Functional Profiling of Enterococcus and Pediococcus Strains: An In Vitro Study on Probiotic and Postbiotic Properties
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Bacterial Strains and Growth Conditions
2.2. Evaluation of Probiotic Properties of LAB Strains
2.2.1. Identification of LAB Strains via 16S rDNA Sequencing
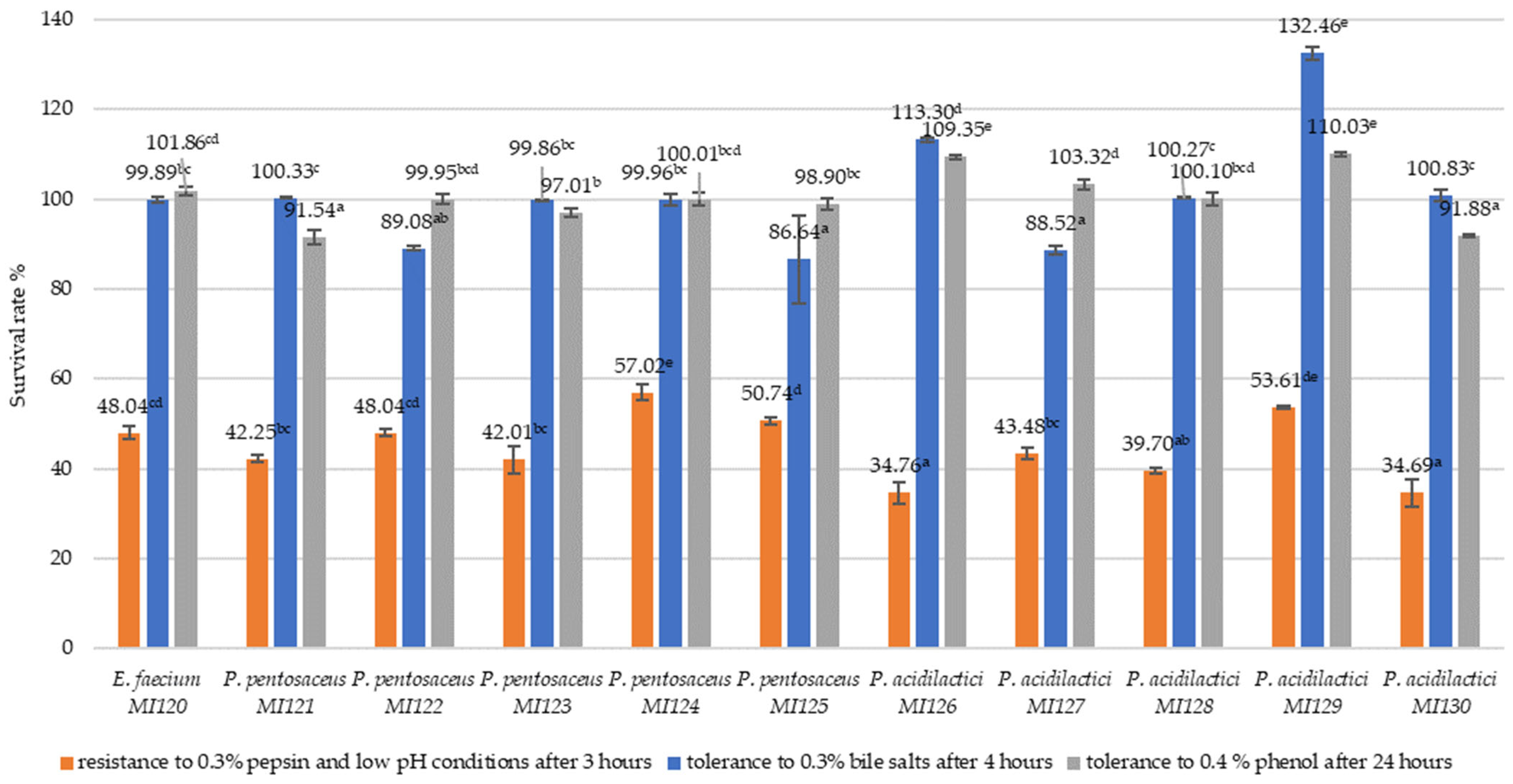
2.2.2. In Vitro Tolerance of LAB Strains to Simulated Gastric and Intestinal Conditions
2.2.3. Phenol Tolerance
2.2.4. Cell Surface Characteristics
Cell Surface Hydrophobicity
Auto-Aggregation Test
2.2.5. Enzymatic Pattern of the LAB Strains
2.3. Safety Assessment
2.3.1. Hemolysis Assay
2.3.2. Antibiotic Susceptibility Testing
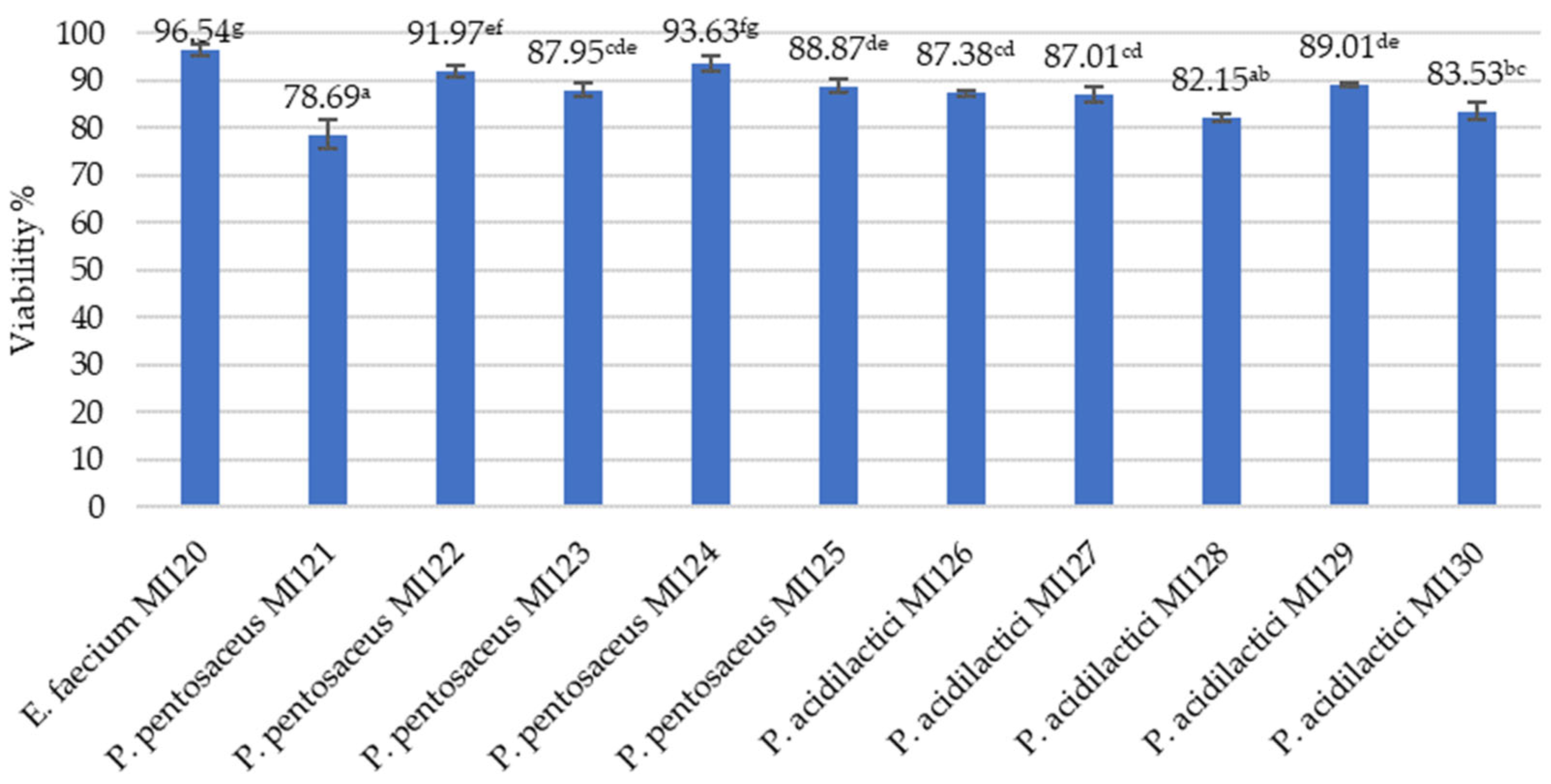
2.4. Preservation of LAB Isolates Through Freeze-Drying
2.5. Evaluation of Postbiotic Properties of CFSs Derived from LAB Strains
2.5.1. Preparation of Cell-Free Supernatants (CFSs) from Probiotic Bacterial Cultures
2.5.2. Antimicrobial Activity of CFSs
Agar Well Diffusion Assay
MBC Evaluations
2.5.3. Assessment of the Biochemical Profile
Total Polyphenol Content
Total Flavonoid Content
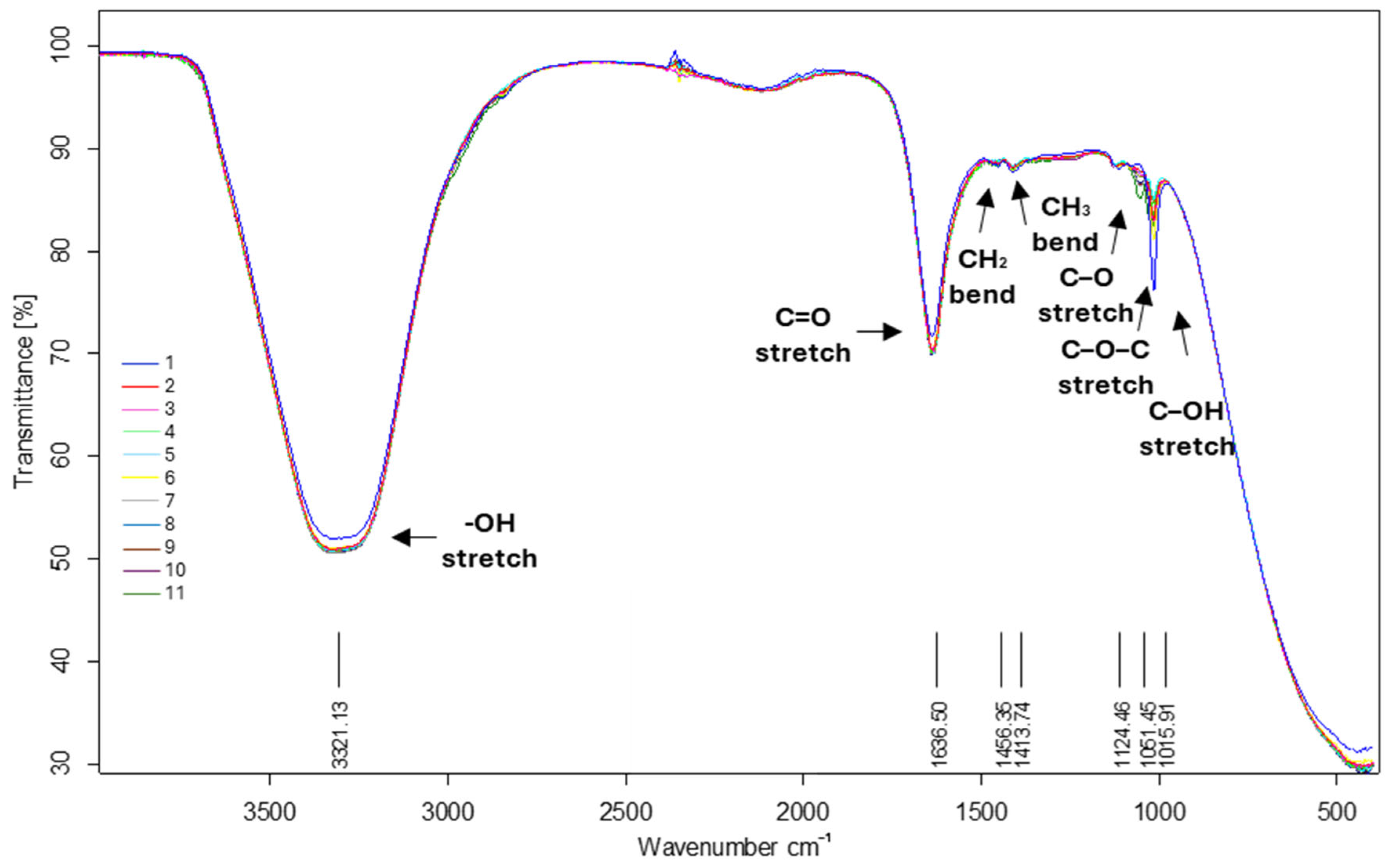
Assessment of Fourier Transform Infrared Spectroscopy of CFS Samples
2.5.4. Antioxidant Activity by DPPH Assay
2.6. Statistical Analysis
3. Results
3.1. Molecular Characterization of the LAB Isolates
3.2. Probiotic Patterns of LAB Strains
3.2.1. Resistance to Simulated Gastrointestinal Conditions
3.2.2. Cell Surface Properties of LAB Strains
3.2.3. Enzymatic Profile
3.2.4. Safety Traits of LAB Strains
3.2.5. Freeze-Drying as a Method for Long-Term Preservation of LAB Strains
3.3. Postbiotic Profiles of LAB Strains
3.3.1. Antimicrobial Patterns of LAB Strain
3.3.2. Biochemical Profile
3.3.3. Antioxidant Activity
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- O’Neill, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. Review on Antimicrobial Resistance; Wellcome Trust: London, UK, 2014; Available online: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf (accessed on 17 March 2025).
- de Kraker, M.E.A.; Stewardson, A.J.; Harbarth, S. Will 10 Million People Die a Year Due to Antimicrobial Resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef] [PubMed]
- Kakoullis, L.; Papachristodoulou, E.; Chra, P.; Panos, G. Mechanisms of Antibiotic Resistance in Important Gram-Positive and Gram-Negative Pathogens and Novel Antibiotic Solutions. Antibiotics 2021, 10, 415. [Google Scholar] [CrossRef] [PubMed]
- Rabetafika, H.N.; Razafindralambo, A.; Ebenso, B.; Razafindralambo, H.L. Probiotics as Antibiotic Alternatives for Human and Animal Applications. Encyclopedia 2023, 3, 561–581. [Google Scholar] [CrossRef]
- FAO; WHO. Guidelines for the Evaluation of Probiotics in Food. In Proceedings of the Report on a Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food, London, ON, Canada, 30 April–1 May 2002. [Google Scholar]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Maftei, N.-M.; Raileanu, C.R.; Balta, A.A.; Ambrose, L.; Boev, M.; Marin, D.B.; Lisa, E.L. The Potential Impact of Probiotics on Human Health: An Update on Their Health-Promoting Properties. Microorganisms 2024, 12, 234. [Google Scholar] [CrossRef]
- Markowiak, P.; Śliżewska, K. Effects of Probiotics, Prebiotics, and Synbiotics on Human Health. Nutrients 2017, 9, 1021. [Google Scholar] [CrossRef]
- Tegegne, B.A.; Kebede, B. Probiotics, their prophylactic and therapeutic applications in human health development: A review of the literature. Heliyon 2022, 8, e09725. [Google Scholar] [CrossRef]
- Zielińska, D.; Kolozyn-Krajewska, D. Food-origin lactic acid bacteria may exhibit probiotic properties: Review. Biomed Res. Int. 2018, 2018, 5063185. [Google Scholar] [CrossRef]
- Mariarosaria, M. Bifidobacteria, Lactobacilli… when, how and why to use them. Glob. Pediatr. 2024, 8, 100139. [Google Scholar]
- Coulibaly, W.H.; Kouadio, N.R.; Camara, F.; Diguță, C.; Matei, F. Functional properties of lactic acid bacteria isolated from Tilapia (Oreochromis niloticus) in Ivory Coast. BMC Microbiol. 2023, 23, 152. [Google Scholar] [CrossRef]
- Kouadio, N.J.; Zady, A.L.O.; Kra, K.A.S.; Diguță, F.C.; Niamke, S.; Matei, F. In Vitro Probiotic Characterization of Lactiplantibacillus plantarum Strains Isolated from Traditional Fermented Dockounou Paste. Fermentation 2024, 10, 264. [Google Scholar] [CrossRef]
- Badea, F.; Diguta, C.F.; Matei, F. The use of lactic acid bacteria and their metabolites to improve the shelf life of perishable fruits and vegetables. Sci. Bull. Ser. F Biotechnol. 2022, XXVI, 117–125. [Google Scholar]
- Zamfir, M.; Angelescu, I.-R.; Voaides, C.; Cornea, C.-P.; Boiu-Sicuia, O.; Grosu-Tudor, S.-S. Non-Dairy Fermented Beverages Produced with Functional Lactic Acid Bacteria. Microorganisms 2022, 10, 2314. [Google Scholar] [CrossRef]
- Voaides, C.; Boiu-Sicuia, O.; Israel-Roming, F.; Zamfir, M.; Grosu-Tudor, S.S.; Angelescu, I.R.; Cornea, C.P. Lactobacillus Strains for Vegetable Juice Fermentation—Quality and Health Aspects. Biomedicines 2022, 10, 2867. [Google Scholar] [CrossRef] [PubMed]
- Badea, F.; Pristavu, M.C.; Aldea, C.A.; Israel-Roming, F.; Matei, F. In vitro screening of lactic acid bacteria as biocontrol agents for biopreservation of perishable agro-food products. Sci. Papers Ser. D Anim. Sci. 2024, LXVII, 427–438. [Google Scholar]
- Pais, P.; Almeida, V.; Yılmaz, M.; Teixeira, M.C. Saccharomyces boulardii: What makes it tick as successful probiotic? J. Fungi 2020, 6, 78. [Google Scholar] [CrossRef]
- Fu, J.; Liu, J.; Wen, X.; Zhang, G.; Cai, J.; Qiao, Z.; An, Z.; Zheng, J.; Li, L. Unique probiotic properties and bioactive metabolites of Saccharomyces boulardii. Probiotics Antimicrob. Proteins 2022, 15, 967–982. [Google Scholar] [CrossRef]
- Diguță, C.F.; Mihai, C.; Toma, R.C.; Cîmpeanu, C.; Matei, F. In Vitro Assessment of Yeasts Strains with Probiotic Attributes for Aquaculture Use. Foods 2023, 12, 124. [Google Scholar] [CrossRef]
- Mogmenga, I.; Somda, M.K.; Ouattara, C.A.T.; Keita, I.; Dabiré, Y.; Diguță, C.F.; Toma, R.C.; Ezeogu, L.I.; Ugwuanyi, J.O.; Ouattara, A.S.; et al. Promising probiotic properties of the yeasts isolated from Rabilé, a traditionally fermented beer produced in Burkina Faso. Microorganisms 2023, 11, 802. [Google Scholar] [CrossRef]
- Jiang, S.; Cai, L.; Lv, L.; Li, L. Pediococcus pentosaceus, a future additive or probiotic candidate. Microb. Cell Factories 2021, 20, 45. [Google Scholar] [CrossRef]
- Qi, Y.; Huang, L.; Zeng, Y.; Li, W.; Zhou, D.; Xie, J.; Xie, J.; Tu, Q.; Deng, D.; Yin, J. Pediococcus pentosaceus: Screening and Application as Probiotics in Food Processing. Front. Microbiol. 2021, 12, 3827. [Google Scholar] [CrossRef] [PubMed]
- Todorov, S.D.; Dioso, C.M.; Liong, M.-T.; Nero, L.A.; Khosravi-Darani, K.; Ivanova, I.V. Beneficial Features of Pediococcus: From Starter Cultures and Inhibitory Activities to Probiotic Benefits. World J. Microbiol. Biotechnol. 2023, 39, 4. [Google Scholar] [CrossRef] [PubMed]
- Hanchi, H.; Mottawea, W.; Sebei, K.; Hammami, R. The genus Enterococcus: Between probiotic potential and safety concerns-an update. Front. Microbiol. 2018, 9, 1791. [Google Scholar] [CrossRef]
- Krawczyk, B.; Wityk, P.; Gałęcka, M.; Michalik, M. The Many Faces of Enterococcus spp.—Commensal, Probiotic and Opportunistic Pathogen. Microorganisms 2021, 9, 1900. [Google Scholar] [CrossRef]
- Tsanasidou, C.; Asimakoula, S.; Sameli, N.; Fanitsios, C.; Vandera, E.; Bosnea, L.; Koukkou, A.-I.; Samelis, J. Safety evaluation, biogenic amine formation, and enzymatic activity profiles of autochthonous enterocin-producing greek cheese isolates of the Enterococcus faecium/durans Group. Microorganisms 2021, 9, 777. [Google Scholar] [CrossRef]
- De Melo Pereira, G.V.; de Oliveira Coelho, B.; Magalhães Júnior, A.I.; Thomaz-Soccol, V.; Soccol, C.R. How to select a probiotic? A review and update of methods and criteria. Biotechnol. Adv. 2018, 36, 2060–2076. [Google Scholar] [CrossRef]
- Ayyash, M.M.; Abdalla, A.K.; AlKalbani, N.S.; Baig, M.A.; Turner, M.S.; Liu, S.-Q.; Shah, N.P. Invited Review: Characterization of new probiotics from dairy and nondairy products-insights into acid tolerance, bile metabolism and tolerance, and adhesion capability. J. Dairy Sci. 2021, 104, 8363–8379. [Google Scholar] [CrossRef]
- Liu, C.; Ma, N.; Feng, Y.; Zhou, M.; Li, H.; Zhang, X.; Ma, X. From probiotics to postbiotics: Concepts and applications. Anim. Res. One Health 2023, 1, 92–114. [Google Scholar] [CrossRef]
- Cicenia, A.; Scirocco, A.; Carabotti, M.; Pallotta, L.; Marignani, M.; Severi, C. Postbiotic activities of lactobacilli-derived factors. J. Clin. Gastroenterol. 2014, 48 (Suppl. S1), S18–S22. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, S.; Li, S.; Jiang, W.; Wang, J.; Xiao, J.; Chen, T.; Ma, J.; Khan, M.Z.; Wang, W.; et al. Unlocking the power of postbiotics: A revolutionary approach to nutrition for humans and animals. Cell Metab. 2024, 36, 725–744. [Google Scholar] [CrossRef]
- Pristavu, M.C.; Diguță, C.; Coulibaly, W.H.; Youte Fanche, S.A.; Dopcea, G.; Matei, F. Review of postbiotics as new health promoters. AgroLife Sci. J. 2022, 11, 142–152. [Google Scholar] [CrossRef]
- Wei, L.; Wang, B.; Bai, J.; Zhang, Y.; Liu, C.; Suo, H.; Wang, C. Postbiotics are a candidate for new functional foods. Food Chem. X 2024, 23, 101650. [Google Scholar] [CrossRef] [PubMed]
- Molina, D.; Marinas, I.C.; Angamarca, E.; Hanganu, A.; Stan, M.; Chifiriuc, M.C.; Tenea, G.N. Postbiotic-based extracts from native probiotic strains: A promising strategy for food preservation and antimicrobial defense. Antibiotics 2025, 14, 318. [Google Scholar] [CrossRef]
- Aguilar-Toalá, J.E.; Garcia-Varela, R.; Garcia, H.S.; Mata-Haro, V.; González-Córdova, A.F.; Vallejo-Cordoba, B.; Hernández-Mendoza, A. Postbiotics: An evolving term within the functional foods field. Trends Food Sci. Technol. 2018, 75, 105–114. [Google Scholar] [CrossRef]
- Ma, L.; Tu, H.; Chen, T. Postbiotics in Human Health: A Narrative Review. Nutrients 2023, 15, 291. [Google Scholar] [CrossRef]
- Hijová, E. Postbiotics as Metabolites and Their Biotherapeutic Potential. Int. J. Mol. Sci. 2024, 25, 5441. [Google Scholar] [CrossRef]
- Teame, T.; Wang, A.; Xie, M.; Zhang, Z.; Yang, Y.; Ding, Q.; Gao, C.; Olsen, R.E.; Ran, C.; Zhou, Z. Paraprobiotics and Postbiotics of Probiotic Lactobacilli, Their Positive Effects on the Host and Action Mechanisms: A Review. Front. Nutr. 2020, 7, 570344. [Google Scholar] [CrossRef]
- Salminen, S.; Collado, M.C.; Endo, A.; Hill, C.; Lebeer, S.; Quigley, E.M.M.; Sanders, M.E.; Shamir, R.; Swann, J.R.; Szajewska, H.; et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 649–667. [Google Scholar] [CrossRef]
- Vinderola, G.; Druart, C.; Gosálbez, L.; Salminen, S.; Vinot, N.; Lebeer, S. Postbiotics in the medical field under the perspective of the ISAPP definition: Scientific, regulatory, and marketing considerations. Front. Pharmacol. 2023, 14, 1239745. [Google Scholar] [CrossRef]
- Wegh, C.A.M.; Geerlings, S.Y.; Knol, J.; Roeselers, G.; Belzer, C. Postbiotics and their potential applications in early life nutrition and beyond. Int. J. Mol. Sci. 2019, 20, 4673. [Google Scholar] [CrossRef]
- Nataraj, B.H.; Ali, S.A.; Behare, P.V.; Yadav, H. Postbiotics-parabiotics: The new horizons in microbial biotherapy and functional foods. Microb. Cell Factories 2020, 19, 168. [Google Scholar] [CrossRef]
- Thorakkattu, P.; Khanashyam, A.C.; Shah, K.; Babu, K.S.; Mundanat, A.S.; Deliephan, A.; Deokar, G.S.; Santivarangkna, C.; Nirmal, N.P. Postbiotics: Current Trends in Food and Pharmaceutical Industry. Foods 2022, 11, 3094. [Google Scholar] [CrossRef]
- Isaac-Bamgboye, F.J.; Mgbechidinma, C.L.; Onyeaka, H.; Isaac-Bamgboye, I.T.; Chukwugozie, D.C. Exploring the potential of postbiotics for food safety and human health improvement. J. Nutr. Metab. 2024, 2024, 1868161. [Google Scholar] [CrossRef]
- Liang, B.; Xing, D. The Current and Future Perspectives of Postbiotics. Probiotics Antimicro. Prot. 2023, 15, 1626–1643. [Google Scholar] [CrossRef]
- Malashree, L.; Angadi, V.; Yadav, K.S.; Prabha, R. “Postbiotics”-one step ahead of probiotics. Int. J. Curr. Microbiol. App. Sci. 2019, 8, 2049–2053. [Google Scholar] [CrossRef]
- Vallejo-Cordoba, B.; Castro-López, C.; García, H.S.; González-Córdova, A.F.; Hernández-Mendoza, A. Postbiotics and Paraprobiotics: A Review of Current Evidence and Emerging Trends. Adv. Food Nutr. Res. 2020, 94, 1–34. [Google Scholar]
- Cuevas-González, P.F.; Liceaga, A.M.; Aguilar-Toalá, J.E. Postbiotics and paraprobiotics: From concepts to applications. Food Res. Int. 2020, 136, 109502. [Google Scholar] [CrossRef]
- Allende, A.; Alvarez-Ordóñez, A.; Bortolaia, V.; Bover-Cid, S.; De Cesare, A.; Dohmen, W.; Guillier, L.; Jacxsens, L.; Nauta, M.; Mughini-Gras, L. Update of the list of qualified presumption of safety (QPS) recommended microbiological agents intentionally added to food or feed as notified to EFSA 21: Suitability of taxonomic units notified to EFSA until September 2024. EFSA J. 2025, 23, e9169. [Google Scholar]
- Hung, Y.-P.; Lee, C.-C.; Lee, J.-C.; Tsai, P.-J.; Hsueh, P.-R.; Ko, W.-C. The potential of probiotics to eradicate gut carriage of pathogenic or antimicrobial-resistant Enterobacterales. Antibiotics 2021, 10, 1086. [Google Scholar] [CrossRef]
- Al-Fakhrany, O.M.; Elekhnawy, E. Next-generation probiotics: The upcoming biotherapeutics. Mol. Biol. Rep. 2024, 51, 505. [Google Scholar] [CrossRef]
- Abouelela, M.E.; Helmy, Y.A. Next-generation probiotics as novel therapeutics for improving human health: Current trends and future perspectives. Microorganisms 2024, 12, 430. [Google Scholar] [CrossRef]
- Almeida-Santos, A.C.; Novais, C.; Peixe, L.; Freitas, A.R. Enterococcus spp. as a producer and target of bacteriocins: A double-edged sword in the antimicrobial resistance crisis context. Antibiotics 2021, 10, 1215. [Google Scholar] [CrossRef]
- Heuer, H.; Krsek, M.; Baker, P.; Smalla, K.; Wellington, E. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 1997, 63, 3233–3241. [Google Scholar] [CrossRef]
- Diguță, C.F.; Nițoi, G.D.; Matei, F.; Luță, G.; Cornea, C.P. The Biotechnological potential of Pediococcus spp. isolated from Kombucha microbial consortium. Foods 2020, 9, 1780. [Google Scholar] [CrossRef]
- Rosenberg, M.; Gutnick, D. and Rosenberg, E. Adherence of bacteria to hydrocarbons: A simple method for measuring cell-surface hydrophobicity. FEMS Microbiol. Lett. 1980, 9, 29–33. [Google Scholar] [CrossRef]
- Proca, I.G.; Diguta, C.F.; Jurcoane, S.; Matei, F. Screening of halotolerant bacteria producing hydrolytic enzymes with biotechnological applications. Sci. Bull. Ser. F Biotechnol. 2020, XXIV, 197–204. [Google Scholar]
- Leclercq, R.; Cantón, R.; Brown, D.F.J.; Giske, C.G.; Heisig, P.; MacGowan, A.P.; Mouton, J.W.; Nordmann, P.; Rodloff, A.C.; Rossolini, G.M.; et al. EUCAST expert rules in antimicrobial susceptibility testing. Clin. Microbiol. Infect. 2013, 19, 141–160. [Google Scholar] [CrossRef]
- CLSI. CLSI Supplement M100S. Performance Standards for Antimicrobial Susceptibility Testing, 26th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016. [Google Scholar]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Asadi, Z.; Abbasi, A.; Ghaemi, A.; Montazeri, E.A.; Akrami, S. Investigating the properties and antibacterial, antioxidant, and cytotoxicity activity of postbiotics derived from Lacticaseibacillus casei on various gastrointestinal pathogens in vitro and in food models. GMS HIC 2024, 19, Doc60. [Google Scholar]
- Hamad, G.M.; Abdelmotilib, N.M.; Darwish, A.M.; Zeitoun, A.M. Commercial probiotic cell-free supernatants for inhibition of Clostridium perfringens poultry meat infection in Egypt. Anaerobe 2020, 62, 102181. [Google Scholar] [CrossRef]
- Qadi, W.S.M.; Mediani, A.; Kasim, Z.M.; Misnan, N.M.; Sani, N.A.; Jamar, N.H. Biological Characterization and Metabolic Variations among Cell-Free Supernatants Produced by Selected Plant-Based Lactic Acid Bacteria. Metabolites 2023, 13, 849. [Google Scholar] [CrossRef]
- Mantzios, P.G.; Spyropoulou, P.; Hatzianastasiou, S.; Efthymiou, D.; Filippopoulos, E.; Mamarelis, C.; Potsios, C.; Filioti, K.; Letsas, C.A. Pediococcus pentosaceus endocarditis in a patient with recent transcatheter aortic valve implantation and liver cirrhosis: A case report and review of the literature. Cureus 2024, 16, e57509. [Google Scholar] [CrossRef] [PubMed]
- Abbasiliasi, S.; Tan, J.S.; Bashokouh, F.; Ibrahim, T.A.T.; Mustafa, S.; Vakhshiteh, F.; Sivasamboo, S.; Ariff, A.B. In vitro assessment of Pediococcus acidilactici Kp10 for its potential use in the food industry. BMC Microbiol. 2017, 17, 121. [Google Scholar] [CrossRef]
- Ayyash, M.; Abushelaibi, A.; Al-Mahadin, S.; Enan, M.; El-Tarabily, K.; Shah, N. In-vitro investigation into probiotic characterisation of Streptococcus and Enterococcus isolated from camel milk. LWT-Food Sci. Technol. 2017, 87, 478–487. [Google Scholar] [CrossRef]
- Bhagat, D.; Raina, N.; Kumar, A.; Katoch, M.; Khajuria, Y.; Slathia, P.S.; Sharma, P. Probiotic properties of a phytase producing Pediococcus acidilactici strain SMVDUDB2 isolated from traditional fermented cheese product, Kalarei. Sci. Rep. 2020, 10, 1926. [Google Scholar] [CrossRef]
- Yi, G.-S.; Jin, X.; Zheng, Q.; Nguyen, T.T.M.; Yang, S.-J.; Yi, T.-H. Antimicrobial Activity of Pediococcus pentosaceus PMY2 Against Multidrug-Resistant Pathogens. Antibiotics 2025, 14, 389. [Google Scholar] [CrossRef] [PubMed]
- Nami, Y.; Bakhshayesh, R.V.; Jalaly, H.M.; Lotfi, H.; Eslami, S.; Hejazi, M.A. Probiotic properties of enterococcus isolated from artisanal dairy products. Front. Microbiol. 2019, 10, 300. [Google Scholar] [CrossRef]
- Yerlikaya, O.; Akbulut, N. In vitro characterisation of probiotic properties of Enterococcus faecium and Enterococcus durans strains isolated from raw milk and traditional dairy products. Int. J. Dairy Technol. 2020, 73, 98–107. [Google Scholar] [CrossRef]
- Hammad, A.M.; Hassan, H.A.; Shimamoto, T. Prevalence, antibiotic resistance and virulence of Enterococcus spp. in Egyptian fresh raw milk cheese. Food Control 2015, 50, 815–820. [Google Scholar] [CrossRef]
- Xia, M.; Mu, S.; Fang, Y.; Zhang, X.; Yang, G.; Hou, X.; He, F.; Zhao, Y.; Huang, Y.; Zhang, W.; et al. Genetic and Probiotic Characteristics of Urolithin A Producing Enterococcus faecium FUA027. Foods 2023, 12, 1021. [Google Scholar] [CrossRef]
- Fugaban, J.I.I.; Holzapfel, W.H.; Todorov, S.D. Probiotic potential and safety assessment of bacteriocinogenic Enterococcus faecium strains with antibacterial activity against Listeria and vancomycin-resistant enterococci. Curr. Res. Microb. Sci. 2021, 2, 100070. [Google Scholar] [CrossRef]
- Sorroche, F.G.; Spesia, M.B.; Zorreguieta, A.; Giordano, W. A positive correlation between bacterial autoaggregation and biofilm formation in native Sinorhizobium meliloti isolates from Argentina. Appl. Environ. Microbiol. 2012, 78, 4092–4101. [Google Scholar] [CrossRef] [PubMed]
- Trunk, T.; Khalil, H.S.; Leo, J.C. Bacterial autoaggregation. AIMS Microbiol. 2018, 4, 140–164. [Google Scholar] [CrossRef]
- Tyfa, A.; Kunicka-Styczyńska, A.; Zabielska, J. Evaluation of hydrophobicity and quantitative analysis of biofilm formation by Alicyclobacillus sp. Acta Biochim. Pol. 2015, 62, 785–790. [Google Scholar]
- Peiren, J.; Hellemans, A.; De Vos, P. Impact of the freeze-drying process on product appearance, residual moisture content, viability, and batch uniformity of freeze-dried bacterial cultures safeguarded at culture collections. Appl. Microbiol. Biotechnol. 2016, 100, 6239–6249. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Han, J.; Sun, Q.; Zhou, Q.; Ye, Z.; Li, P.; Gu, Q. Research Progress on Improving the Freeze-Drying Resistance of Probiotics: A Review. Trends Food Sci. Technol. 2024, 147, 104425. [Google Scholar] [CrossRef]
- Ganesan, G.; Selvam, G.; Varatharaju, A.; Annamalai, P. Chapter 41-Preservation of postbiotics. In Postbiotics: Health and Industry; Academic Press: Cambridge, MA, USA, 2025; pp. 703–707. [Google Scholar]
- Nicolae, G.; Dopcea, I.; Nicolae, A.-M.; Stelian, P.; Matei, F. Conservation methods used for yeast isolated from vineyards—Lyophilisation advantage. Sci. Bull. Biotechnol. 2010, 14, 31–36. [Google Scholar]
- Tóth, A.G.; Csabai, I.; Judge, M.F.; Maróti, G.; Becsei, Á.; Spisák, S.; Solymosi, N. Mobile Antimicrobial Resistance Genes in Probiotics. Antibiotics 2021, 10, 1287. [Google Scholar] [CrossRef]
- FAO/WHO Expert Consultation. Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Lactic Acid Bacteria; FAO/WHO: Córdoba, Argentina, 2001. [Google Scholar]
- Rosales Cavaglieri, L.A.; Corti Isgro, M.; Aminahuel, C.; Parada, J.; Poloni, V.L.; Montenegro, M.A.; Alonso, V.; Falcone, R.D.; Cavaglieri, L.R. Exploring the potential of lactic acid bacteria to produce postbiotics with antimicrobial and antioxidant properties: Focus on the probiotic strain Pediococcus pentosaceus RC007 for industrial-scale production. IJFST 2025, 60, vvae003. [Google Scholar] [CrossRef]
- Megur, A.; Ambrutaitytė, K.; Šimoliūnas, E.; Lastauskienė, E.; Burokas, A. Whole-genome sequencing and in vitro probiotic characterization of Pediococcus pentosaceus ELAB 60WB isolated from fermented cherry tomatoes. LWT 2025, 220, 117547. [Google Scholar] [CrossRef]
- Zhang, Q.; Xu, B.; Min, Y.; Liu, J.; Shang, Y.; Lan, X.; Xiang, W.; Tang, J. Evaluation of the safety and probiotic properties of GABA-producing Enterococcus faecium AB157 based on whole genome and phenotype analysis. LWT 2025, 215, 117242. [Google Scholar] [CrossRef]
- İncili, G.K.; Karatepe, P.; Akgöl, M.; Güngören, A.; Koluman, A.; İlhak, O.; Kanmaz, H.; Kaya, B.; Hayaloğlu, A.A. Characterization of lactic acid bacteria postbiotics, evaluation of in-vitro antibacterial effect, microbial and chemical quality on chicken drumsticks. Food Microbiol. 2022, 104, 104001. [Google Scholar] [CrossRef] [PubMed]
- Vodnar, D.C.; Paucean, A.; Dulf, F.V.; Socaciu, C. HPLC Characterization of Lactic Acid Formation and FTIR Fingerprint of Probiotic Bacteria during Fermentation Processes. Not. Bot. Hort. Agrobot. Cluj. 2010, 38, 109–113. [Google Scholar]
- Pinilla, C.M.B.; Galland, F.; Pacheco, M.T.B.; Bócoli, P.J.; Borges, D.F.; Alvim, I.D.; Spadoti, L.M.; Alves, A.T.S. Unraveling the Real Potential of Liquid Whey as Media Culture and Microencapsulation Material for Lactic Acid Bacteria. Innov. Food Sci. Emerg. Technol. 2025, 100, 103885. [Google Scholar] [CrossRef]
- Guan, M.; Che, P.; Wu, M.; Liu, X.; Qian, S.; Xiao, F. Performance and economical evaluation of rotary peeling veneer from logs fermented by lactic acid bacteria: A low-cost, green log pretreatment method. Ind. Crops Prod. 2025, 227, 120841. [Google Scholar] [CrossRef]
- Yamamoto, N.; Shoji, M.; Hoshigami, H.; Watanabe, K.; Watanabe, K.; Takatsuzu, T.; Yasuda, S.; Igoshi, K.; Kinoshita, H. Antioxidant capacity of soymilk yogurt and exopolysaccharides produced by lactic acid bacteria. Biosci. Microbiota Food Health 2019, 38, 97–104. [Google Scholar] [CrossRef]
- Ayyash, M.; Abu-Jdayil, B.; Olaimat, A.; Esposito, G.; Itsaranuwat, P.; Osaili, T.; Obaid, R.; Kizhakkayil, J.; Liu, S. Physicochemical, bioactive and rheological properties of an exopolysaccharide produced by a probiotic Pediococcus pentosaceus M41. Carbohydr. Polym. 2020, 229, 115462. [Google Scholar] [CrossRef]
- Łepecka, A.; Szymański, P.; Okoń, A.; Zielińska, D. Antioxidant activity of environmental lactic acid bacteria strains isolated from organic raw fermented meat products. LWT 2023, 174, 114440. [Google Scholar] [CrossRef]
- Ranjan, A. The Use of Probiotics, Prebiotics, and Synbiotics as an Alternative to Antibiotics. In Alternatives to Antibiotics: Recent Trends and Future Prospects; Springer Nature: Singapore, 2022; pp. 449–465. [Google Scholar]
- Sarita, B.; Samadhan, D.; Hassan, M.Z.; Kovaleva, E.G. A comprehensive review of probiotics and human health-current prospective and applications. Front. Microbiol. 2025, 15, 1487641. [Google Scholar] [CrossRef]
- Sachdeva, A.; Tomar, T.; Malik, T.; Bains, A.; Karnwal, A. Exploring probiotics as a sustainable alternative to antimicrobial growth promoters: Mechanisms and benefits in animal health. Front. Sustain. Food Syst. 2025, 8, 1523678. [Google Scholar] [CrossRef]
- Tian, Q.; Ye, H.; Zhou, X.; Wang, J.; Zhang, L.; Sun, W.; Duan, C.; Fan, M.; Zhou, W.; Bi, C.; et al. Evaluating the health risk of probiotic supplements from the perspective of antimicrobial resistance. Microbiol. Spectr. 2025, 13, e0001924. [Google Scholar] [CrossRef] [PubMed]
- Spacova, I.; Binda, S.; Ter Haar, J.A.; Henoud, S.; Legrain-Raspaud, S.; Dekker, J.; Espadaler-Mazo, J.; Langella, P.; Martín, R.; Pane, M. Comparing technology and regulatory landscape of probiotics as food, dietary supplements and live biotherapeutics. Front. Microbiol. 2023, 14, 1272754. [Google Scholar] [CrossRef]
- Homayouni-Rad, A.; Pouragha, B.; Houshyar, J.; Soleimani, R.A.; Kazemi, S.; Keisan, S.; Akhlaghi, A. Postbiotic application: A review on extraction, purification, and characterization methods. Food Bioprocess Tech. 2025, 18, 4153–4174. [Google Scholar] [CrossRef]
- Moradi, M.; Mardani, K.; Tajik, H. Characterization and application of postbiotics of Lactobacillus spp. on Listeria monocytogenes in vitro and in food models. LWT 2019, 111, 457–464. [Google Scholar] [CrossRef]
- Barzegar, H.; Behbahani, B.A.; Taki, M. Development of active packaging coating with Lallemantia iberica mucilage and Lacticaseibacillus rhamnosus SMHA30 postbiotics: Preparation, characterization, application in the preservation of beef slices, and modelling. J. Agr. Food Res. 2025, 21, 101964. [Google Scholar] [CrossRef]
- Jalali, S.; Mojgani, N.; Haghighat, S.; Sanjabi, M.R.; Sarem-Nezhad, S. Investigation of antimicrobial and antioxidant properties of postbiotics produced by Lactobacillus rhamnosus and Limosilactobacillus reuteri and their potential application in surface decontamination of red meat. LWT 2024, 209, 116758. [Google Scholar] [CrossRef]
- Chakravarty, K.; Gaur, S.; Kumar, R.; Jha, N.K.; Gupta, P.K. Exploring the multifaceted therapeutic potential of probiotics: A review of current insights and applications. Probiotics Antimicrob. Proteins 2024, 17, 341–363. [Google Scholar] [CrossRef]
- Gurunathan, S.; Thangaraj, P.; Kim, J.-H. Postbiotics: Functional Food Materials and Therapeutic Agents for Cancer, Diabetes, and Inflammatory Diseases. Foods 2024, 13, 89. [Google Scholar] [CrossRef]
- Viazis, N.; Argyriou, K.; Kotzampassi, K.; Christodoulou, D.K.; Apostolopoulos, P.; Georgopoulos, S.D.; Liatsos, C.; Giouleme, O.; Koustenis, K.; Veretanos, C.; et al. A Four-Probiotics Regimen Combined with A Standard Helicobacter pylori-Eradication Treatment Reduces Side Effects and Increases Eradication Rates. Nutrients 2022, 14, 632. [Google Scholar] [CrossRef]
| LAB Strains | Codes | Accession Number |
|---|---|---|
| Enterococcus faecium | MI120 | PV400812 |
| Pediococcus pentosaceus | MI121 | PV400813 |
| MI122 | PV400814 | |
| MI123 | PV400815 | |
| MI124 | PV400816 | |
| MI125 | PV400817 | |
| Pediococcus acidilactici | MI126 | PV400818 |
| MI127 | PV400819 | |
| MI128 | PV400820 | |
| MI129 | PV400821 | |
| MI130 | PV400822 |
| LAB Strains | Codes | Auto-Aggregation % | Hydrophobicity (H) (%) | |
|---|---|---|---|---|
| Hexane | Xylene | |||
| E. faecium | MI120 | 39.49 ± 1.72 a | 15.39 ± 1.12 f | 14.54 ± 2.15 b |
| P. pentosaceus | MI121 | 70.53 ± 0.93 d | 24.11 ± 1.68 g | 9.76 ± 0.96 b |
| MI122 | 58.19 ± 5.24 b | 7.41 ± 1.22 bc | 31.84 ± 1.69 d | |
| MI123 | 63.25 ± 0.62 c | 22.19 ± 0.46 g | 21.05 ± 1.04 c | |
| MI124 | 81.88 ± 1.01 d | 11.23 ± 0.82 e | 22.55 ± 4.02 c | |
| MI125 | 69.00 ± 1.13 cd | 3.98 ± 1.25 a | 22.94 ± 0.69 c | |
| P. acidilactici | MI126 | 78.52 ± 0.70 d | 2.99 ± 0.47 a | 25.04 ± 2.22 c |
| MI127 | 80.61 ± 1.51 d | 5.01 ± 0.91 ab | 55.04 ± 0.97 e | |
| MI128 | 78.50 ± 0.58 | 8.07 ± 0.60 cd | 10.05 ± 0.97 b | |
| MI129 | 41.77 ± 2.77 a | 17.54 ± 0.88 f | 31.17 ± 1.39 d | |
| MI130 | 54.16 ± 0.74 b | 10.33 ± 0.84 de | 3.82 ± 1.12 a | |
| LAB Strains | Codes | Enzymatic Activities | |||||
|---|---|---|---|---|---|---|---|
| Amylases | Cellulases | Catalase | Gelatinase | Phytase | Proteases | ||
| E. faecium | MI120 | − | − | − | − | + | + |
| P. pentosaceus | MI121 | − | − | − | − | + | + |
| MI122 | − | − | − | − | + | + | |
| MI123 | − | − | − | − | + | + | |
| MI124 | − | − | − | − | + | + | |
| MI125 | − | − | − | − | + | + | |
| P. acidilactici | MI126 | − | − | − | − | + | + |
| MI127 | − | − | − | − | + | + | |
| MI128 | − | − | − | − | + | + | |
| MI129 | − | − | − | − | + | + | |
| MI130 | − | − | − | − | + | + | |
| Enzymes | E. faecium | P. pentosaceus Strains | P. acidilactici Strains | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MI120 | MI121 | MI122 | MI123 | MI124 | MI125 | MI126 | MI127 | MI128 | MI129 | MI130 | |
| Alkaline phosphatase | + | − | − | − | − | − | − | − | − | − | − |
| Esterase (C4) | ++ | + | + | + | + | + | + | + | + | − | + |
| Esterase lipase (C8) | ++ | + | + | + | + | + | + | + | + | − | + |
| Lipase (C14) | + | + | + | + | + | + | + | + | + | − | + |
| Leucine arylamidase | ++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Valine arylamidase | − | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Cystine arylamidase | − | + | + | − | + | − | + | + | + | + | − |
| Trypsin | − | − | − | − | − | − | − | + | + | + | − |
| α-chymotrypsin | − | − | − | − | + | − | − | − | − | − | − |
| Acid phosphatase | +++ | + | + | + | + | − | + | + | + | + | − |
| Naftol-AS-BI-phosphohydrolase | + | + | ++ | ++ | ++ | +++ | + | ++ | ++ | ++ | + |
| α-galactosidase | − | − | − | + | − | − | − | − | − | − | − |
| β-galactosidase | − | +++ | +++ | +++ | +++ | +++ | + | + | − | − | ++ |
| β-glucuronidase | − | − | − | − | + | − | − | − | − | − | |
| α-glucosidase | − | − | − | − | − | − | − | − | − | − | − |
| β-glucosidase | − | +++ | +++ | +++ | +++ | +++ | + | + | − | − | +++ |
| N-acetyl-β-glucosaminidase | − | +++ | +++ | +++ | +++ | +++ | ++ | − | − | − | +++ |
| α-mannosidase | − | − | − | − | + | − | − | ++ | + | − | − |
| LAB Strains | Codes | Antibiotics Susceptibility | Hemolytic Activity | ||||||
|---|---|---|---|---|---|---|---|---|---|
| TE30 | C30 | CT10 | VA30 | CXM30 | CN30 | P10 | |||
| E. faecium | MI120 | S | S | R | S | R | R | R | γ |
| P. pentosaceus | MI121 | I | S | R | R | R | R | R | γ |
| MI122 | I | S | R | R | R | R | R | γ | |
| MI123 | I | S | R | R | R | R | R | γ | |
| MI124 | I | S | R | R | R | R | R | γ | |
| MI125 | I | S | R | R | R | R | R | γ | |
| P. acidilactici | MI126 | S | S | R | R | R | R | R | γ |
| MI127 | S | S | R | R | R | R | R | γ | |
| MI128 | S | S | R | R | R | R | R | γ | |
| MI129 | S | S | R | R | R | R | R | γ | |
| MI130 | S | S | R | R | R | R | R | γ | |
| Pathogenic Bacteria | E. faecium | Pediococcus pentosaceus Strains | Pediococcus acidilactici Strains | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MI120 | MI121 | MI122 | MI123 | MI124 | MI125 | MI126 | MI127 | MI128 | MI129 | MI130 | |
| B. cereus ATCC 11778 | + | ++ | ++ | ++ | +++ | ++ | ++ | ++ | ++ | ++ | ++ |
| E. coli ATCC 8739 | + | ++ | ++ | ++ | +++ | ++ | ++ | +++ | +++ | +++ | ++ |
| K. kristianae MI 20 | + | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| L. monocytogenes ATCC 13932 | ++ | ++ | ++ | ++ | ++ | +++ | ++ | ++ | ++ | ++ | ++ |
| P. aeruginosa ATCC 15442 | + | + | ++ | ++ | ++ | ++ | + | ++ | + | + | + |
| R. equi ATCC 6939 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | + | + | ++ |
| P. vulgaris ATCC 13315 | ++ | +++ | +++ | ++ | ++ | +++ | +++ | +++ | +++ | ++ | ++ |
| S. Typhimurium ATCC 14028 | + | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| S. Enteretidis ATCC 13076 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| S. marcescens ATCC 14756 | + | + | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | + |
| S. aureus ATCC 33592 | ++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| S. aureus ATCC 6538 | ++ | +++ | +++ | ++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ |
| S. epidermidis ATCC 551625 | ++ | ++ | +++ | +++ | +++ | ++ | ++ | +++ | ++ | +++ | ++ |
| S. epidermidis ATCC 12228 | ++ | ++ | +++ | +++ | +++ | +++ | ++ | +++ | ++ | +++ | ++ |
| Pathogenic Bacteria | E. faecium | P. pentosaceus Strains | P. acidilactici Strains | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MI120 | MI121 | MI122 | MI123 | MI124 | MI125 | MI126 | MI127 | MI128 | MI129 | MI130 | |
| B. cereus ATCC 11778 | 100.00 ± 0.00 | 50.00 ± 0.00 | 50.00 ± 0.00 | 25.00 ± 0.00 | 25.00 ± 0.00 | 25.00 ± 0.00 | 50.00 ± 0.00 | 50.00 ± 0.00 | 25.00 ± 0.00 | 25.00 ± 0.00 | 25.00 ± 0.00 |
| E. coli ATCC 8739 | 100.00 ± 0.00 | 50.00 ± 0.00 | 50.00 ± 0.00 | 25.00 ± 0.00 | 25.00 ± 0.00 | 25.00 ± 0.00 | 50.00 ± 0.00 | 50.00 ± 0.00 | 25.00 ± 0.00 | 25.00 ± 0.00 | 25.00 ± 0.00 |
| Kocuria cristianae MI20 | 3.13 ± 0.00 | 3.13 ± 0.00 | 100 ± 0.00 | 50.00 ± 0.00 | 50.00 ± 0.00 | 50.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 50.00 ± 0.00 | 50.00 ± 0.00 | 100.00 ± 0.00 |
| L. monocytogenes ATCC 13932 | 12.50 ± 0.00 | 50.00 ± 0.00 | 25.00 ± 0.00 | 50.00 ± 0.00 | 25.00 ± 0.00 | 50.00 ± 0.00 | 50.00 ± 0.00 | 50.00 ± 0.00 | 50.00 ± 0.00 | 50.00 ± 0.00 | 50.00 ± 0.00 |
| P. aeruginosa ATCC 15442 | 50.00 ± 0.00 | 12.5 ± 0.00 | 12.50 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 | 12.50 ± 0.00 |
| R. equi ATCC 6939 | 50.00 ± 0.00 | 50.00 ± 0.00 | 25.00 ± 0.00 | 12.50 ± 0.00 | 25.00 ± 0.00 | 25.00 ± 0.00 | 50.00 ± 0.00 | 25.00 ± 0.00 | 12.50 ± 0.00 | 25.00 ± 0.00 | 25.00 ± 0.00 |
| P. vulgaris ATCC 13315 | 25.00 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 | 25.00 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 | 3.13 ± 0.00 | 3.13 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 |
| S. Typhimurium ATCC 14028 | 100.00 ± 0.00 | 25.00 ± 0.00 | 50.00 ± 0.00 | 12.50 ± 0.00 | 25.00 ± 0.00 | 12.50 ± 0.00 | 25.00 ± 0.00 | 12.50 ± 0.00 | 25.00 ± 0.00 | 25.00 ± 0.00 | 25.00 ± 0.00 |
| S. Enteritidis ATCC 13076 | 50.00 ± 0.00 | 12.50 ± 0.00 | 12.50 ± 0.00 | 12.50 ± 0.00 | 12.50 ± 0.00 | 12.50 ± 0.00 | 12.50 ± 0.00 | 12.50 ± 0.00 | 6.25 ± 0.00 | 12.50 ± 0.00 | 12.50 ± 0.00 |
| S. marcescens ATCC 14756 | 50.00 ± 0.00 | 25.00 ± 0.00 | 25.00 ± 0.00 | 12.50 ± 0.00 | 12.50 ± 0.00 | 12.5 ± 0.00 | 12.5 ± 0.00 | 12.5 ± 0.00 | 12.50 ± 0.00 | 12.50 ± 0.00 | 12.50 ± 0.00 |
| S. aureus ATCC 33592 | 50.00 ± 0.00 | 50.00 ± 0.00 | 12.50 ± 0.00 | 12.50 ± 0.00 | 25.00 ± 0.00 | 12.50 ± 0.00 | 12.50 ± 0.00 | 12.50 ± 0.00 | 12.50 ± 0.00 | 25.00 ± 0.00 | 25.00 ± 0.00 |
| S. aureus ATCC 6538 | 25.00 ± 0.00 | 12.50 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 | 12.50 ± 0.00 | 12.50 ± 0.00 | 12.50 ± 0.00 | 6.25 ± 0.00 | 12.50 ± 0.00 | 12.50 ± 0.00 | 12.50 ± 0.00 |
| S. epidermidis ATCC 51625 | 50.00 ± 0.00 | 12.50 ± 0.00 | 12.50 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 | 12.5 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 |
| S. epidermidis ATCC 12228 | 50.00 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 | 6.25 ± 0.00 | 3.13 ± 0.00 |
| LAB Strains | CFS Samples | TPC (μg·mL−1) | TFC (μg·mL−1) |
|---|---|---|---|
| E. faecium | MI120 | 159.99 ± 1.35 ab | 21.18 ± 0.72 a |
| P. pentosaceus | MI121 | 162.18 ± 2.05 a | 21.04 ± 0.44 a |
| MI122 | 158.12 ± 2.01 abc | 20.56 ± 0.22 a | |
| MI123 | 157.11 ± 1.65 ad | 17.57 ± 0.17 b | |
| MI124 | 151.95 ± 1.57 df | 20.93 ± 0.23 a | |
| MI125 | 153.19 ± 1.89 cde | 21.69 ± 0.72 a | |
| P. acidilactici | MI126 | 152.73 ± 1.85 df | 20.28 ± 0.21 a |
| MI127 | 151.77 ± 2.11 ef | 21.52 ± 0.75 a | |
| MI128 | 156.79 ± 2.22 bde | 21.14 ± 0.69 a | |
| MI129 | 155.42 ± 2.02 bde | 21.79 ± 0.81 a | |
| MI130 | 156.34 ± 1.68 bde | 20.87 ± 0.18 a |
| LAB Strains | CFS Samples | Antioxidant Activity by DPPH | ||
|---|---|---|---|---|
| Inhibition Percent (%) | IC50 (df) | TE (μg·mL−1) | ||
| E. faecium | MI120 | 75.19 ± 0.37 a | 1/12 | 21.37 ± 0.10 a |
| P. pentosaceus | MI121 | 70.99 ± 0.21 bc | 1/8 | 20.23 ± 0.06 bc |
| MI122 | 69.75 ± 0.21 bc | 1/7 | 19.89 ± 0.06 cd | |
| MI123 | 70.12 ± 0.21 bc | 1/7 | 19.99 ± 0.06 bd | |
| MI124 | 66.05 ± 2.35 d | 1/4 | 18.88 ± 0.46 e | |
| MI125 | 70.62 ± 0.43 bc | 1/9 | 20.12 ± 0.12 bd | |
| P. acidilactici | MI126 | 68.64 ± 1.71 cd | 1/9 | 19.58 ± 0.47 d |
| MI127 | 70.25 ± 0.43 bc | 1/9 | 20.02 ± 0.12 bd | |
| MI128 | 68.77 ± 0.21 cd | 1/9 | 19.62 ± 0.06 cd | |
| MI129 | 70.99 ± 0.27 bc | 1/11 | 20.23 ± 0.07 bc | |
| MI130 | 72.10 ± 0.57 b | 1/11 | 20.53 ± 0.15 b | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pristavu, M.-C.; Diguță, F.C.; Aldea, A.C.; Badea, F.; Dragoi Cudalbeanu, M.; Ortan, A.; Matei, F. Functional Profiling of Enterococcus and Pediococcus Strains: An In Vitro Study on Probiotic and Postbiotic Properties. Microorganisms 2025, 13, 1348. https://doi.org/10.3390/microorganisms13061348
Pristavu M-C, Diguță FC, Aldea AC, Badea F, Dragoi Cudalbeanu M, Ortan A, Matei F. Functional Profiling of Enterococcus and Pediococcus Strains: An In Vitro Study on Probiotic and Postbiotic Properties. Microorganisms. 2025; 13(6):1348. https://doi.org/10.3390/microorganisms13061348
Chicago/Turabian StylePristavu, Mircea-Cosmin, Filofteia Camelia Diguță, Alexandru Constantin Aldea, Florentina Badea, Mihaela Dragoi Cudalbeanu, Alina Ortan, and Florentina Matei. 2025. "Functional Profiling of Enterococcus and Pediococcus Strains: An In Vitro Study on Probiotic and Postbiotic Properties" Microorganisms 13, no. 6: 1348. https://doi.org/10.3390/microorganisms13061348
APA StylePristavu, M.-C., Diguță, F. C., Aldea, A. C., Badea, F., Dragoi Cudalbeanu, M., Ortan, A., & Matei, F. (2025). Functional Profiling of Enterococcus and Pediococcus Strains: An In Vitro Study on Probiotic and Postbiotic Properties. Microorganisms, 13(6), 1348. https://doi.org/10.3390/microorganisms13061348






