Phytoplasma on the Community Structure of Endophytes in Jujube. Microorganisms. 2025; 13(6):1371. https://doi.org/10.3390/microorganisms13061371
Chicago/Turabian StyleWang, Nian, Mengli Wang, Ziming Jiang, Wenzhe Zhang, Ziyang You, Xueru Zhao, Jia Yao, Chenrui Gong, Assunta Bertaccini, and Jidong Li. 2025. "The Impact of Jujube Witches’ Broom Phytoplasma on the Community Structure of Endophytes in Jujube" Microorganisms 13, no. 6: 1371. https://doi.org/10.3390/microorganisms13061371
APA StyleWang, N., Wang, M., Jiang, Z., Zhang, W., You, Z., Zhao, X., Yao, J., Gong, C., Bertaccini, A., & Li, J. (2025). The Impact of Jujube Witches’ Broom Phytoplasma on the Community Structure of Endophytes in Jujube. Microorganisms, 13(6), 1371. https://doi.org/10.3390/microorganisms13061371







