Shotgun Metagenomic Sequencing Analysis as a Diagnostic Strategy for Patients with Lower Respiratory Tract Infections
Abstract
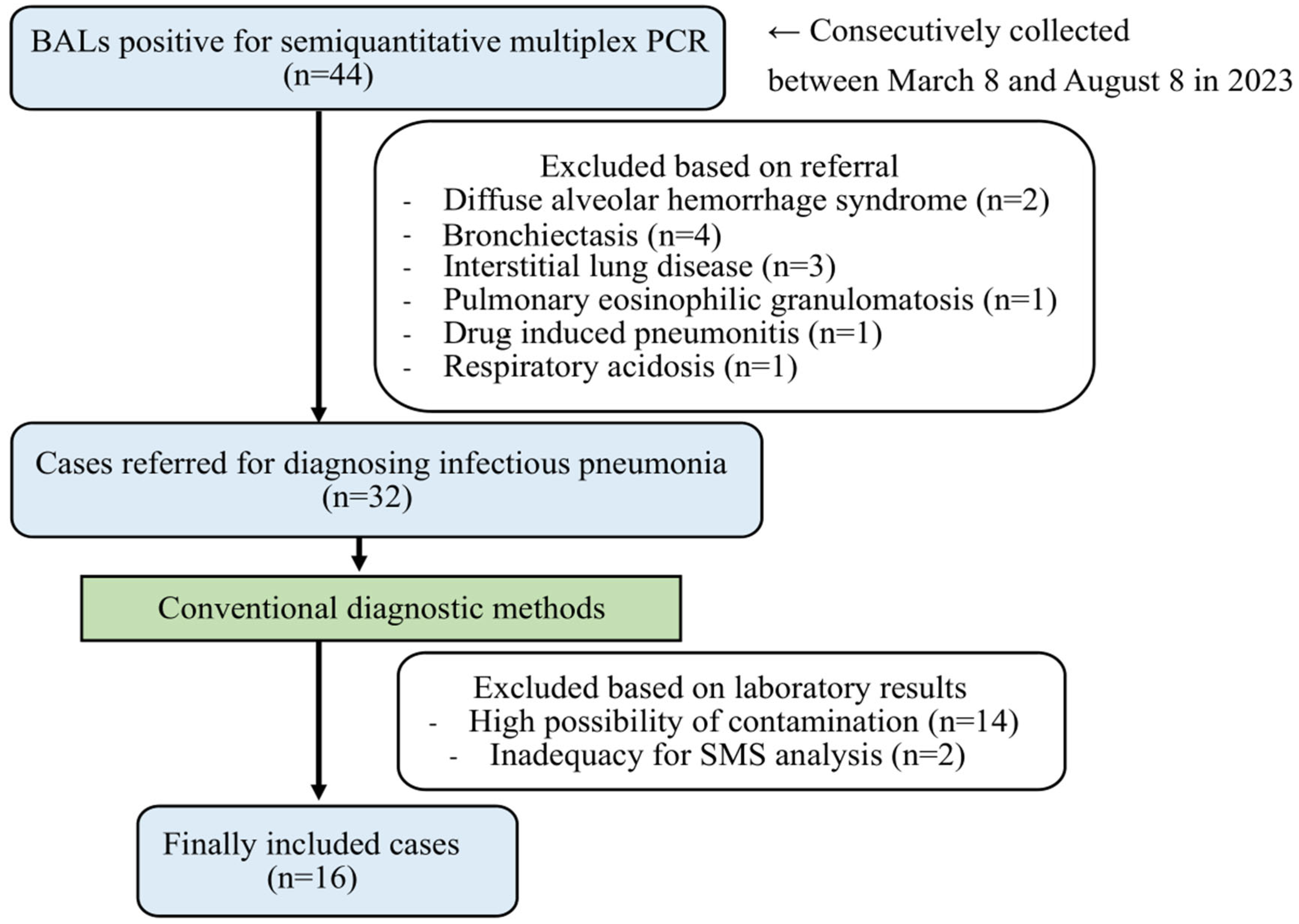
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Processing
- (1)
- Specimens with a high presence of oropharyngeal normal flora, such as Streptococcus mitis, Streptococcus australis, Streptococcus parasanguinis, Streptococcus sanguinis, Streptococcus clone, Streotpcoccus gordonii, Streptococcus intermedius, Gemella haemolysans, Gemella sanguinis, Granulicatella adiacens, Granulicatella elegans, Abiotrophia defective, and Rothia dentocariosa, were excluded. These microorganisms were identified and presumed to be contaminants during the BAL procedure [30,31,32].
- (2)
- Specimens with the identification of cutaneous normal flora, such as Cutibacteriium acnes, Cutibacterium granulosum, Corynebacterium striatum, Staphylococcus epidermidis, Staphylococcus hominis, Staphylococcus haemolyticus, Staphylococcus captis, Staphylococcus warneri, Staphylococcus saprophyticus, Staphylococcus cohnnii, Staphylococcus xylosus, Staphylococcus simulans, Micrococcus luteus, and Micrococcus varians, were excluded. These microorganisms were identified and presumed to pose a risk of contamination during specimen collection and processing [31,33,34,35,36].
- (3)
2.2. Extraction of Nucleic Acids and Sequencing
2.3. SMS Procedures
3. Results
3.1. Identification of Microbes by SMS
3.2. Metagenomic Results of Antibiotic Resistance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Amati, F.; Bindo, F.; Stainer, A.; Gramegna, A.; Mantero, M.; Nigro, M.; Bussini, L.; Bartoletti, M.; Blasi, F.; Aliberti, S. Identify Drug-Resistant Pathogens in Patients with Community-Acquired Pneumonia. Adv. Respir. Med. 2023, 91, 224–238. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.Y.; Chen, Y.J.; Er, T.K. Retrospective Study of Lower Respiratory Tract Infections in the Intensive Care Unit Detected by the FilmArray Pneumonia Panel. Clin. Lab. 2023, 69. Available online: https://pubmed.ncbi.nlm.nih.gov/37436371/ (accessed on 6 June 2025). [CrossRef] [PubMed]
- Jain, S.; Self, W.H.; Wunderink, R.G.; Fakhran, S.; Balk, R.; Bramley, A.M.; Reed, C.; Grijalva, C.G.; Anderson, E.J.; Courtney, D.M.; et al. Community-Acquired Pneumonia Requiring Hospitalization among U.S. Adults. N. Engl. J. Med. 2015, 373, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Tarsia, P.; Aliberti, S.; Pappalettera, M.; Blasi, F. Mixed community-acquired lower respiratory tract infections. Curr. Infect. Dis. Rep. 2007, 9, 14–20. [Google Scholar] [CrossRef]
- Cilloniz, C.; Ewig, S.; Ferrer, M.; Polverino, E.; Gabarrus, A.; Puig de la Bellacasa, J.; Mensa, J.; Torres, A. Community-acquired polymicrobial pneumonia in the intensive care unit: Aetiology and prognosis. Crit. Care 2011, 15, R209. [Google Scholar] [CrossRef]
- de Roux, A.; Ewig, S.; Garcia, E.; Marcos, M.A.; Mensa, J.; Lode, H.; Torres, A. Mixed community-acquired pneumonia in hospitalised patients. Eur. Respir. J. 2006, 27, 795–800. [Google Scholar] [CrossRef]
- Gutierrez, F.; Masia, M.; Rodriguez, J.C.; Mirete, C.; Soldan, B.; Padilla, S.; Hernandez, I.; Royo, G.; Martin-Hidalgo, A. Community-acquired pneumonia of mixed etiology: Prevalence, clinical characteristics, and outcome. Eur. J. Clin. Microbiol. Infect. Dis. 2005, 24, 377–383. [Google Scholar] [CrossRef]
- Holter, J.C.; Muller, F.; Bjorang, O.; Samdal, H.H.; Marthinsen, J.B.; Jenum, P.A.; Ueland, T.; Froland, S.S.; Aukrust, P.; Husebye, E.; et al. Etiology of community-acquired pneumonia and diagnostic yields of microbiological methods: A 3-year prospective study in Norway. BMC Infect. Dis. 2015, 15, 64. [Google Scholar] [CrossRef]
- Lieberman, D.; Schlaeffer, F.; Boldur, I.; Lieberman, D.; Horowitz, S.; Friedman, M.G.; Leiononen, M.; Horovitz, O.; Manor, E.; Porath, A. Multiple pathogens in adult patients admitted with community-acquired pneumonia: A one year prospective study of 346 consecutive patients. Thorax 1996, 51, 179–184. [Google Scholar] [CrossRef]
- GBD 2016 Lower Respiratory Infections Collaborators. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990-2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 2018, 18, 1191–1210. [Google Scholar] [CrossRef]
- Carroll, K.C.; Adams, L.L. Lower Respiratory Tract Infections. Microbiol. Spectr. 2016, 4. Available online: https://pubmed.ncbi.nlm.nih.gov/27726814/ (accessed on 6 June 2025). [CrossRef] [PubMed]
- Doern, G.V. Detection of selected fastidious bacteria. Clin. Infect. Dis. 2000, 30, 166–173. [Google Scholar] [CrossRef]
- Kelly, B.T.; Pennington, K.M.; Limper, A.H. Advances in the diagnosis of fungal pneumonias. Expert Rev. Respir. Med. 2020, 14, 703–714. [Google Scholar] [CrossRef]
- Schmitz, J.E.; Stratton, C.W.; Persing, D.H.; Tang, Y.W. Forty Years of Molecular Diagnostics for Infectious Diseases. J. Clin. Microbiol. 2022, 60, e0244621. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Ruan, S.Y.; Pan, S.C.; Lee, T.F.; Chien, J.Y.; Hsueh, P.R. Performance of a multiplex PCR pneumonia panel for the identification of respiratory pathogens and the main determinants of resistance from the lower respiratory tract specimens of adult patients in intensive care units. J. Microbiol. Immunol. Infect. 2019, 52, 920–928. [Google Scholar] [CrossRef]
- Schlaberg, R.; Chiu, C.Y.; Miller, S.; Procop, G.W.; Weinstock, G.; Professional Practice Committee and Committee on Laboratory Practices of the American Society for Microbiology; Microbiology Resource Committee of the College of American Pathologists. Validation of Metagenomic Next-Generation Sequencing Tests for Universal Pathogen Detection. Arch. Pathol. Lab. Med. 2017, 141, 776–786. [Google Scholar] [CrossRef]
- Chen, Y.; Feng, W.; Ye, K.; Guo, L.; Xia, H.; Guan, Y.; Chai, L.; Shi, W.; Zhai, C.; Wang, J.; et al. Application of Metagenomic Next-Generation Sequencing in the Diagnosis of Pulmonary Infectious Pathogens From Bronchoalveolar Lavage Samples. Front. Cell. Infect. Microbiol. 2021, 11, 541092. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Jiang, E.; Yang, D.; Wei, J.; Zhao, M.; Feng, J.; Cao, J. Metagenomic Next-Generation Sequencing versus Traditional Pathogen Detection in the Diagnosis of Peripheral Pulmonary Infectious Lesions. Infect. Drug Resist. 2020, 13, 567–576. [Google Scholar] [CrossRef]
- Wang, J.; Han, Y.; Feng, J. Metagenomic next-generation sequencing for mixed pulmonary infection diagnosis. BMC Pulm. Med. 2019, 19, 252. [Google Scholar] [CrossRef]
- Zhang, X.; Qin, Y.; Lei, W.; Huang, J.A. Metagenomic next-generation sequencing of BALF for the clinical diagnosis of severe community-acquired pneumonia in immunocompromised patients: A single-center study. Exp. Ther. Med. 2023, 25, 178. [Google Scholar] [CrossRef]
- Saladie, M.; Caparros-Martin, J.A.; Agudelo-Romero, P.; Wark, P.A.B.; Stick, S.M.; O’Gara, F. Microbiomic Analysis on Low Abundant Respiratory Biomass Samples; Improved Recovery of Microbial DNA From Bronchoalveolar Lavage Fluid. Front. Microbiol. 2020, 11, 572504. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhou, C.E.; Wei, S.C.; Wang, L.N.; Shi, M.W.; Sun, C.P.; Lin, L.J.; Liu, X.M. Diagnostic Value of Metagenomic Next-Generation Sequencing for Pneumonia in Immunocompromised Patients. Can. J. Infect. Dis. Med. Microbiol. 2022, 2022, 5884568. [Google Scholar] [CrossRef]
- Lin, P.; Chen, Y.; Su, S.; Nan, W.; Zhou, L.; Zhou, Y.; Li, Y. Diagnostic value of metagenomic next-generation sequencing of bronchoalveolar lavage fluid for the diagnosis of suspected pneumonia in immunocompromised patients. BMC Infect. Dis. 2022, 22, 416. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Tu, X.; Zhao, J.; Huang, L.; Dai, X.; Chen, X.; Xu, Y.; Li, W.; Wang, Y.; Lou, J.; et al. Microbiological diagnostic performance of metagenomic next-generation sequencing compared with conventional culture for patients with community-acquired pneumonia. Front. Cell. Infect. Microbiol. 2023, 13, 1136588. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, Y.; Chen, G.; Sun, S.; Wang, J.; Chen, F.; Liu, C.; Zhuang, Q. Diagnostic Significance of Metagenomic Next-Generation Sequencing for Community-Acquired Pneumonia in Southern China. Front. Med. 2022, 9, 807174. [Google Scholar] [CrossRef]
- Sun, T.; Wu, X.; Cai, Y.; Zhai, T.; Huang, L.; Zhang, Y.; Zhan, Q. Metagenomic Next-Generation Sequencing for Pathogenic Diagnosis and Antibiotic Management of Severe Community-Acquired Pneumonia in Immunocompromised Adults. Front. Cell. Infect. Microbiol. 2021, 11, 661589. [Google Scholar] [CrossRef]
- Leber, A.L. Clinical Microbiology Procedures Handbook; American Society for Microbiology Press: New York, NY, USA, 2023. [Google Scholar]
- Schulthess, B.; Bloemberg, G.V.; Zbinden, A.; Mouttet, F.; Zbinden, R.; Bottger, E.C.; Hombach, M. Evaluation of the Bruker MALDI Biotyper for Identification of Fastidious Gram-Negative Rods. J. Clin. Microbiol. 2016, 54, 543–548. [Google Scholar] [CrossRef]
- Schulthess, B.; Bloemberg, G.V.; Zbinden, R.; Bottger, E.C.; Hombach, M. Evaluation of the Bruker MALDI Biotyper for identification of Gram-positive rods: Development of a diagnostic algorithm for the clinical laboratory. J. Clin. Microbiol. 2014, 52, 1089–1097. [Google Scholar] [CrossRef]
- Aas, J.A.; Paster, B.J.; Stokes, L.N.; Olsen, I.; Dewhirst, F.E. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 2005, 43, 5721–5732. [Google Scholar] [CrossRef]
- Davis, C.P. Normal Flora. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996. Available online: https://pubmed.ncbi.nlm.nih.gov/21413249/ (accessed on 6 June 2025).
- Pang, J.A.; Cheng, A.F.; Chan, H.S.; French, G.L. Special precautions reduce oropharyngeal contamination in bronchoalveolar lavage for bacteriologic studies. Lung 1989, 167, 261–267. [Google Scholar] [CrossRef]
- Chiller, K.; Selkin, B.A.; Murakawa, G.J. Skin microflora and bacterial infections of the skin. J. Investig. Dermatol. Symp. Proc. 2001, 6, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Mushtaq, A.; Chen, D.J.; Strand, G.J.; Dylla, B.L.; Cole, N.C.; Mandrekar, J.; Patel, R. Clinical significance of coryneform Gram-positive rods from blood identified by MALDI-TOF mass spectrometry and their susceptibility profiles—A retrospective chart review. Diagn. Microbiol. Infect. Dis. 2016, 85, 372–376. [Google Scholar] [CrossRef]
- Noble, W.C. Skin flora of the normal and immune compromised host. Curr. Probl. Dermatol. 1989, 18, 37–41. [Google Scholar] [CrossRef]
- Roth, R.R.; James, W.D. Microbiology of the skin: Resident flora, ecology, infection. J. Am. Acad. Dermatol. 1989, 20, 367–390. [Google Scholar] [CrossRef]
- Lambotte, O.; Timsit, J.F.; Garrouste-Orgeas, M.; Misset, B.; Benali, A.; Carlet, J. The significance of distal bronchial samples with commensals in ventilator-associated pneumonia: Colonizer or pathogen? Chest 2002, 122, 1389–1399. [Google Scholar] [CrossRef][Green Version]
- Rasmussen, T.R.; Korsgaard, J.; Moller, J.K.; Sommer, T.; Kilian, M. Quantitative culture of bronchoalveolar lavage fluid in community-acquired lower respiratory tract infections. Respir. Med. 2001, 95, 885–890. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mohsen, A.H.; McKendrick, M. Varicella pneumonia in adults. Eur. Respir. J. 2003, 21, 886–891. [Google Scholar] [CrossRef]
- Simoons-Smit, A.M.; Kraan, E.M.; Beishuizen, A.; Strack van Schijndel, R.J.; Vandenbroucke-Grauls, C.M. Herpes simplex virus type 1 and respiratory disease in critically-ill patients: Real pathogen or innocent bystander? Clin. Microbiol. Infect. 2006, 12, 1050–1059. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Steinegger, M.; Breitwieser, F.; Soding, J.; Levy Karin, E. Fast and sensitive taxonomic assignment to metagenomic contigs. Bioinformatics 2021, 37, 3029–3031. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef]
- Wang, H.; Lu, Z.; Bao, Y.; Yang, Y.; de Groot, R.; Dai, W.; de Jonge, M.I.; Zheng, Y. Clinical diagnostic application of metagenomic next-generation sequencing in children with severe nonresponding pneumonia. PLoS ONE 2020, 15, e0232610. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef]
- Arango-Argoty, G.; Garner, E.; Pruden, A.; Heath, L.S.; Vikesland, P.; Zhang, L. DeepARG: A deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome 2018, 6, 23. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Simner, P.J.; Miller, S.; Carroll, K.C. Understanding the Promises and Hurdles of Metagenomic Next-Generation Sequencing as a Diagnostic Tool for Infectious Diseases. Clin. Infect. Dis. 2018, 66, 778–788. [Google Scholar] [CrossRef]
- Shi, C.L.; Han, P.; Tang, P.J.; Chen, M.M.; Ye, Z.J.; Wu, M.Y.; Shen, J.; Wu, H.Y.; Tan, Z.Q.; Yu, X.; et al. Clinical metagenomic sequencing for diagnosis of pulmonary tuberculosis. J. Infect. 2020, 81, 567–574. [Google Scholar] [CrossRef]
- Zhou, X.; Wu, H.; Ruan, Q.; Jiang, N.; Chen, X.; Shen, Y.; Zhu, Y.M.; Ying, Y.; Qian, Y.Y.; Wang, X.; et al. Clinical Evaluation of Diagnosis Efficacy of Active Mycobacterium tuberculosis Complex Infection via Metagenomic Next-Generation Sequencing of Direct Clinical Samples. Front. Cell. Infect. Microbiol. 2019, 9, 351. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Cao, L.J.; Xia, H.L.; Ji, Z.M.; Hu, N.N.; Leng, Z.J.; Xie, W.; Fang, Y.; Zhang, J.Q.; Xia, D.Q. The performance of detecting Mycobacterium tuberculosis complex in lung biopsy tissue by metagenomic next-generation sequencing. BMC Pulm. Med. 2022, 22, 288. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, H.; Li, Y.; Yu, Z. Clinical application of metagenomic next-generation sequencing in tuberculosis diagnosis. Front. Cell. Infect. Microbiol. 2022, 12, 984753. [Google Scholar] [CrossRef]
- Kok, N.A.; Peker, N.; Schuele, L.; de Beer, J.L.; Rossen, J.W.A.; Sinha, B.; Couto, N. Host DNA depletion can increase the sensitivity of Mycobacterium spp. detection through shotgun metagenomics in sputum. Front. Microbiol. 2022, 13, 949328. [Google Scholar] [CrossRef]
- Kaser, M.; Ruf, M.T.; Hauser, J.; Pluschke, G. Optimized DNA preparation from mycobacteria. Cold Spring Harb. Protoc. 2010, 2010, pdb.prot5408. [Google Scholar] [CrossRef] [PubMed]
- Mun, B.S.; Yoon, J.; Yoon, S.Y. Optimized Method for Mycobacteria DNA Extraction from Sputum for Isothermal Amplification. Ann. Clin. Lab. Sci. 2023, 53, 476–481. [Google Scholar] [PubMed]
- Ratnam, S.; March, S.B. Effect of relative centrifugal force and centrifugation time on sedimentation of mycobacteria in clinical specimens. J. Clin. Microbiol. 1986, 23, 582–585. [Google Scholar] [CrossRef]
- Bouso, J.M.; Planet, P.J. Complete nontuberculous mycobacteria whole genomes using an optimized DNA extraction protocol for long-read sequencing. BMC Genom. 2019, 20, 793. [Google Scholar] [CrossRef]
- Tiew, P.Y.; Mac Aogain, M.; Ali, N.; Thng, K.X.; Goh, K.; Lau, K.J.X.; Chotirmall, S.H. The Mycobiome in Health and Disease: Emerging Concepts, Methodologies and Challenges. Mycopathologia 2020, 185, 207–231. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Mills, D.A. Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl. Environ. Microbiol. 2013, 79, 2519–2526. [Google Scholar] [CrossRef]
- Jiang, J.; Bai, L.; Yang, W.; Peng, W.; An, J.; Wu, Y.; Pan, P.; Li, Y. Metagenomic Next-Generation Sequencing for the Diagnosis of Pneumocystis jirovecii Pneumonia in Non-HIV-Infected Patients: A Retrospective Study. Infect. Dis. Ther. 2021, 10, 1733–1745. [Google Scholar] [CrossRef] [PubMed]
- Limper, A.H.; Offord, K.P.; Smith, T.F.; Martin, W.J., 2nd. Pneumocystis carinii pneumonia. Differences in lung parasite number and inflammation in patients with and without AIDS. Am. Rev. Respir. Dis. 1989, 140, 1204–1209. [Google Scholar] [CrossRef]
- Yang, J.; Yang, F.; Ren, L.; Xiong, Z.; Wu, Z.; Dong, J.; Sun, L.; Zhang, T.; Hu, Y.; Du, J.; et al. Unbiased parallel detection of viral pathogens in clinical samples by use of a metagenomic approach. J. Clin. Microbiol. 2011, 49, 3463–3469. [Google Scholar] [CrossRef]
- Wylie, K.M.; Weinstock, G.M.; Storch, G.A. Emerging view of the human virome. Transl. Res. 2012, 160, 283–290. [Google Scholar] [CrossRef]
- Modi, A.; Vai, S.; Caramelli, D.; Lari, M. The Illumina Sequencing Protocol and the NovaSeq 6000 System. Methods Mol. Biol. 2021, 2242, 15–42. [Google Scholar] [CrossRef] [PubMed]
- Locher, K.; Roscoe, D.; Jassem, A.; Wong, T.; Hoang, L.M.N.; Charles, M.; Bryce, E.; Grant, J.; Stefanovic, A. FilmArray respiratory panel assay: An effective method for detecting viral and atypical bacterial pathogens in bronchoscopy specimens. Diagn. Microbiol. Infect. Dis. 2019, 95, 114880. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Sugimura, N.; Burgermeister, E.; Ebert, M.P.; Zuo, T.; Lan, P. The gut virome: A new microbiome component in health and disease. EBioMedicine 2022, 81, 104113. [Google Scholar] [CrossRef]
- Mirzaei, M.K.; Maurice, C.F. Menage a trois in the human gut: Interactions between host, bacteria and phages. Nat. Rev. Microbiol. 2017, 15, 397–408. [Google Scholar] [CrossRef]
- Moon, K.; Cho, J.C. Metaviromics coupled with phage-host identification to open the viral ‘black box’. J. Microbiol. 2021, 59, 311–323. [Google Scholar] [CrossRef]
- Singh, P.; Pandey, A. Community-acquired lower respiratory tract infection due to Serratia rubidaea: A rare and opportunistic pathogen. MRIMS J. Health Sci. 2025, 13, 35–38. [Google Scholar] [CrossRef]
- Mehdi, A.; Trifi, A.; Abbes, S.; Seghir, E.; Tlili, B.; Masseoud, L.; Noussair, A.; Ouhibi, A.; Battikh, H.; Zribi, M.; et al. Bacteremia due to Serratia rubidaea in intensive care unit: A case series. J. Med. Case Rep. 2023, 17, 482. [Google Scholar] [CrossRef]
- Buchan, B.W.; Windham, S.; Balada-Llasat, J.M.; Leber, A.; Harrington, A.; Relich, R.; Murphy, C.; Dien Bard, J.; Naccache, S.; Ronen, S.; et al. Practical Comparison of the BioFire FilmArray Pneumonia Panel to Routine Diagnostic Methods and Potential Impact on Antimicrobial Stewardship in Adult Hospitalized Patients with Lower Respiratory Tract Infections. J. Clin. Microbiol. 2020, 58. Available online: https://pubmed.ncbi.nlm.nih.gov/32350045/ (accessed on 6 June 2025).
- Ambroa, A.; Blasco, L.; Lopez-Causape, C.; Trastoy, R.; Fernandez-Garcia, L.; Bleriot, I.; Ponce-Alonso, M.; Pacios, O.; Lopez, M.; Canton, R.; et al. Temperate Bacteriophages (Prophages) in Pseudomonas aeruginosa Isolates Belonging to the International Cystic Fibrosis Clone (CC274). Front. Microbiol. 2020, 11, 556706. [Google Scholar] [CrossRef]
- Secor, P.R.; Burgener, E.B.; Kinnersley, M.; Jennings, L.K.; Roman-Cruz, V.; Popescu, M.; Van Belleghem, J.D.; Haddock, N.; Copeland, C.; Michaels, L.A.; et al. Pf Bacteriophage and Their Impact on Pseudomonas Virulence, Mammalian Immunity, and Chronic Infections. Front. Immunol. 2020, 11, 244. [Google Scholar] [CrossRef]
- Schmidt, A.K.; Fitzpatrick, A.D.; Schwartzkopf, C.M.; Faith, D.R.; Jennings, L.K.; Coluccio, A.; Hunt, D.J.; Michaels, L.A.; Hargil, A.; Chen, Q.; et al. A Filamentous Bacteriophage Protein Inhibits Type IV Pili To Prevent Superinfection of Pseudomonas aeruginosa. mBio 2022, 13, e0244121. [Google Scholar] [CrossRef]
- Huang, L.; Weng, B.; Gu, X.; Wang, Y.; Wang, M.; Weng, J.; Ju, Y.; Zhong, X.; Tong, X.; Li, Y. Performance of various pneumonia severity models for predicting adverse outcomes in elderly inpatients with community-acquired pneumonia. Clin. Microbiol. Infect. 2024, 30, 1426–1432. [Google Scholar] [CrossRef] [PubMed]
- Govender, K.N.; Street, T.L.; Sanderson, N.D.; Eyre, D.W. Metagenomic Sequencing as a Pathogen-Agnostic Clinical Diagnostic Tool for Infectious Diseases: A Systematic Review and Meta-analysis of Diagnostic Test Accuracy Studies. J. Clin. Microbiol. 2021, 59, e0291620. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Liu, D.; Han, P.; Wang, H.; Wang, S.; Gao, J.; Chen, F.; Zhou, X.; Deng, K.; Luo, J.; et al. Rapid inference of antibiotic resistance and susceptibility for Klebsiella pneumoniae by clinical shotgun metagenomic sequencing. Int. J. Antimicrob. Agents 2024, 64, 107252. [Google Scholar] [CrossRef]
- Hu, X.; Zhao, Y.; Han, P.; Liu, S.; Liu, W.; Mai, C.; Deng, Q.; Ren, J.; Luo, J.; Chen, F.; et al. Novel Clinical mNGS-Based Machine Learning Model for Rapid Antimicrobial Susceptibility Testing of Acinetobacter baumannii. J. Clin. Microbiol. 2023, 61, e0180522. [Google Scholar] [CrossRef]
- Liu, B.; Gao, J.; Liu, X.F.; Rao, G.; Luo, J.; Han, P.; Hu, W.; Zhang, Z.; Zhao, Q.; Han, L.; et al. Direct prediction of carbapenem resistance in Pseudomonas aeruginosa by whole genome sequencing and metagenomic sequencing. J. Clin. Microbiol. 2023, 61, e0061723. [Google Scholar] [CrossRef]
- Ruppe, E.; Cherkaoui, A.; Lazarevic, V.; Emonet, S.; Schrenzel, J. Establishing Genotype-to-Phenotype Relationships in Bacteria Causing Hospital-Acquired Pneumonia: A Prelude to the Application of Clinical Metagenomics. Antibiotics 2017, 6, 30. [Google Scholar] [CrossRef]
- Caroll, K.C.; Pfaller, M.A.; Landry, M.L. Manual of Clinical Microbiology; American Society for Microbiology Press: New York, NY, USA, 2019; Volume 1. [Google Scholar]
- Ni, J.; Yan, Q.; Yu, Y. How much metagenomic sequencing is enough to achieve a given goal? Sci. Rep. 2013, 3, 1968. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.L.; Konstantinidis, K.T. Estimating coverage in metagenomic data sets and why it matters. ISME J. 2014, 8, 2349–2351. [Google Scholar] [CrossRef]
- Vollmers, J.; Wiegand, S.; Kaster, A.K. Comparing and Evaluating Metagenome Assembly Tools from a Microbiologist’s Perspective—Not Only Size Matters! PLoS ONE 2017, 12, e0169662. [Google Scholar] [CrossRef]
- Liu, D.; Zhou, H.; Xu, T.; Yang, Q.; Mo, X.; Shi, D.; Ai, J.; Zhang, J.; Tao, Y.; Wen, D.; et al. Multicenter assessment of shotgun metagenomics for pathogen detection. EBioMedicine 2021, 74, 103649. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, G.; Lau, H.C.; Yu, J. Metagenomic Sequencing for Microbial DNA in Human Samples: Emerging Technological Advances. Int. J. Mol. Sci. 2022, 23, 2181. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ye, J.; Yang, L.; Chen, X.; Fang, H.; Liu, Z.; Xia, G.; Zhang, Y.; Zhang, Z. Inconsistency analysis between metagenomic next-generation sequencing results of cerebrospinal fluid and clinical diagnosis with suspected central nervous system infection. BMC Infect. Dis. 2022, 22, 764. [Google Scholar] [CrossRef] [PubMed]
- Van Driessche, L.; Bokma, J.; Deprez, P.; Haesebrouck, F.; Boyen, F.; Pardon, B. Rapid identification of respiratory bacterial pathogens from bronchoalveolar lavage fluid in cattle by MALDI-TOF MS. Sci. Rep. 2019, 9, 18381. [Google Scholar] [CrossRef]
- Murphy, C.N.; Fowler, R.; Balada-Llasat, J.M.; Carroll, A.; Stone, H.; Akerele, O.; Buchan, B.; Windham, S.; Hopp, A.; Ronen, S.; et al. Multicenter Evaluation of the BioFire FilmArray Pneumonia/Pneumonia Plus Panel for Detection and Quantification of Agents of Lower Respiratory Tract Infection. J. Clin. Microbiol. 2020, 58. Available online: https://pubmed.ncbi.nlm.nih.gov/32350043/ (accessed on 6 June 2025). [CrossRef]
- Webber, D.M.; Wallace, M.A.; Burnham, C.A.; Anderson, N.W. Evaluation of the BioFire FilmArray Pneumonia Panel for Detection of Viral and Bacterial Pathogens in Lower Respiratory Tract Specimens in the Setting of a Tertiary Care Academic Medical Center. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef]
- Cho, W.H.; Won, E.J.; Choi, H.J.; Kee, S.J.; Shin, J.H.; Ryang, D.W.; Suh, S.P. Comparison of AdvanSure TB/NTM PCR and COBAS TaqMan MTB PCR for Detection of Mycobacterium tuberculosis Complex in Routine Clinical Practice. Ann. Lab. Med. 2015, 35, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Yang, S.; Deighton, M.A.; Qu, Y.; Hong, L.; Su, F. A Comprehensive Evaluation of Xpert MTB/RIF Assay with Bronchoalveolar Lavage Fluid as a Single Test or Combined With Conventional Assays for Diagnosis of Pulmonary Tuberculosis in China: A Two-Center Prospective Study. Front. Microbiol. 2018, 9, 444. [Google Scholar] [CrossRef]
- Sow, D.; Fall, B.; Ndiaye, M.; Ba, B.S.; Sylla, K.; Tine, R.; Lo, A.C.; Abiola, A.; Wade, B.; Dieng, T.; et al. Usefulness of MALDI-TOF Mass Spectrometry for Routine Identification of Candida Species in a Resource-Poor Setting. Mycopathologia 2015, 180, 173–179. [Google Scholar] [CrossRef]
- Fan, L.C.; Lu, H.W.; Cheng, K.B.; Li, H.P.; Xu, J.F. Evaluation of PCR in bronchoalveolar lavage fluid for diagnosis of Pneumocystis jirovecii pneumonia: A bivariate meta-analysis and systematic review. PLoS ONE 2013, 8, e73099. [Google Scholar] [CrossRef]
- Imbert, S.; Meyer, I.; Palous, M.; Brossas, J.Y.; Uzunov, M.; Touafek, F.; Gay, F.; Trosini-Desert, V.; Fekkar, A. Aspergillus PCR in Bronchoalveolar Lavage Fluid for the Diagnosis and Prognosis of Aspergillosis in Patients With Hematological and Non-hematological Conditions. Front. Microbiol. 2018, 9, 1877. [Google Scholar] [CrossRef]
- Zeng, H.Q.; Zhang, X.B.; Cai, X.Y.; Yang, D.Y.; Lin, L.; Chen, M.J.; Guo, W.F.; Luo, X. Diagnostic value of bronchoalveolar lavage fluid cryptococcal antigen-lateral flow immunochromatographic assay for pulmonary cryptococcosis in non-HIV patients. Diagn. Microbiol. Infect. Dis. 2021, 99, 115276. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, H.H.; Lautenschlager, I.; Pinsky, B.A.; Cardenoso, L.; Aslam, S.; Cobb, B.; Vilchez, R.A.; Valsamakis, A. An international multicenter performance analysis of cytomegalovirus load tests. Clin. Infect. Dis. 2013, 56, 367–373. [Google Scholar] [CrossRef] [PubMed]
| No | Results of CDM | Results of SMS * | |||
|---|---|---|---|---|---|
| Culture (CFU/mL) | Filmarray Pneumonia Panel PCR (Copies/mL) | Singleplex Tests † | Taxon Above Threshold ‡ | Subdominant Taxon | |
| 1 | Escherichia coli (60,000) | Escherichia coli (106) Klebsiella pneumoniae (105) | MTB complex | None | Streptococcus salivarius (111, 25.58%) |
| 2 | Klebsiella pneumoniae (30,000) Corynebacterium striatum (≥100,000) MTB complex | Klebsiella aerogenes (106) Staphylococcus aureus (106) | MTB complex CMV (128,345) Pneumocystis jirovecii | None | Corynebacterium striatum (110, 26.44%) |
| 3 | Candida albicans (50,000) | Pseudomonas aeruginosa (106) Escherichia coli (104) Rhinovirus/Enterovirus | CMV (4395) | Pseudomonas aeruginosa (35, 87.5%) | Lactobacillus fermentum (2, 5%) |
| 4 | Candida tropicalis (10,000) | Enterobacter cloacae complex (104) | CMV (1,343,600) | Candida tropicalis (5) | Corynebacterium striatum (1, 100%) |
| 5 | NRF | Adenovirus | CMV (9665) Pneumocystis jirovecii | None | Staphylococcus kloosii (2, 11.11%) |
| 6 | NRF | Parainfluenza virus | CMV (200,175) Aspergillus(7.68) | None | Geobacillus stearothermophilus (1, 25%) |
| 7 | Klebsiella pneumoniae (10,000) | Klebsiella pneumoniae (105) | None | Klebsiella pneumoniae (13, 30.23%) | Serratia marcescens (3, 6.98%) |
| 8 | NRF | Klebsiella pneumoniae (105) | CMV (1015) | None | Klebsiella pneumoniae (8, 17.78%) |
| 9 | NRF | Klebsiella pneumoniae (105) | None | None | None |
| 10 | NRF | Haemophilus influenzae (106) Metapneumovirus | None | Haemophilus influenzae (90, 59.60%) | Haemophilus parasuis (2, 1.33%) |
| 11 | NRF | Haemophilus influenzae (105) Influenza A | CMV (<257) | Haemophilus influenzae (5, 31.25%) | Pasteurella multocida (2, 12.5%) |
| 12 | Pseudomonas aeruginosa (≥100,000) | Pseudomonas aeruginosa (106) | CMV (<257) Pneumocystis jirovecii Aspergillus (1.08) | Pseudomonas aeruginosa (59, 75.64%) | Pseudomonas sp. (6, 7.69%) |
| 13 | Acinetobacter baumannii (≥100,000) | Acinetobacter calcoaceticus- baumannii complex (106) | Aspergillus (2.27) | Acinetobacter baumannii (141, 66.82%) | Acinetobacter nosocomialis (11, 5.21%) Candida albicans (6) |
| 14 | Pseudomonas aeruginosa (≥100,000) | Acinetobacter calcoaceticus- baumannii complex (≥107) Pseudomonas aeruginosa (≥107) Escherichia coli (106) Serratia marcescens (106) Klebsiella pneumoniae 105) | CMV (<257) | Pseudomonas aeruginosa (23,869, 86.63%) | Acinetobacter baumannii (347, 1.26%) |
| 15 | Pseudomonas aeruginosa (≥100,000) | Pseudomonas aeruginosa (106) | CMV (2065) Aspergillus (1.90) | Pseudomonas aeruginosa (24, 75%) | Pseudomonas sp. (2, 6.25%) |
| 16 | Stenotrophomonas maltophilia (≥100,000) | Parainfluenza virus | CMV (10,780) | Stenotrophomonas maltophilia (147, 44.41%) | Stenotrophomonas pavanii (26, 7.85%) |
| Antibiotic Group | Resistance Gene | Gene Detection Status | Antibiotic Susceptibility | Consistency * |
|---|---|---|---|---|
| Carbapenem | None | Not detected | Doripenem (S), Ertapenem (S), Imipenem (S), Meropenem (S) | Consistent |
| Aminoglycoside | AAC(6′)-Ia | Detected | Amikacin (S), Gentamicin (S) | Inconsistent |
| Sulfonamide | Sul1 | Detected | Trimethoprim/Sulfamethoxazole (R) | Consistent |
| Antibiotic Group | Resistance Gene | Gene Detection Status | Antibiotic Susceptibility | Consistency * |
|---|---|---|---|---|
| Carbapenem | AXC-1, OXA-217 | Detected | Doripenem (S), Imipenem (S), Meropenem (S) | Inconsistent |
| Aminoglycoside | AAC(6′)-Ia, cpxA, smeR | Detected | Amikacin (S), Tobramycin (I) | Inconsistent |
| Fluoroquinolone and Tetracycline | MexI, H-NS | Detected | Levofloxacin (R), Tetracycline (R) | Consistent |
| Penicillin derivatives and Cephalosporins | OXA-217, smeR, H-NS, CTX-M-15 | Detected | Ampicillin (R), Piperacillin (R), Cefotaxime (R), Cefepime (S), Ceftazidime (S) | Inconsistent |
| Disease | Sequencing | Principles of Setting Thresholds | Thresholds for Identifying Pathogen by SMS | Study | |||
|---|---|---|---|---|---|---|---|
| Bacteria | Mycobacteria | Fungi | Viruses | ||||
| Suspected pulmonary infection | BGISEQ (BGI, Shenzhen, China) | ≥50 unique reads from a single species | ≥1 unique read from MTBC | ≥50 unique reads from a single species | ≥50 unique reads from a single species | [19] | |
| Suspected pulmonary infection | BGISEQ-100 (BGI, China) | Reads number ≥ 50 and pathogen detected by traditional method | >30% relative abundance at the genus level | ≥1 unique read from MTB | SMRN ≥ 3 | [18] | |
| Suspected pulmonary infection | MiniSeq (Illumina, USA) | >30% relative abundance at the genus level, or histopathological examination and/or culture positive with ≥50 unique reads of a single species | ≥1 unique read from MTB | >30% relative abundance at the genus level, or histopathological examination and/or culture positive with ≥50 unique reads of a single species | >30% relative abundance at the genus level | [17] | |
| Suspected pneumonia (immunocompromised) | Agilent 2100 Bioanalyzer (Thermo Fisher Scientific, USA) | Regardless of coverage rate, oral commensals were not defined as CSMs unless they were deemed to be significant by the physicians or proven otherwise | Coverage rate ≥ 10 times any other microbes | ≥1 unique read from MTB Mapping read number in the top 10 in the bacteria list of NTM | Coverage rate ≥ 5 times any other fungus | Coverage rate ≥ 10 times any other microbes | [23] |
| Pneumonia | Bioelectron Seq 4000 (CapitalBio Corporation, Beijing, China) | Clinically pathologic microorganism is defined; -Definite: SMS result is consistent with results from CDMs (culture, nucleic acid-based testing, and pathological examination) Probable: SMS pathogen is likely the cause of pneumonia according to clinical, radiologic, or laboratory findings, but the SMS result was consistent with CDMs. | Coverage rate ≥ 10 times any other microbes | ≥1 unique read from MTB | Coverage rate ≥ 5 times any other fungus | Coverage rate of species level ≥ 10 times any other microbes | [22] |
| Severe pneumonia (immunocompromised) | KAPA Library Quantification 75-cycle sequencing kit (Illumina, USA) | >30% Relative abundance at the genus level, Reads number ≥ 50 from a single species and pathogen detected by culture | [20] | ||||
| Community-acquired pneumonia | Nextseq 550Dx (Illumina, USA) | RPM counts ≥ 5 times the values of the NEC | Coverage of ≥3 non-overlapping regions on the genome | [25] | |||
| Community-acquired pneumonia | NextSeq CN500 (Illumina, USA) | Reads number ≥ 50 or pathogen detected by culture | ≥1 unique read from MTB Mapping read number in the top 10 in the bacteria list of NTM | ≥3 reads mapped to pathogen species, or supported by clinical culture | ≥3 reads mapped to pathogen species, or supported by clinical culture | [24] | |
| Severe community-acquired pneumonia (immunocompromised) | N/A | Reads number ≥ 50 from a single species and pathogen detected by culture | >30% Relative abundance at the genus level, Coverage rate ≥ 10 times any other bacteria | ≥1 unique read from MTB | >30% relative abundance at the genus level; Coverage rate ≥ 5 times any other fungus | [26] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, H.-e.; Kim, M.J.; Choi, J.; Sohn, Y.-H.; Lee, J.J.; Park, K.S.; Cho, S.Y.; Park, K.-H.; Kim, Y.J. Shotgun Metagenomic Sequencing Analysis as a Diagnostic Strategy for Patients with Lower Respiratory Tract Infections. Microorganisms 2025, 13, 1338. https://doi.org/10.3390/microorganisms13061338
Cho H-e, Kim MJ, Choi J, Sohn Y-H, Lee JJ, Park KS, Cho SY, Park K-H, Kim YJ. Shotgun Metagenomic Sequencing Analysis as a Diagnostic Strategy for Patients with Lower Respiratory Tract Infections. Microorganisms. 2025; 13(6):1338. https://doi.org/10.3390/microorganisms13061338
Chicago/Turabian StyleCho, Ha-eun, Min Jin Kim, Jongmun Choi, Yong-Hak Sohn, Jae Joon Lee, Kyung Sun Park, Sun Young Cho, Ki-Ho Park, and Young Jin Kim. 2025. "Shotgun Metagenomic Sequencing Analysis as a Diagnostic Strategy for Patients with Lower Respiratory Tract Infections" Microorganisms 13, no. 6: 1338. https://doi.org/10.3390/microorganisms13061338
APA StyleCho, H.-e., Kim, M. J., Choi, J., Sohn, Y.-H., Lee, J. J., Park, K. S., Cho, S. Y., Park, K.-H., & Kim, Y. J. (2025). Shotgun Metagenomic Sequencing Analysis as a Diagnostic Strategy for Patients with Lower Respiratory Tract Infections. Microorganisms, 13(6), 1338. https://doi.org/10.3390/microorganisms13061338






