Screening and Isolation of Bacterial Strains Able to Degrade Trimethylamine
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Collection and Bacterium Isolation
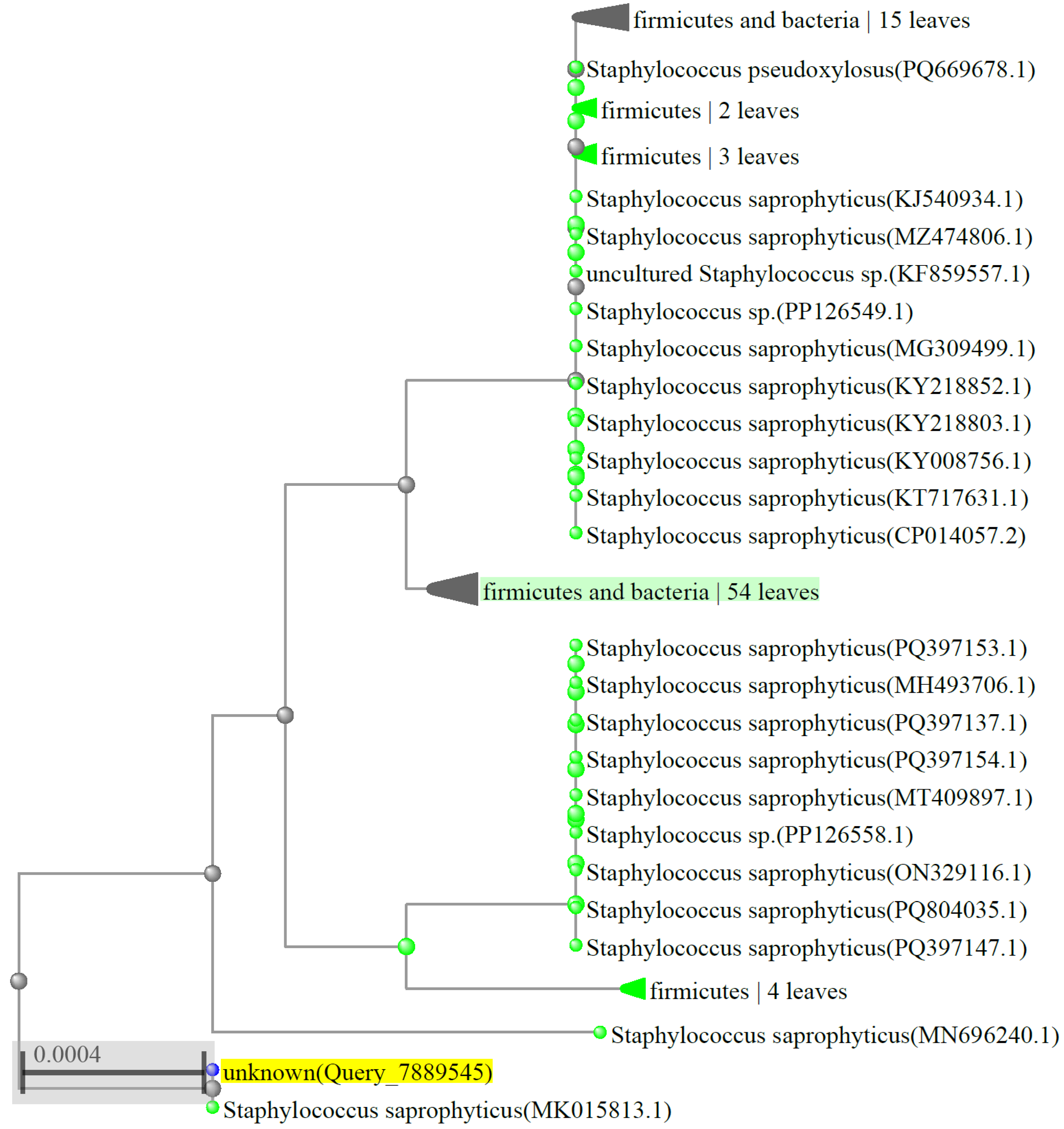
2.2. Bacterial Identification
2.3. Bacterial Growth
2.4. TMA Quantification
2.5. Bioreactor System
3. Results and Discussion
3.1. Isolation of Strains Able to Convert TMA
3.2. Growth Conditions with Trimethylamine as a Carbon Source
3.2.1. Effect of the Medium Used to Grow the Inoculum
3.2.2. Effect of Temperature, Stirring Speed and Salinity
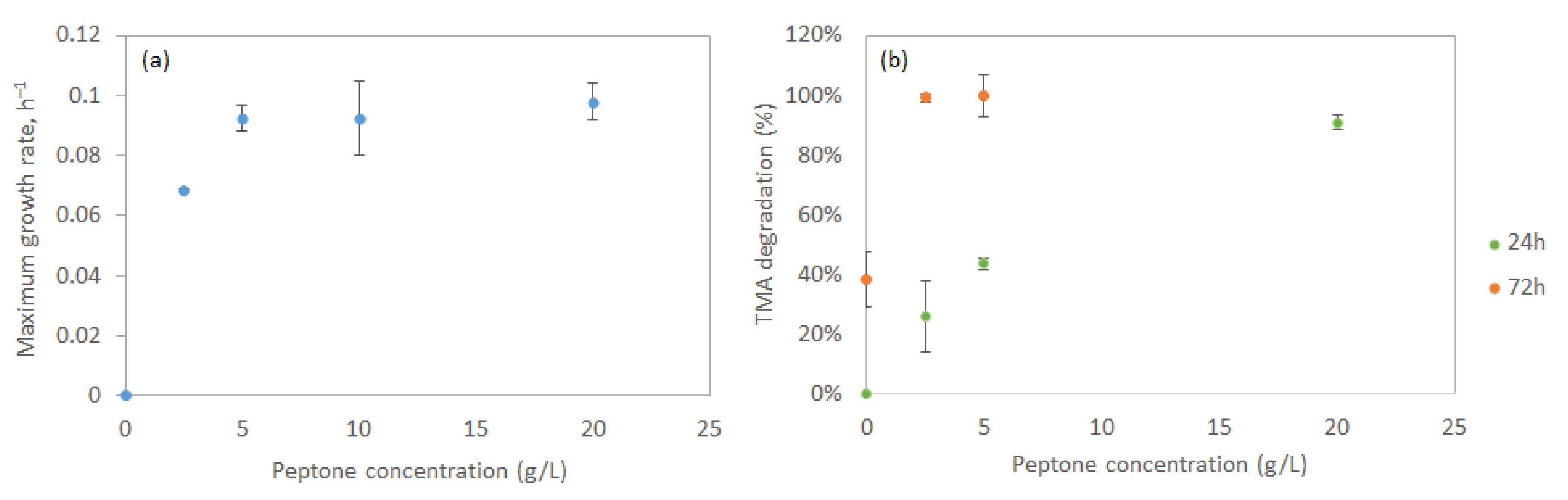
3.2.3. Nitrogen Source
3.3. Application of Trimethylamine Degradation in Bioreactor
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Du Preez, R.; Majzoub, M.E.; Thomas, T.; Panchal, S.K.; Brown, L. Nannochloropsis oceanica as a microalgal food intervention in diet-induced metabolic syndrome in rats. Nutrients 2021, 13, 3991. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, D.J. Chapter One—Microalgae for biofuel production. In Advance in Applied Microbiolpgy; Gadd, G.M., Sariaslani, S., Eds.; Academic Press: Cambridge, MA, USA, 2019; Volume 109, pp. 1–30. [Google Scholar]
- Napolitano, G.; Fasciolo, G.; Salbitani, G.; Venditti, P. Chlorella sorokiniana dietary supplementation increases antioxidant capacities and reduces ROS release in mitochondria of hyperthyroid rat liver. Antioxidants 2020, 9, 883. [Google Scholar] [CrossRef]
- Stiefvatter, L.; Lehnert, K.; Frick, K.; Montoya-Arroyo, A.; Frank, J.; Vetter, W.; Schmid-Staiger, U.; Bischoff, S.C. Oral bioavailability of omega-3 fatty acids and carotenoids from the microalgae Phaeodactylum tricornutum in healthy young adults. Mar. Drugs 2021, 19, 700. [Google Scholar] [CrossRef]
- De Carvalho, C.C.C.R.; Neves, K.; de Almeida, S.V.T.F.; Steinrücken, P.; Kleinegris, D.M.M. Profiles of volatiles in microalgae depend on the extraction and analytical methods. Front. Bioeng. Biotechnol. 2025, 13, 1589776. [Google Scholar] [CrossRef]
- Pascual, C.; Akmirza, I.; Pérez, R.; Arnaiz, E.; Muñoz, R.; Lebrero, R. Trimethylamine abatement in algal-bacterial photobioreactors. Environ. Sci. Pollut. Res. 2020, 27, 9028–9037. [Google Scholar] [CrossRef]
- Chang, C.-T.; Chen, B.-Y.; Shiu, I.S.; Jeng, F.-T. Biofiltration of trimethylamine-containing waste gas by entrapped mixed microbial cells. Chemosphere 2004, 55, 751–756. [Google Scholar] [CrossRef]
- Mehta, R.S.; Bassette, R.; Ward, G. Trimethylamine responsible for fishy flavor in milk from cows on wheat pasture. J. Dairy Sci. 1974, 57, 285–289. [Google Scholar] [CrossRef]
- Namieśnik, J.; Jastrzębska, A.; Zygmunt, B. Determination of volatile aliphatic amines in air by solid-phase microextraction coupled with gas chromatography with flame ionization detection. J. Chromatogr. A 2003, 1016, 1–9. [Google Scholar] [CrossRef]
- Lidbury, I.; Mausz, M.A.; Scanlan, D.J.; Chen, Y. Identification of dimethylamine monooxygenase in marine bacteria reveals a metabolic bottleneck in the methylated amine degradation pathway. ISME J. 2017, 11, 1592–1601. [Google Scholar] [CrossRef]
- Lidbury, I.D.E.A.; Murrell, J.C.; Chen, Y. Trimethylamine and trimethylamine N-oxide are supplementary energy sources for a marine heterotrophic bacterium: Implications for marine carbon and nitrogen cycling. ISME J. 2015, 9, 760–769. [Google Scholar] [CrossRef]
- Yan, C.; Zhong, M.; Han, J.; Lu, X.; Zhang, Y.; Liu, L. Efficient degradation of trimethylamine in gas phase by petal-shaped Co-MoS2 catalyst in the photo-electrochemical system. Chem. Eng. J. 2021, 405, 127034. [Google Scholar] [CrossRef]
- Seth, M.; Mondal, P.; Ghosh, D.; Mukhopadhyay, S.K. The foul play of two dietary metabolites trimethylamine (TMA) and trimethylamine N-oxide (TMAO) on human health and the role of microbes in mitigating their effects. Nutrire 2023, 48, 52. [Google Scholar] [CrossRef]
- Perillo, P.M.; Rodríguez, D.F. Low temperature trimethylamine flexible gas sensor based on TiO2 membrane nanotubes. J. Alloys Compd. 2016, 657, 765–769. [Google Scholar] [CrossRef]
- Ho, K.-L.; Chung, Y.-C.; Lin, Y.-H.; Tseng, C.-P. Biofiltration of trimethylamine, dimethylamine, and methylamine by immobilized Paracoccus sp. CP2 and Arthrobacter sp. CP1. Chemosphere 2008, 72, 250–256. [Google Scholar] [CrossRef]
- Estrada, J.M.; Kraakman, N.J.R.B.; Muñoz, R.; Lebrero, R. A Comparative Analysis of Odour Treatment Technologies in Wastewater Treatment Plants. Environ. Sci. Technol. 2011, 45, 1100–1106. [Google Scholar] [CrossRef]
- Sun, J.; Mausz, M.A.; Chen, Y.; Giovannoni, S.J. Microbial trimethylamine metabolism in marine environments. Environ. Microbiol. 2019, 21, 513–520. [Google Scholar] [CrossRef]
- Goris, M.; Puntervoll, P.; Rojo, D.; Claussen, J.; Larsen, Ø.; Garcia-Moyano, A.; Almendral, D.; Barbas, C.; Ferrer, M.; Bjerga Gro Elin, K. Use of Flavin-Containing Monooxygenases for Conversion of Trimethylamine in Salmon Protein Hydrolysates. Appl. Environ. Microbiol. 2020, 86, e02105–e02120. [Google Scholar] [CrossRef]
- Lidbury, I.; Murrell, J.C.; Chen, Y. Trimethylamine N-oxide metabolism by abundant marine heterotrophic bacteria. Proc. Natl. Acad. Sci. USA 2014, 111, 2710–2715. [Google Scholar] [CrossRef]
- Yancey, P.H.; Gerringer, M.E.; Drazen, J.C.; Rowden, A.A.; Jamieson, A. Marine fish may be biochemically constrained from inhabiting the deepest ocean depths. Proc. Natl. Acad. Sci. USA 2014, 111, 4461–4465. [Google Scholar] [CrossRef]
- Huijbers, M.M.E.; Montersino, S.; Westphal, A.H.; Tischler, D.; van Berkel, W.J.H. Flavin dependent monooxygenases. Arch. Biochem. Biophys. 2014, 544, 2–17. [Google Scholar] [CrossRef]
- Chen, Y.; Patel, N.A.; Crombie, A.; Scrivens, J.H.; Murrell, J.C. Bacterial flavin-containing monooxygenase is trimethylamine monooxygenase. Proc. Natl. Acad. Sci. USA 2011, 108, 17791–17796. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.S.; Kim, J.K.; Cho, E.H.; Kim, Y.C.; Kim, J.I.; Kim, S.W. A novel flavin-containing monooxygenase from Methylophaga sp. strain SK1 and its indigo synthesis in Escherichia coli. Biochem. Biophys. Res. Commun. 2003, 306, 930–936. [Google Scholar] [CrossRef]
- Li, C.-Y.; Chen, X.-L.; Zhang, D.; Wang, P.; Sheng, Q.; Peng, M.; Xie, B.-B.; Qin, Q.-L.; Li, P.-Y.; Zhang, X.-Y.; et al. Structural mechanism for bacterial oxidation of oceanic trimethylamine into trimethylamine-oxide. Mol. Microbiol. 2017, 103, 992–1003. [Google Scholar] [CrossRef]
- Ameria, S.P.L.; Jung, H.S.; Kim, H.S.; Han, S.S.; Kim, H.S.; Lee, J.H. Characterization of a flavin-containing monooxygenase from Corynebacterium glutamicum and its application to production of indigo and indirubin. Biotechnol. Lett. 2015, 37, 1637–1644. [Google Scholar] [CrossRef]
- Lončar, N.; Fiorentini, F.; Bailleul, G.; Savino, S.; Romero, E.; Mattevi, A.; Fraaije, M.W. Characterization of a thermostable flavin-containing monooxygenase from Nitrincola lacisaponensis (NiFMO). Appl. Microbiol. Biotechnol. 2019, 103, 1755–1764. [Google Scholar] [CrossRef]
- De Carvalho, C.C.C.R. Whole cell biocatalysts: Essential workers from Nature to the industry. Microb. Biotechnol. 2017, 10, 250–263. [Google Scholar] [CrossRef]
- Rodrigues, C.J.C.; Pereira, R.F.S.; Fernandes, P.; Cabral, J.M.S.; de Carvalho, C.C.C.R. Cultivation-based strategies to find efficient marine biocatalysts. Biotechnol. J. 2017, 12, 1700036. [Google Scholar] [CrossRef]
- Kunitsky, C.; Osterhout, G.; Sasser, M. Identification of microorganisms using fatty acid methyl ester (FAME) analysis and the MIDI Sherlock Microbial Identification System. In Encyclopedia of Rapid Microbiological Methods; PDA/DHI: Bethesda, MD, USA, 2006; Volume 3, pp. 1–17. [Google Scholar]
- Cortes, M.A.L.R.M.; de Carvalho, C.C.C.R. Effect of carbon sources on lipid accumulation in Rhodococcus cells. Biochem. Eng. J. 2015, 94, 100–105. [Google Scholar] [CrossRef]
- Rodrigues, C.J.C.; de Carvalho, C.C.C.R. Cultivating marine bacteria under laboratory conditions: Overcoming the “unculturable” dogma. Front. Bioeng. Biotechnol. 2022, 10, 964589. [Google Scholar] [CrossRef]
- Meyrath, J. Influence of the size of inoculum on various growth phases inAspergillus oryzae. Antonie Van Leeuwenhoek 1963, 29, 57–78. [Google Scholar] [CrossRef]
- Hamill, P.G.; Stevenson, A.; McMullan, P.E.; Williams, J.P.; Lewis, A.D.R.; Sudharsan, S.; Stevenson, K.E.; Farnsworth, K.D.; Khroustalyova, G.; Takemoto, J.Y.; et al. Microbial lag phase can be indicative of, or independent from, cellular stress. Sci. Rep. 2020, 10, 5948. [Google Scholar] [CrossRef] [PubMed]
- Swift, E., Jr.; Hochanadel, H.P. The Vapor Pressure of Trimethylamine from 0 to 40°. J. Am. Chem. Soc. 1945, 67, 880–881. [Google Scholar] [CrossRef]
- Mohri, S.; Kanauchi, M. Isolation of Lactic Acid Bacteria Eliminating Trimethylamine (TMA) for Application to Fishery Processing. In Lactic Acid Bacteria: Methods and Protocols; Kanauchi, M., Ed.; Springer: New York, NY, USA, 2019; pp. 109–117. [Google Scholar]
- Leng, C.; Kish, J.D.; Roberts, J.E.; Dwebi, I.; Liu, Y. Temperature-dependent Henry’s law constants of atmospheric amines. J. Phys. Chem. A 2015, 119, 8884–8891. [Google Scholar] [CrossRef] [PubMed]
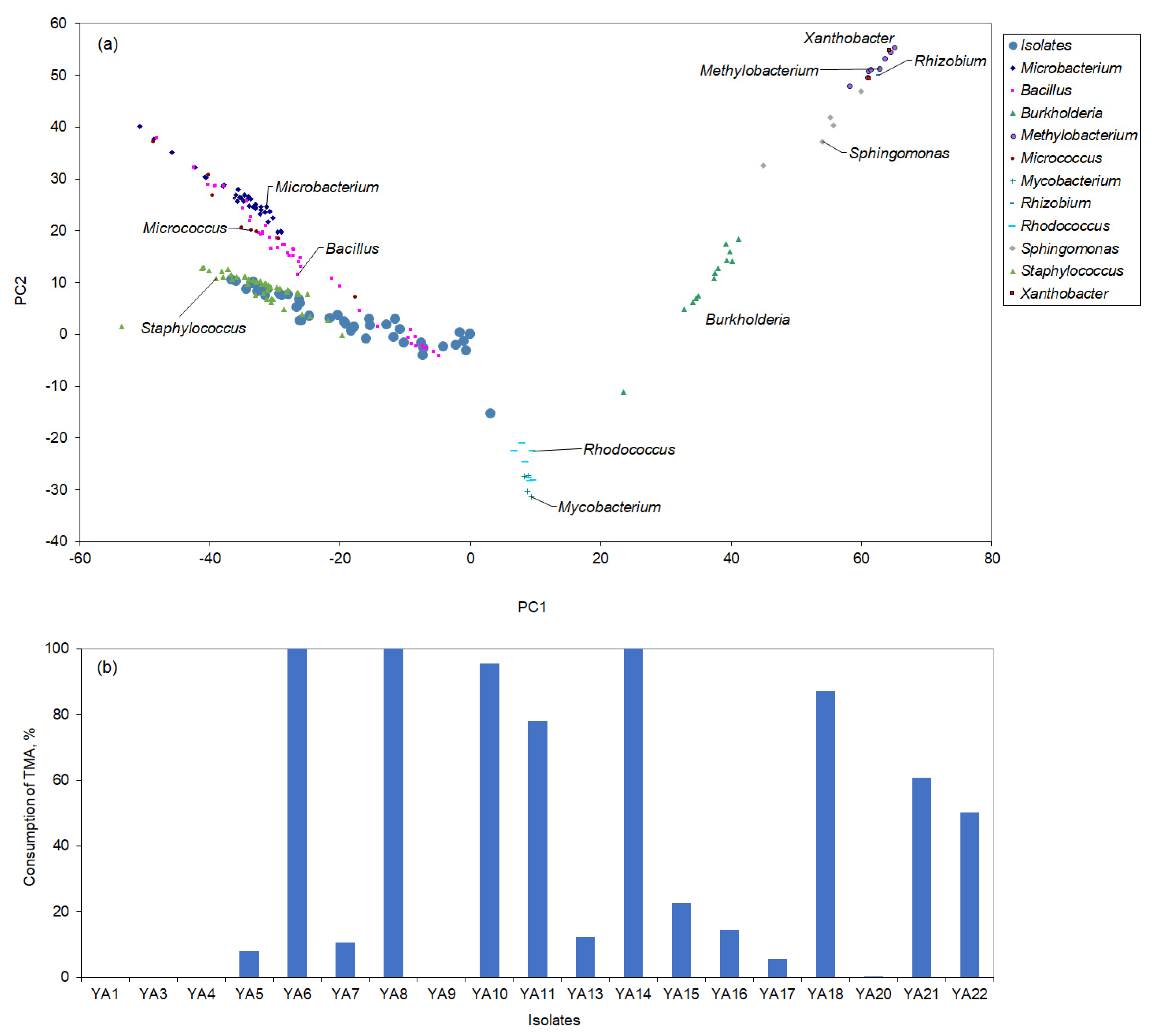
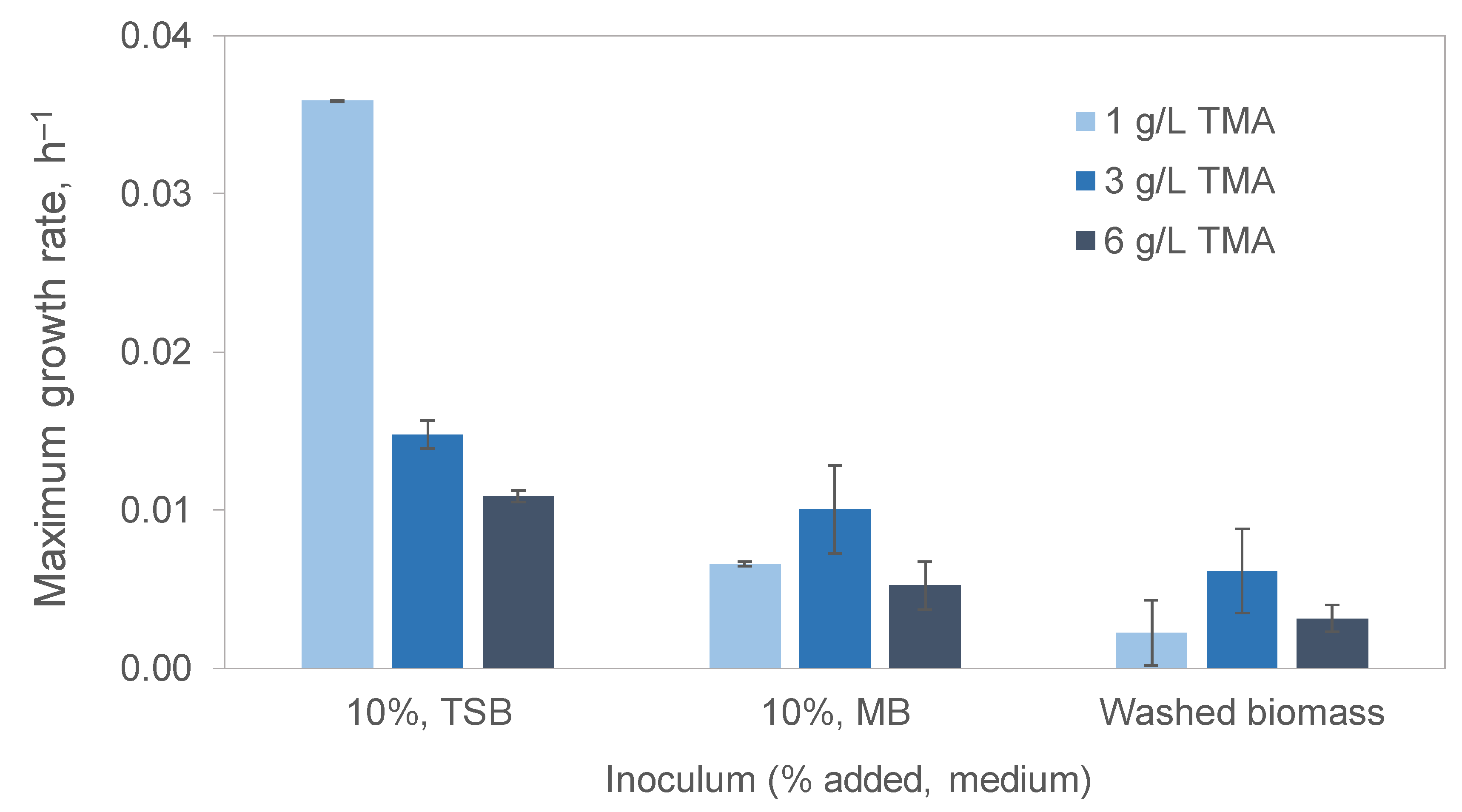
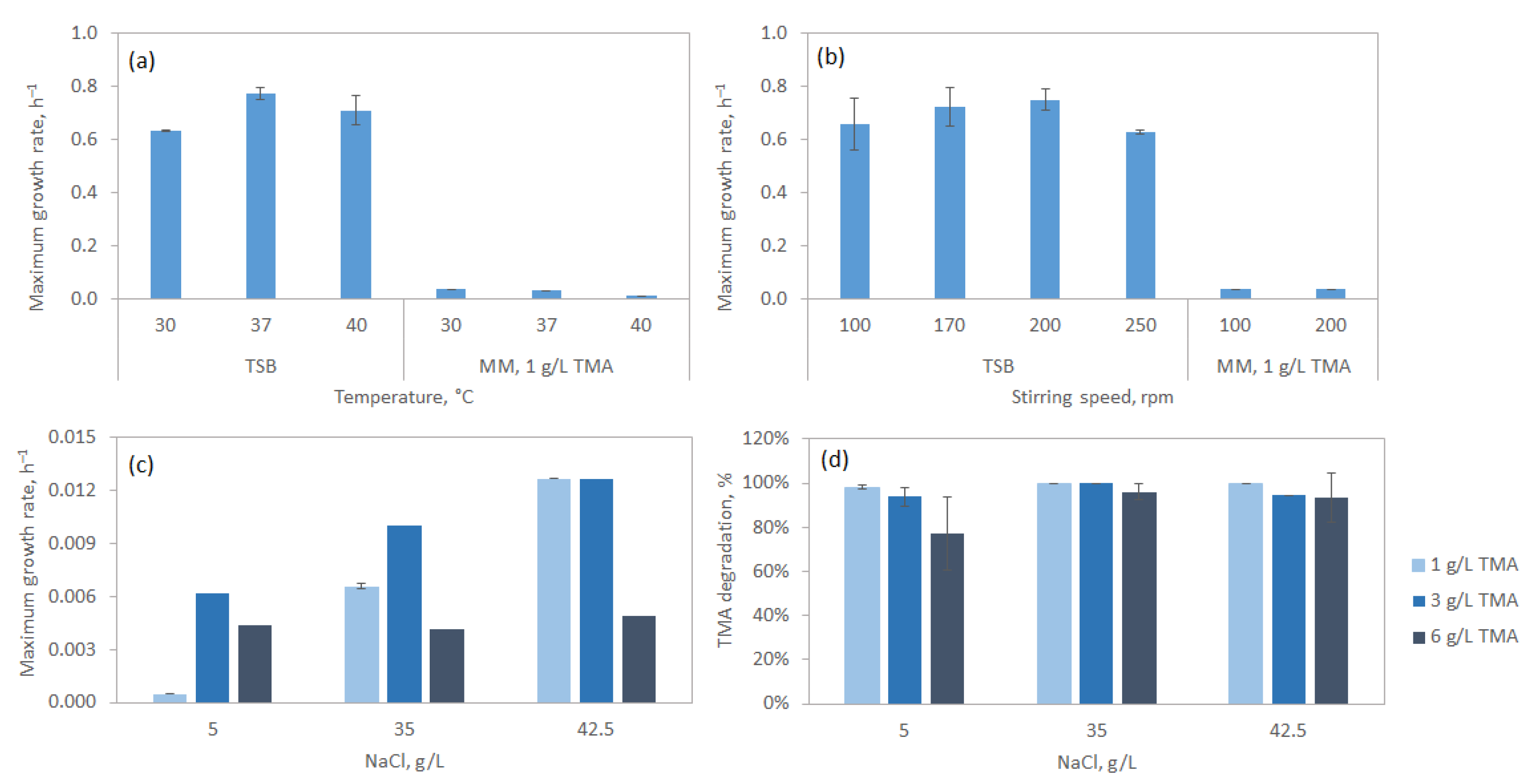


| TMA (g/L) | Temperature (°C) | Agitation (rpm) | Salinity (g/L) | Peptone (g/L) | µmax, (h−1) | TMA Degradation (%) |
|---|---|---|---|---|---|---|
| 1 | 30 | 100 | 35 | 5 | 0.04 | - |
| 1 | 30 | 100 | 35 | 5 | 0.04 | - |
| 1 | 37 | 100 | 35 | 5 | 0.03 | - |
| 1 | 40 | 100 | 35 | 5 | 0.01 | - |
| 1 | 30 | 200 | 5 | 0 | 0.0005 | 98.2 |
| 1 | 30 | 200 | 35 | 0 | 0.007 | 100 |
| 1 | 30 | 200 | 35 | 5 | 0.04 | - |
| 1 | 30 | 200 | 42.5 | 0 | 0.013 | 100 |
| 1 | 30 | 200 | 42.5 | 2.5 | 0.068 | 99 |
| 1 | 30 | 200 | 42.5 | 5 | 0.092 | 100 |
| 1 | 30 | 200 | 42.5 | 10 | 0.092 | 100 |
| 1 | 30 | 200 | 42.5 | 20 | 0.098 | - |
| 3 | 30 | 200 | 5 | 0 | 0.006 | 93.8 |
| 3 | 30 | 200 | 35 | 0 | 0.01 | 99.9 |
| 3 | 30 | 200 | 42.5 | 0 | 0.013 | 94.5 |
| 6 | 30 | 200 | 5 | 0 | 0.003 | 77.1 |
| 6 | 30 | 200 | 35 | 0 | 0.004 | 95.9 |
| 6 | 30 | 200 | 42.5 | 0 | 0.005 | 47 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Almeida, S.V.T.F.; Neves, K.; de Carvalho, C.C.C.R. Screening and Isolation of Bacterial Strains Able to Degrade Trimethylamine. Microorganisms 2025, 13, 1369. https://doi.org/10.3390/microorganisms13061369
de Almeida SVTF, Neves K, de Carvalho CCCR. Screening and Isolation of Bacterial Strains Able to Degrade Trimethylamine. Microorganisms. 2025; 13(6):1369. https://doi.org/10.3390/microorganisms13061369
Chicago/Turabian Stylede Almeida, Sebastião V. T. F., Kilian Neves, and Carla C. C. R. de Carvalho. 2025. "Screening and Isolation of Bacterial Strains Able to Degrade Trimethylamine" Microorganisms 13, no. 6: 1369. https://doi.org/10.3390/microorganisms13061369
APA Stylede Almeida, S. V. T. F., Neves, K., & de Carvalho, C. C. C. R. (2025). Screening and Isolation of Bacterial Strains Able to Degrade Trimethylamine. Microorganisms, 13(6), 1369. https://doi.org/10.3390/microorganisms13061369







