Overview of Microorganisms: Bacterial Microbiome, Mycobiome, Virome Identified Using Next-Generation Sequencing, and Their Application to Ophthalmic Diseases
Abstract
1. Introduction
2. Human Bacterial Microbiome
3. Human Mycobiome
4. Human Virome
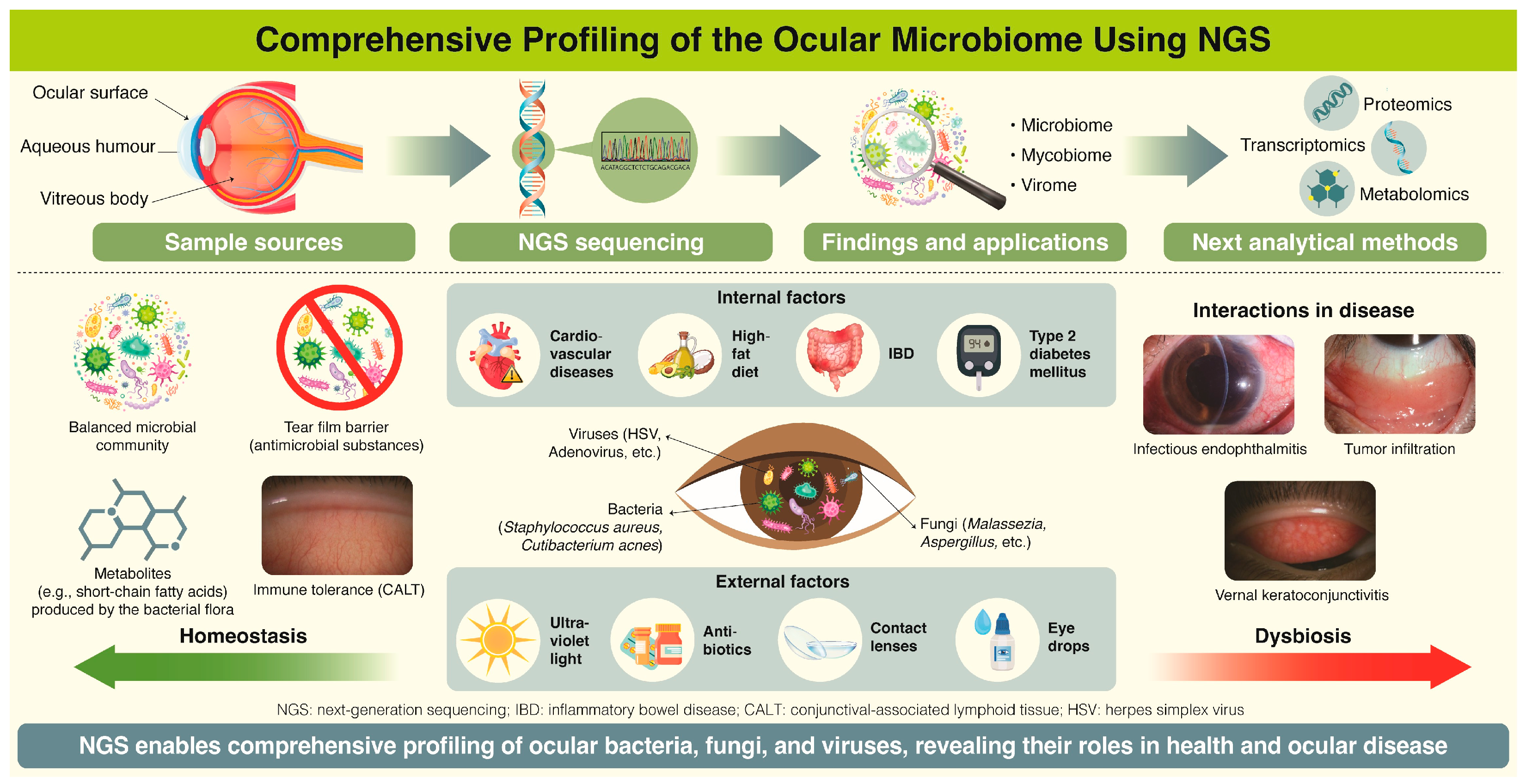
5. Clinical Studies of the Ocular Microbiome
5.1. Bacterial Microbiome
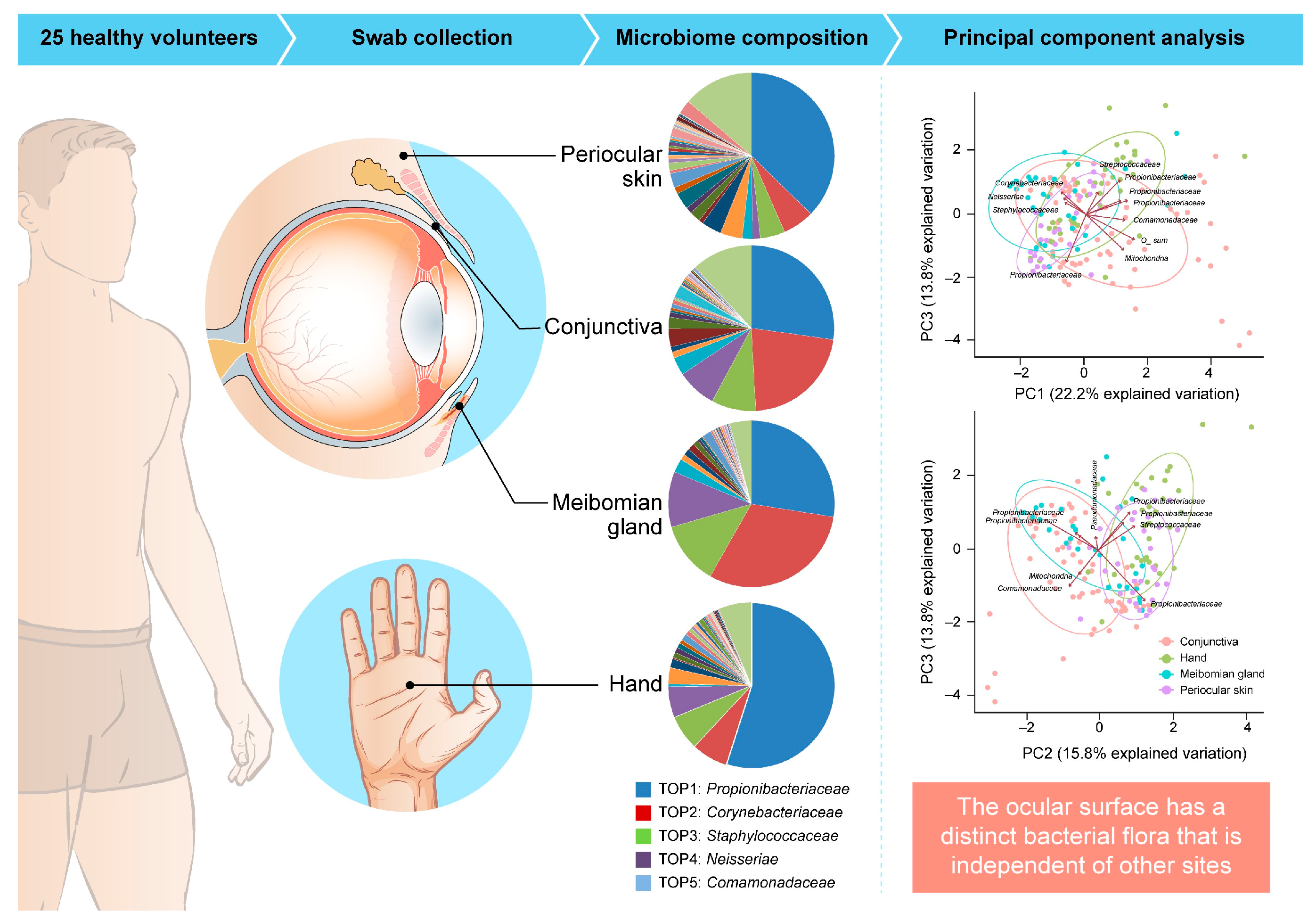
5.1.1. Ocular Surface
5.1.2. Aqueous Humor
5.1.3. Vitreous Body
5.2. Mycobiome
5.2.1. Ocular Surface
5.2.2. Aqueous Humor and Vitreous Body
5.3. Virome
5.3.1. Ocular Surface
5.3.2. Aqueous Humor and Vitreous Body
6. Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Clements, F.E. The development and structure of biotic communities. J. Ecol. 1917, 5, 120–121. [Google Scholar]
- Clements, F.E.; Shelford, V.E. Bio-Ecology; John Wiley and Sons: Hoboken, NJ, USA, 1939. [Google Scholar]
- Mohr, J.L. Protozoa as indicators of pollution. Sci Mon. 1952, 74, 7–9. [Google Scholar]
- Whipps, J.M. Fungi biol control Syst. In Mycoparasitism and Plant Disease Control; Burge, N., Ed.; Manchester University Press: Manchester, UK, 1988; pp. 161–187. [Google Scholar]
- Costello, E.K.; Lauber, C.L.; Hamady, M.; Fierer, N.; Gordon, J.I.; Knight, R. Bacterial community variation in human body habitats across space and time. Science 2009, 326, 1694–1697. [Google Scholar] [CrossRef]
- Eisenstein, B.I. The polymerase chain reaction. A new method of using molecular genetics for medical diagnosis. N. Engl. J. Med. 1990, 322, 178–183. [Google Scholar] [CrossRef]
- Relman, D.A. The search for unrecognized pathogens. Science 1999, 284, 1308–1310. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R. Bacterial evolution. Microbiol. Res. 1987, 51, 221–271. [Google Scholar] [CrossRef]
- Ludwig, W.; Klenk, H.-P. Overview: A Phylogenetic Backbone and Taxonomic Framework for Procaryotic Systematics. In Bergey’s Manual® of Systematic Bacteriology; Springer: New York, NY, USA, 2001. [Google Scholar]
- Kameoka, S.; Motooka, D.; Watanabe, S.; Kubo, R.; Jung, N.; Midorikawa, Y.; Shinozaki, N.O.; Sawai, Y.; Takeda, A.K.; Nakamura, S. Benchmark of 16S rRNA gene amplicon sequencing using Japanese gut microbiome data from the V1-V2 and V3-V4 primer sets. BMC Genom. 2021, 22, 527. [Google Scholar] [CrossRef]
- Handelsman, J.; Rondon, M.R.; Brady, S.F.; Clardy, J.; Goodman, R.M. Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chem. Biol. 1998, 5, R245–R249. [Google Scholar] [CrossRef] [PubMed]
- Bunnik, E.M.; Le Roch, K.G. An introduction to functional genomics and systems biology. Adv. Wound Care 2013, 2, 490–498. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, H.; Gesteland, R. Capillary gel electrophoresis for rapid, high resolution DNA sequencing. Nucleic Acids Res. 1990, 18, 1415–1419. [Google Scholar] [CrossRef] [PubMed]
- Kambara, H.; Nishikawa, T.; Katayama, Y.; Yamaguchi, T. Optimization of parameters in a DNA sequenator using fluorescence detection. Nat. Biotechnol. 1988, 6, 816–821. [Google Scholar] [CrossRef]
- Hunkapiller, T.; Kaiser, R.J.; Koop, B.F.; Hood, L. Large scale and automated DNA sequence determination. Science 1991, 254, 59–67. [Google Scholar] [CrossRef]
- Miller, J.R.; Koren, S.; Sutton, G. Assembly algorithms for next-generation sequencing data. Genomics 2010, 95, 315–327. [Google Scholar] [CrossRef]
- Dönmez, D.; Şimşek, Ö.; Ak Yeni Nesil, Y. DNA dizileme teknolojileri ve bitkilerde kullanımı. Türk Bilim. Derlemeler Derg. 2015, 8, 30. [Google Scholar]
- Wang, J.; Lin, M.; Crenshaw, A.; Hutchinson, A.; Hicks, B.; Yeager, M.; Berndt, S.; Huang, W.Y.; Hayes, R.B.; Chanock, S.J.; et al. High-throughput single nucleotide polymorphism genotyping using nanofluidic dynamic arrays. BMC Genom. 2009, 10, 561. [Google Scholar] [CrossRef]
- Peters, E.J.; McLeod, H.L. Ability of whole-genome SNP arrays to capture “must have” pharmacogenomic variants. Pharmacogenomics 2008, 9, 1573–1577. [Google Scholar] [CrossRef]
- Liu, L.; Li, Y.; Li, S.; Hu, N.; He, Y.; Pong, R.; Lin, D.; Lu, L.; Law, M. Comparison of next-generation sequencing systems. J. Biomed. Biotechnol. 2012, 2012, 251364. [Google Scholar] [CrossRef]
- Shendure, J.; Porreca, G.J.; Reppas, N.B.; Lin, X.; McCutcheon, J.P.; Rosenbaum, A.M.; Wang, M.D.; Zhang, K.; Mitra, R.D.; Church, G.M. Accurate multiplex polony sequencing of an evolved bacterial genome. Science 2005, 309, 1728–1732. [Google Scholar] [CrossRef]
- Timp, W.; Mirsaidov, U.M.; Wang, D.; Comer, J.; Aksimentiev, A.; Timp, G. Nanopore sequencing: Electrical measurements of the code of life. IEEE Trans. Nanotechnol. 2010, 9, 281–294. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Liu, D.; Zhang, X.; Li, W.; Liu, H.; Hong, W.; Jiang, C.; Guan, N.; Ma, C.; Zeng, H.; et al. SLAF-seq: An efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS ONE 2013, 8, e58700. [Google Scholar] [CrossRef]
- Rothberg, J.M.; Hinz, W.; Rearick, T.M.; Schultz, J.; Mileski, W.; Davey, M.; Leamon, J.H.; Johnson, K.; Milgrew, M.J.; Edwards, M.; et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature 2011, 475, 348–352. [Google Scholar] [CrossRef] [PubMed]
- Voelkerding, K.V.; Dames, S.A.; Durtschi, J.D. Next-generation sequencing: From basic research to diagnostics. Clin. Chem. 2009, 55, 641–658. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, Y.; Komiya, S.; Yasumizu, Y.; Yasuoka, Y.; Mizushima, K.; Takagi, T.; Kryukov, K.; Fukuda, A.; Morimoto, Y.; Naito, Y.; et al. Full-length 16S rRNA gene amplicon analysis of human gut microbiota using MinION™ nanopore sequencing confers species-level resolution. BMC Microbiol. 2021, 21, 35. [Google Scholar] [CrossRef]
- Burke, C.M.; Darling, A.E. A method for high precision sequencing of near full-length 16S rRNA genes on an Illumina MiSeq. PeerJ 2016, 4, e2492. [Google Scholar] [CrossRef]
- Roberts, R.J.; Carneiro, M.O.; Schatz, M.C. The advantages of SMRT sequencing. Genome Biol. 2013, 14, 405. [Google Scholar] [CrossRef]
- PacBio. Whole Genome Sequencing. Available online: https://www.pacb.com/products-and-services/applications/whole-genome-sequencing/ (accessed on 15 January 2025).
- Eid, J.; Fehr, A.; Gray, J.; Luong, K.; Lyle, J.; Otto, G.; Peluso, P.; Rank, D.; Baybayan, P.; Bettman, B.; et al. Real-time DNA sequencing from single polymerase molecules. Science 2009, 323, 133–138. [Google Scholar] [CrossRef]
- Jain, M.; Fiddes, I.T.; Miga, K.H.; Olsen, H.E.; Paten, B.; Akeson, M. Improved data analysis for the MinION nanopore sequencer. Nat. Methods 2015, 12, 351–356. [Google Scholar] [CrossRef]
- Leggett, R.M.; Alcon-Giner, C.; Heavens, D.; Caim, S.; Brook, T.C.; Kujawska, M.; Martin, S.; Peel, N.; Acford-Palmer, H.; Hoyles, L.; et al. Rapid MinION profiling of preterm microbiota and antimicrobial-resistant pathogens. Nat. Microbiol. 2020, 5, 430–442. [Google Scholar] [CrossRef]
- Moon, J.; Kim, N.; Kim, T.J.; Jun, J.S.; Lee, H.S.; Shin, H.R.; Lee, S.T.; Jung, K.H.; Park, K.I.; Jung, K.Y.; et al. Rapid diagnosis of bacterial meningitis by nanopore 16S amplicon sequencing: A pilot study. Int. J. Med. Microbiol. 2019, 309, 151338. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Wadden, J.; Erb-Downward, J.R.; Ranjan, P.; Zhou, W.; McDonald, T.L.; Mills, R.E.; Boyle, A.P.; Dickson, R.P.; Blaauw, D.; et al. SquiggleNet: Real-time, direct classification of nanopore signals. Genome Biol. 2021, 22, 298. [Google Scholar] [CrossRef]
- Richterich, P. Estimation of errors in “raw” DNA sequences: A validation study. Genome Res. 1998, 8, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, A.; Nakagawa, H.; Hosono, N.; Nakano, K.; Abe, T.; Boroevich, K.A.; Nagasaki, M.; Yamaguchi, R.; Shibuya, T.; Kubo, M.; et al. Whole-genome sequencing and comprehensive variant analysis of a Japanese individual using massively parallel sequencing. Nat. Genet. 2010, 42, 931–936. [Google Scholar] [CrossRef]
- Olson, N.D.; Wagner, J.; McDaniel, J.; Stephens, S.H.; Westreich, S.T.; Prasanna, A.G.; Johanson, E.; Boja, E.; Maier, E.J.; Serang, O.; et al. PrecisionFDA Truth Challenge V2: Calling variants from short and long reads in difficult-to-map regions. Cell Genom. 2022, 2, 100129. [Google Scholar] [CrossRef]
- Cheng, C.; Fei, Z.; Xiao, P. Methods to improve the accuracy of next-generation sequencing. Front. Bioeng. Biotechnol. 2023, 11, 982111. [Google Scholar] [CrossRef]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Dusko Ehrlich, S.; MetaHIT consortium. Metagenomics of the intestinal microbiota: Potential applications. Gastroenterol. Clin. Biol. 2010, 34 (Suppl. 1), S23–S28. [Google Scholar] [CrossRef]
- Morgan, X.C.; Segata, N.; Huttenhower, C. Biodiversity and functional genomics in the human microbiome. Trends Genet. 2013, 29, 51–58. [Google Scholar] [CrossRef]
- Aggarwal, N.; Kitano, S.; Puah, G.R.Y.; Kittelmann, S.; Hwang, I.Y.; Chang, M.W. Microbiome and human health: Current understanding, engineering, and enabling technologies. Chem. Rev. 2023, 123, 31–72. [Google Scholar] [CrossRef] [PubMed]
- Grazul, H.; Kanda, L.L.; Gondek, D. Impact of probiotic supplements on microbiome diversity following antibiotic treatment of mice. Gut Microbes 2016, 7, 101–114. [Google Scholar] [CrossRef]
- Khalil, M.; Di Ciaula, A.; Mahdi, L.; Jaber, N.; Di Palo, D.M.; Graziani, A.; Baffy, G.; Portincasa, P. Unraveling the role of the human gut microbiome in health and diseases. Microorganisms 2024, 12, 2333. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Fan, J.G. Gut microbiota in gastrointestinal diseases: Insights and therapeutic strategies. World J. Gastroenterol. 2024, 30, 4329–4332. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wu, J.; Dong, X.; Yin, H.; Shi, X.; Su, S.; Che, B.; Li, Y.; Yang, J. Gut flora-targeted photobiomodulation therapy improves senile dementia in an Aß-induced Alzheimer’s disease animal model. J. Photochem. Photobiol. B 2021, 216, 112152. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Qian, Z.; Yin, J.; Xu, W.; Zhou, X. The role of intestinal microbiota in cardiovascular disease. J. Cell. Mol. Med. 2019, 23, 2343–2350. [Google Scholar] [CrossRef]
- Maciel-Fiuza, M.F.; Muller, G.C.; Campos, D.M.S.; do Socorro Silva Costa, P.; Peruzzo, J.; Bonamigo, R.R.; Veit, T.; Vianna, F.S.L. Role of gut microbiota in infectious and inflammatory diseases. Front. Microbiol. 2023, 14, 1098386. [Google Scholar] [CrossRef]
- DeGruttola, A.K.; Low, D.; Mizoguchi, A.; Mizoguchi, E. Current understanding of dysbiosis in disease in human and animal models. Inflamm. Bowel Dis. 2016, 22, 1137–1150. [Google Scholar] [CrossRef]
- Markowiak-Kopeć, P.; Śliżewska, K. Effect of probiotics on the production of short-chain fatty acids by human intestinal microbiome. Nutrients 2020, 12, 1107. [Google Scholar] [CrossRef]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Liu, Y.; Xu, Q.; Mao, L. Short chain fatty acids: Essential weapons of traditional medicine in treating inflammatory bowel disease. Molecules 2024, 29, 379. [Google Scholar] [CrossRef]
- Kovtonyuk, L.V.; McCoy, K.D. Microbial metabolites and immunotherapy: Basic rationale and clinical indications. Semin. Immunol. 2023, 67, 101755. [Google Scholar] [CrossRef]
- Lorefice, L.; Zoledziewska, M. Propionic acid impact on multiple sclerosis: Evidence and challenges. Nutrients 2024, 16, 3887. [Google Scholar] [CrossRef] [PubMed]
- Gisevius, B.; Duscha, A.; Poschmann, G.; Stühler, K.; Motte, J.; Fisse, A.L.; Augustyniak, S.; Rehm, A.; Renk, P.; Böse, C.; et al. Propionic acid promotes neurite recovery in damaged multiple sclerosis neurons. Brain Commun. 2024, 6, fcae182. [Google Scholar] [CrossRef] [PubMed]
- Scott, N.; Whittle, E.; Jeraldo, P.; Chia, N. A systemic review of the role of enterotoxic Bacteroides fragilis in colorectal cancer. Neoplasia 2022, 29, 100797. [Google Scholar] [CrossRef] [PubMed]
- Valciukiene, J.; Strupas, K.; Poskus, T. Tissue vs. fecal-derived bacterial dysbiosis in precancerous colorectal lesions: A systematic review. Cancers 2023, 15, 1602. [Google Scholar] [CrossRef]
- DeDecker, L.; Coppedge, B.; Avelar-Barragan, J.; Karnes, W.; Whiteson, K. Microbiome distinctions between the CRC carcinogenic pathways. Gut Microbes 2021, 13, 1854641. [Google Scholar] [CrossRef]
- Pleguezuelos-Manzano, C.; Puschhof, J.; Rosendahl Huber, A.; van Hoeck, A.; Wood, H.M.; Nomburg, J.; Gurjao, C.; Manders, F.; Dalmasso, G.; Stege, P.B.; et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 2020, 580, 269–273. [Google Scholar] [CrossRef]
- Mohseni, A.H.; Taghinezhad-S, S.; Casolaro, V.; Lv, Z.; Li, D. Potential links between the microbiota and T cell immunity determine the tumor cell fate. Cell Death Dis. 2023, 14, 154. [Google Scholar] [CrossRef]
- Ma, C.; Han, M.; Heinrich, B.; Fu, Q.; Zhang, Q.; Sandhu, M.; Agdashian, D.; Terabe, M.; Berzofsky, J.A.; Fako, V.; et al. Gut microbiome–mediated bile acid metabolism regulates liver cancer via NKT cells. Science 2018, 360, eaan5931. [Google Scholar] [CrossRef]
- Guo, W.; Zhang, Y.; Guo, S.; Mei, Z.; Liao, H.; Dong, H.; Wu, K.; Ye, H.; Zhang, Y.; Zhu, Y.; et al. Tumor microbiome contributes to an aggressive phenotype in the basal-like subtype of pancreatic cancer. Commun. Biol. 2021, 4, 1019. [Google Scholar] [CrossRef]
- Revankar, N.A.; Negi, P.S. Biotics: An emerging food supplement for health improvement in the era of immune modulation. Nutr. Clin. Pract. 2024, 39, 311–329. [Google Scholar] [CrossRef]
- Hazra, R.; Chattopadhyay, S.; Mallick, A.; Gayen, S.; Roy, S. Revealing the therapeutic properties of gut microbiota: Transforming cancer immunotherapy from basic to clinical approaches. Med. Oncol. 2024, 41, 175. [Google Scholar] [CrossRef] [PubMed]
- Faith, J.J. Assessing live microbial therapeutic transmission. Gut Microbes 2025, 17, 2447836. [Google Scholar] [CrossRef]
- Berzack, S.; Galor, A. Microbiome-based therapeutics for ocular diseases. Clin. Exp. Optom. 2024, 108, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Tavakoli, A.; Markoulli, M.; Papas, E.; Flanagan, J. The impact of probiotics and prebiotics on dry eye disease signs and symptoms. J. Clin. Med. 2022, 11, 4889. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Gu, X.; Zhao, Q.; Wang, C.; Liu, X.; Chen, Y.; Zhao, X. Causal effects of gut microbiota on chalazion: A two-sample Mendelian randomization study. Front. Med. 2024, 11, 1411271. [Google Scholar] [CrossRef]
- Morandi, S.C.; Herzog, E.L.; Munk, M.; Kreuzer, M.; Largiadèr, C.R.; Wolf, S.; Zinkernagel, M.; Zysset-Burri, D.C. The gut microbiome and HLA-B27-associated anterior uveitis: A case-control study. J. Neuroinflammation 2024, 21, 120. [Google Scholar] [CrossRef]
- Xiao, J.; Zhang, J.Y.; Luo, W.; He, P.C.; Skondra, D. The emerging role of gut microbiota in age-related macular degeneration. Am. J. Pathol. 2023, 193, 1627–1637. [Google Scholar] [CrossRef]
- Limon, J.J.; Skalski, J.H.; Underhill, D.M. Commensal fungi in health and disease. Cell Host Microbe 2017, 22, 156–165. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Zmora, N.; Levy, M.; Elinav, E. The microbiome and innate immunity.CA. Nature 2016, 535, 65–74. [Google Scholar] [CrossRef]
- Nagao-Kitamoto, H.; Kamada, N. Host-microbial cross-talk in inflammatory bowel disease. Immune Netw. 2017, 17, 1–12. [Google Scholar] [CrossRef]
- Lass-Flörl, C.; Mutschlechner, W.; Aigner, M.; Grif, K.; Marth, C.; Girschikofsky, M.; Grander, W.; Greil, R.; Russ, G.; Cerkl, P.; et al. Utility of PCR in diagnosis of invasive fungal infections: Real-life data from a multicenter study. Clin. Microbiol. 2013, 51, 863–868. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Wu, J.; Cheng, M.; Zhu, X.; Du, M.; Chen, C.; Liao, W.; Zhi, K.; Pan, W. Diagnosis of invasive fungal infections: Challenges and recent developments. J. Biomed. Sci. 2023, 30, 42. [Google Scholar] [CrossRef] [PubMed]
- Finkelman, M.A. Specificity influences in (1→3)-β-d-glucan-supported diagnosis of invasive fungal disease. J. Fungi 2020, 7, 14. [Google Scholar] [CrossRef]
- Huang, Q.Y.; Li, P.C.; Yue, J.R. Diagnostic performance of serum galactomannan and β-D-glucan for invasive aspergillosis in suspected patients: A meta-analysis. Medicine 2024, 103, e37067. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Fungal Bar Coding Consortium; Fungal Barcoding Consortium Author List. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Seifert, K.A. Progress towards DNA barcoding of fungi. Mol. Ecol. Resour. 2009, 9 (Suppl. S1), 83–89. [Google Scholar] [CrossRef] [PubMed]
- Hillis, D.M.; Dixon, M.T. Ribosomal DNA: Molecular evolution and phylogenetic inference. Q. Rev. Biol. 1991, 66, 411–453. [Google Scholar] [CrossRef]
- White, T.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR-Protocols a Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninski, J.J., White, T.J., Eds.; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar]
- Nilsson, R.H.; Anslan, S.; Bahram, M.; Wurzbacher, C.; Baldrian, P.; Tedersoo, L. Mycobiome diversity: High-throughput sequencing and identification of fungi. Nat. Rev. Microbiol. 2019, 17, 95–109. [Google Scholar] [CrossRef]
- Motooka, D.; Fujimoto, K.; Tanaka, R.; Yaguchi, T.; Gotoh, K.; Maeda, Y.; Furuta, Y.; Kurakawa, T.; Goto, N.; Yasunaga, T.; et al. ITS1 deep-sequencing strategies to reconstruct the composition of a 26-species community and evaluation of the gut mycobiota of healthy Japanese individuals. Front. Microbiol. 2017, 8, 238. [Google Scholar] [CrossRef]
- Liu, J.; Yu, Y.; Cai, Z.; Bartlam, M.; Wang, Y. Comparison of ITS and 18S rDNA for estimating fungal diversity using PCR-DGGE. World J. Microbiol. Biotechnol. 2015, 31, 1387–1395. [Google Scholar] [CrossRef]
- Ghannoum, M.A.; Jurevic, R.J.; Mukherjee, P.K.; Cui, F.; Sikaroodi, M.; Naqvi, A.; Gillevet, P.M.P. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010, 6, e1000713. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Perez-Perez, G.I.; Chen, Y.; Blaser, M.J. Quantitation of major human cutaneous bacterial and fungal populations. J. Clin. Microbiol. 2010, 48, 3575–3581. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.D.N.; Viscogliosi, E.; Delhaes, L. The lung mycobiome: An emerging field of the human respiratory microbiome. Front. Microbiol. 2015, 13, 89. [Google Scholar] [CrossRef]
- Mok, N.; Knox, N.C.; Zhu, F.; Arnold, D.L.; Bar-Or, A.; Bernstein, C.N.; Bonner, C.; Forbes, J.D.; Graham, M.; Marrie, R.A.; et al. The fungal gut microbiota in pediatric-onset multiple sclerosis. Front. Microbiol. 2024, 15, 1258978. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Zhang, Y.; Zhang, W.; Zhang, A.; Wang, G.; Chen, C.; Ullah, H.; Ayaz, T.; Li, S.; Zhaxi, D.; et al. Characterizations of gut bacteriome, mycobiome, and virome of healthy individuals living in sea-level and high-altitude areas. Int. Microbiol. 2025, 28, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Ge, A.; Lydia, B.; Huang, C.; Wang, Q.; Yu, Y. Gut mycobiome dysbiosis contributes to the development of hypertension and its response to immunoglobulin light chains. Front. Immunol. 2022, 13, 1089295. [Google Scholar] [CrossRef] [PubMed]
- Jung, W.H. Alteration in skin mycobiome due to atopic dermatitis and seborrheic dermatitis. Biophys. Rev. 2023, 4, 011309. [Google Scholar] [CrossRef]
- Vavreckova, M.; Galanova, N.; Kostovcik, M.; Krystynik, O.; Ivanovova, E.; Roubalova, R.; Jiraskova Zakostelska, Z.; Friedecky, D.; Friedecka, J.; Haluzik, M.; et al. Specific gut bacterial and fungal microbiota pattern in the first half of pregnancy is linked to the development of gestational diabetes mellitus in the cohort including obese women. Front. Endocrinol. 2022, 13, 970825. [Google Scholar] [CrossRef] [PubMed]
- Dalamaga, M.; Zheng, L.; Liu, J. Gut mycobiome as a promising preventive and therapeutic target for metabolic disorders. Metab. Open 2022, 13, 100168. [Google Scholar] [CrossRef]
- Lang, S.; Duan, Y.; Liu, J.; Torralba, M.G.; Kuelbs, C.; Ventura-Cots, M.; Abraldes, J.G.; Bosques-Padilla, F.; Verna, E.C.; Brown, R.S.; et al. Intestinal fungal dysbiosis and systemic immune response to fungi in patients with alcoholic hepatitis. Hepatology 2020, 71, 522–538. [Google Scholar] [CrossRef]
- Aykut, B.; Pushalkar, S.; Chen, R.; Li, Q.; Abengozar, R.; Kim, J.I.; Shadaloey, S.A.; Wu, D.; Preiss, P.; Verma, N.; et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature 2019, 574, 264–267. [Google Scholar] [CrossRef] [PubMed]
- Saftien, A.; Puschhof, J.; Elinav, E. Fungi and cancer. Gut 2023, 72, 1410–1425. [Google Scholar] [CrossRef]
- Narunsky-Haziza, L.; Sepich-Poore, G.D.; Livyatan, I.; Asraf, O.; Martino, C.; Nejman, D.; Gavert, N.; Stajich, J.E.; Amit, G.; González, A.; et al. Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions. Cell 2022, 185, 3789–3806.E17. [Google Scholar] [CrossRef]
- Proctor, D.M.; Drummond, R.A.; Lionakis, M.S.; Segre, J.A. One population, multiple lifestyles: Commensalism and pathogenesis in the human mycobiome. Cell Host Microbe 2023, 31, 539–553. [Google Scholar] [CrossRef] [PubMed]
- Tsui, C.; Kong, E.F.; Jabra-Rizk, M.A. Pathogenesis of Candida albicans biofilm. Pathog. Dis. 2016, 74, ftw018. [Google Scholar] [CrossRef]
- Li, X.V.; Leonardi, I.; Putzel, G.G.; Semon, A.; Fiers, W.D.; Kusakabe, T.; Lin, W.Y.; Gao, I.H.; Doron, I.; Gutierrez-Guerrero, A.; et al. Immune regulation by fungal strain diversity in inflammatory bowel disease. Nature 2022, 603, 672–678. [Google Scholar] [CrossRef] [PubMed]
- Alshanta, O.A.; Albashaireh, K.; McKloud, E.; Delaney, C.; Kean, R.; McLean, W.; Ramage, G. Candida albicans and Enterococcus faecalis biofilm frenemies: When the relationship sours. Biofilm 2022, 4, 100072. [Google Scholar] [CrossRef]
- Sharma, S.; Mohler, J.; Mahajan, S.D.; Schwartz, S.A.; Bruggemann, L.; Aalinkeel, R. Microbial biofilm: A review on formation, infection, antibiotic resistance, control measures, and innovative treatment. Microorganisms 2023, 11, 1614. [Google Scholar] [CrossRef]
- Markus, V. Gut bacterial quorum sensing molecules and their association with inflammatory bowel disease: Advances and future perspectives. Biochem. Biophys. Res. Commun. 2024, 724, 150243. [Google Scholar] [CrossRef] [PubMed]
- Eichelberger, K.R.; Paul, S.; Peters, B.M.; Cassat, J.E. Candida-bacterial cross-kingdom interactions. Trends Microbiol. 2023, 31, 1287–1299. [Google Scholar] [CrossRef]
- Schmid, B.; Künstner, A.; Fähnrich, A.; Bersuch, E.; Schmid-Grendelmeier, P.; Busch, H.; Glatz, M.; Bosshard, P.P. Dysbiosis of skin microbiota with increased fungal diversity is associated with severity of disease in atopic dermatitis. J. Eur. Acad. Dermatol. Venereol. 2022, 36, 1811–1819. [Google Scholar] [CrossRef] [PubMed]
- Sparber, F.; De Gregorio, C.; Steckholzer, S.; Ferreira, F.M.; Dolowschiak, T.; Ruchti, F.; Kirchner, F.R.; Mertens, S.; Prinz, I.; Joller, N.; et al. The skin commensal yeast malassezia triggers a type 17 response that coordinates anti-fungal immunity and exacerbates skin inflammation. Cell Host Microbe 2019, 25, 389–403.E6. [Google Scholar] [CrossRef]
- Cadwell, K. The virome in host health and disease. Immunity 2015, 42, 805–813. [Google Scholar] [CrossRef]
- Sussman, M.; Topley, W.W.C.; Wilson, G.K.; Collier, L.H.; Balows, A. Topley and Wilson’s Microbiology and Microbial Infections; Arnold: London, UK, 1998; p. 4. ISBN 0-340-66316-2. [Google Scholar]
- Horsfall, F.L. Thomas Milton Rivers, September 3, 1888–May 12, 1962 (PDF). Biogr. Mem. Natl. Acad. Sci. 1965, 38, 263–294. [Google Scholar]
- Ackermann, H.-W. The first phage electron micrographs. Bacteriophage 2011, 1, 225–227. [Google Scholar] [CrossRef] [PubMed]
- Leland, D.S.; Ginocchio, C.C. Role of cell culture for virus detection in the age of technology. Clin. Microbiol. Rev. 2007, 20, 49–78. [Google Scholar] [CrossRef] [PubMed]
- Dolskiy, A.A.; Grishchenko, I.V.; Yudkin, D.V. Cell cultures for virology: Usability, advantages, and prospects. Int. J. Mol. Sci. 2020, 21, 7978. [Google Scholar] [CrossRef]
- Dronina, J.; Samukaite-Bubniene, U.; Ramanavicius, A. Advances and insights in the diagnosis of viral infections. J. Nanobiotechnology 2021, 19, 348. [Google Scholar] [CrossRef]
- Sahel, D.K.; Giriprasad, G.; Jatyan, R.; Guha, S.; Korde, A.; Mittal, A.; Bhand, S.; Chitkara, D. Next-generation CRISPR/Cas-based ultrasensitive diagnostic tools: Current progress and prospects. RSC Adv. 2024, 14, 32411–32435. [Google Scholar] [CrossRef]
- Malekshoar, M.; Azimi, S.A.; Kaki, A.; Mousazadeh, L.; Motaei, J.; Vatankhah, M. CRISPR-Cas9 targeted enrichment and next-generation sequencing for mutation detection. J. Mol. Diagn. 2023, 25, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, M.; Hewson, I.; Felts, B.; Mahaffy, J.M.; Nulton, J.; Salamon, P.; Rohwer, F. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 2003, 185, 6220–6223. [Google Scholar] [CrossRef]
- Breitbart, M.; Haynes, M.; Kelley, S.; Angly, F.; Edwards, R.A.; Felts, B.; Mahaffy, J.M.; Mueller, J.; Nulton, J.; Rayhawk, S.; et al. Viral diversity and dynamics in an infant gut. Res. Microbiol. 2008, 159, 367–373. [Google Scholar] [CrossRef]
- Zhang, T.; Breitbart, M.; Lee, W.H.; Run, J.Q.; Wei, C.L.; Soh, S.W.L.; Hibberd, M.L.; Liu, E.T.; Rohwer, F.; Ruan, Y. RNA viral community in human feces: Prevalence of plant pathogenic viruses. PLoS Biol. 2006, 4, e3. [Google Scholar] [CrossRef]
- Rosenberg, R.; Johansson, M.A.; Powers, A.M.; Miller, B.R. Search strategy has influenced the discovery rate of human viruses. Proc. Natl. Acad. Sci. USA 2013, 110, 13961–13964. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.G.; Gerin, J.L.; Anderson, N.L. Global Screening for Human Viral Pathogens. Emerg. Infect Dis. 2003, 9, 768–773. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Bushman, F.D. The human virome: Assembly, composition and host interactions. Nat. Rev. Microbiol. 2021, 19, 514–527. [Google Scholar] [CrossRef]
- Yadegar, A.; Bar-Yoseph, H.; Monaghan, T.M.; Pakpour, S.; Severino, A.; Kuijper, E.J.; Smits, W.K.; Terveer, E.M.; Neupane, S.; Nabavi-Rad, A.; et al. Fecal microbiota transplantation: Current challenges and future landscapes. Clin. Microbiol. Rev. 2024, 37, e0006022. [Google Scholar] [CrossRef] [PubMed]
- Sasa, N.; Kojima, S.; Koide, R.; Hasegawa, T.; Namkoong, H.; Hirota, T.; Watanabe, R.; Nakamura, Y.; Oguro-Igashira, E.; Ogawa, K.; et al. Blood DNA virome associates with autoimmune diseases and COVID-19. Nat. Genet. 2025, 57, 65–79. [Google Scholar] [CrossRef]
- Huang, H.; Yang, Y.; Wang, X.; Wen, B.; Yang, X.; Zhong, W.; Wang, Q.; He, F.; Li, J. Gut virome dysbiosis impairs antitumor immunity and reduces 5-fluorouracil treatment efficacy for colorectal cancer. Front. Oncol. 2024, 14, 1501981. [Google Scholar] [CrossRef] [PubMed]
- Pyöriä, L.; Pratas, D.; Toppinen, M.; Simmonds, P.; Hedman, K.; Sajantila, A.; Perdomo, M.F. Intra-host genomic diversity and integration landscape of human tissue-resident DNA virome. Nucleic Acids Res. 2024, 52, 13073–13093. [Google Scholar] [CrossRef]
- Chen, W.; Liang, F.; Zhang, Y.; Zhang, Y.; Lv, J.; Jin, X.; Ran, Y.; Li, S.; Sun, W. Metagenome-based characterization of the gut bacteriome, mycobiome, and virome in patients with chronic hepatitis B-related liver fibrosis. Front. Microbiol. 2024, 15, 1449090. [Google Scholar] [CrossRef] [PubMed]
- Natarelli, N.; Gahoonia, N.; Sivamani, R.K. Bacteriophages and the microbiome in dermatology: The role of the Phageome and a potential therapeutic strategy. Int. J. Mol. Sci. 2023, 24, 2695. [Google Scholar] [CrossRef] [PubMed]
- Kumata, R.; Ito, J.; Takahashi, K.; Suzuki, T.; Sato, K. A tissue level atlas of the healthy human virome. BMC Biol. 2020, 18, 55. [Google Scholar] [CrossRef]
- Dal Lago, S.; Brani, P.; Ietto, G.; Dalla Gasperina, D.; Gianfagna, F.; Giaroni, C.; Bosi, A.; Drago Ferrante, F.; Genoni, A.; Manzoor, H.Z.; et al. Torque Teno Virus: A Promising Biomarker in Kidney Transplant Recipients. Int. J. Mol. Sci. 2024, 25, 7744. [Google Scholar] [CrossRef]
- Kuczaj, A.; Przybyłowski, P.; Hrapkowicz, T. Torque Teno Virus (TTV)—A potential marker of immunocompetence in solid organ recipients. Viruses 2023, 16, 17. [Google Scholar] [CrossRef]
- Buddle, S.; Forrest, L.; Akinsuyi, N.; Martin Bernal, L.M.; Brooks, T.; Venturini, C.; Miller, C.; Brown, J.R.; Storey, N.; Atkinson, L.; et al. Evaluating metagenomics and targeted approaches for diagnosis and surveillance of viruses. Genome Med. 2024, 16, 111. [Google Scholar] [CrossRef]
- Ren, J.; Song, K.; Deng, C.; Ahlgren, N.A.; Fuhrman, J.A.; Li, Y.; Xie, X.; Poplin, R.; Sun, F. Identifying viruses from metagenomic data using deep learning. Quant. Biol. 2020, 8, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Artesi, M.; Hahaut, V.; Cole, B.; Lambrechts, L.; Ashrafi, F.; Marçais, A.; Hermine, O.; Griebel, P.; Arsic, N.; van der Meer, F.; et al. PCIP-seq: Simultaneous sequencing of integrated viral genomes and their insertion sites with long reads. Genome Biol. 2021, 22, 97. [Google Scholar] [CrossRef]
- Lapidus, A.L.; Korobeynikov, A.I. Metagenomic data assembly—The way of decoding unknown microorganisms. Front. Microbiol. 2021, 12, 613791. [Google Scholar] [CrossRef]
- Asao, K.; Hashida, N.; Ando, S.; Motooka, D.; Kurakami, H.; Nakamura, S.; Yamashita, D.; Maruyama, K.; Kawasaki, S.; Yamada, T.; et al. Conjunctival dysbiosis in mucosa-associated lymphoid tissue lymphoma. Sci. Rep. 2019, 9, 8424. [Google Scholar] [CrossRef]
- McNatt, J.; Allen, S.D.; Wilson, L.A.; Dowell, V.R., Jr. Anaerobic flora of the normal human conjunctival sac. Arch. Ophthalmol. 1978, 96, 1448–1450. [Google Scholar] [CrossRef]
- Dong, Q.; Brulc, J.M.; Iovieno, A.; Bates, B.; Garoutte, A.; Miller, D.; Revanna, K.V.; Gao, X.; Antonopoulos, D.A.; Slepak, V.Z.; et al. Diversity of bacteria at healthy human conjunctiva. Investig. Ophthalmol. Vis. Sci. 2011, 52, 5408–5413. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Holland, M.J.; Makalo, P.; Joof, H.; Roberts, C.H.; Mabey, D.C.; Bailey, R.L.; Burton, M.J.; Weinstock, G.M.; Burr, S.E. The conjunctival microbiome in health and trachomatous disease: A case control study. Genome Med. 2014, 6, 99. [Google Scholar] [CrossRef]
- Patel, V.; Cavuoto, K.; Galor, A. The “normal’ ocular microbiome and human eye health. In Microbiome and the Eye; Galor, A., Sun, J., Eds.; Academic Press: Cambridge, MA, USA, 2023; Chapter 3; pp. 87–107. [Google Scholar] [CrossRef]
- Böhm, E.W.; Buonfiglio, F.; Voigt, A.M.; Bachmann, P.; Safi, T.; Pfeiffer, N.; Gericke, A. Oxidative stress in the eye and its role in the pathophysiology of ocular diseases. Redox Biol. 2023, 68, 102967. [Google Scholar] [CrossRef] [PubMed]
- Mudgil, P. Antimicrobial tear lipids in the ocular surface defense. Front. Cell. Infect. Microbiol. 2022, 12, 866900. [Google Scholar] [CrossRef] [PubMed]
- Asao, K.; Hashida, N.; Maruyama, K.; Motooka, D.; Tsukamoto, T.; Usui, Y.; Nakamura, S.; Nishida, K. Comparative evaluation of 16S rRNA metagenomic sequencing in the diagnosis and understanding of bacterial endophthalmitis. BMJ Open Ophthalmol. 2023, 8, e001342. [Google Scholar] [CrossRef] [PubMed]
- Asao, K.; Hashida, N.; Motooka, D.; Tsukamoto, T.; Nakamura, S.; Maruyama, K.; Nishida, K. Fungal dysbiosis and decreased tear mucin at the conjunctiva in patients with conjunctival mucosa-associated lymphoid tissue lymphoma. BMJ Open Ophthalmol. 2023, 8, e001360. [Google Scholar] [CrossRef]
- Asao, K.; Hashida, N.; Maruyama, K.; Motooka, D.; Nakamura, S.; Nishida, K. Cases of endophthalmitis caused by Candida albicans and Candida dubliniensis identified via internal transcribed spacer deep sequencing. BMC Ophthalmol. 2024, 24, 444. [Google Scholar] [CrossRef]
- Wu, J.; Chen, N.; Grau, E.; Johnson, L.; Liu, Y.; Li, C.; Scott, P.A.; Kim, C.; Sun, D.; Kaplan, H.J.; et al. Short chain fatty acids inhibit corneal inflammatory responses to TLR ligands via the ocular G-protein coupled receptor 43. Ocul. Surf. 2024, 32, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Ciurariu, E.; Tirziu, A.T.; Varga, N.I.; Hirtie, B.; Alexandru, A.; Ivan, C.S.; Nicolescu, L. Short-chain fatty acids and the gut-retina connection: A systematic review. Int. J. Mol. Sci. 2025, 26, 2470. [Google Scholar] [CrossRef] [PubMed]
- Schlegel, I.; De Goüyon Matignon de Pontourade, C.M.F.; Lincke, J.-B.; Keller, I.; Zinkernagel, M.S.; Zysset-Burri, D.C. The human ocular surface microbiome and its associations with the tear proteome in dry eye disease. Int. J. Mol. Sci. 2023, 24, 14091. [Google Scholar] [CrossRef] [PubMed]
- Ting, D.S.J.; Mohammed, I.; Lakshminarayanan, R.; Beuerman, R.W.; Dua, H.S. Host defense peptides at the ocular surface: Roles in health and major diseases, and therapeutic potentials. Front. Med. 2022, 9, 835843. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.J.; Winn, B.J. Perturbations of the ocular surface microbiome and their effect on host immune function. Curr. Opin. Ophthalmol. 2023, 34, 181–188. [Google Scholar] [CrossRef]
- Doularamani, M.; Murthy, S.I. Role of ocular surface microbiome in health and disease. Indian J. Ophthalmol. 2023, 71, 2595. [Google Scholar] [CrossRef] [PubMed]
- Azari, A.A.; Arabi, A. Conjunctivitis: A systematic review. J. Ophthalmic Vis. Res. 2020, 15, 372–395. [Google Scholar] [CrossRef]
- Zhang, Z.; Cao, K.; Liu, J.; Wei, Z.; Xu, X.; Liang, Q. Pathogens and antibiotic susceptibilities of global bacterial keratitis: A meta-analysis. Antibiotics 2022, 11, 238. [Google Scholar] [CrossRef]
- Shorbatli, L.A.; Koranyi, K.I.; Nahata, M.C. Effectiveness of antibiotic therapy in pediatric patients with cat scratch disease. Int. J. Clin. Pharm. 2018, 40, 1458–1461. [Google Scholar] [CrossRef]
- Luo, B.; Li, M.; Xiang, N.; Hu, W.; Liu, R.; Yan, X. The microbiologic spectrum of dacryocystitis. BMC Ophthalmol. 2021, 21, 29. [Google Scholar] [CrossRef] [PubMed]
- Cirks, B.T.; Claunch, K.M.; DePerrior, S.; Poitras, B.; Adams, D.J. Microbiology and epidemiology of orbital cellulitis in pediatric and young adult patients. Mil. Med. 2025, 190, e593–e600. [Google Scholar] [CrossRef]
- Delbeke, H.; Younas, S.; Casteels, I.; Joossens, M. Current knowledge on the human eye microbiome: A systematic review of available amplicon and metagenomic sequencing data. Acta Ophthalmol. 2021, 99, 16–25. [Google Scholar] [CrossRef]
- Chowdhary, A.; Van Gelder, R.N.; Sundararajan, M. Methodologic considerations for studying the ocular surface microbiome. Ophthalmol. Sci. 2023, 3, 100408. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Yi, S.; Wei, L. Ocular microbiota and intraocular inflammation. Front. Immunol. 2020, 11, 609765. [Google Scholar] [CrossRef]
- Ozkan, J.; Majzoub, M.E.; Coroneo, M.; Thomas, T.; Willcox, M. Ocular microbiome changes in dry eye disease and Meibomian gland dysfunction. Exp. Eye Res. 2023, 235, 109615. [Google Scholar] [CrossRef]
- Clougher, S.B.; Foschi, C.; Moramarco, A.; Fontana, L.; Lazzarotto, T.; Marangoni, A.; Versura, P. Critical insights into the ocular surface microbiome: The need to standardize. New Microbiol. 2024, 47, 201–216. [Google Scholar]
- Wang, C.; Dou, X.; Li, J.; Wu, J.; Cheng, Y.; An, N. Composition and diversity of the ocular surface microbiota in patients with blepharitis in Northwestern China. Front. Med. 2021, 8, 768849. [Google Scholar] [CrossRef]
- Ueta, M.; Hosomi, K.; Park, J.; Mizuguchi, K.; Sotozono, C.; Kinoshita, S.; Kunisawa, J. Categorization of the ocular microbiome in Japanese Stevens-Johnson syndrome patients with severe ocular complications. Front. Cell. Infect. Microbiol. 2021, 11, 741654. [Google Scholar] [CrossRef] [PubMed]
- Doan, T.; Akileswaran, L.; Andersen, D.; Johnson, B.; Ko, N.; Shrestha, A.; Shestopalov, V.; Lee, C.S.; Lee, A.Y.; Van Gelder, R.N. Paucibacterial microbiome and resident DNA virome of the healthy conjunctiva. Investig. Ophthalmol. Vis. Sci. 2016, 57, 5116–5126. [Google Scholar] [CrossRef]
- Zilliox, M.J.; Bouchard, C.S. The microbiome, ocular surface, and corneal disorders. Am. J. Pathol. 2023, 193, 1648–1661. [Google Scholar] [CrossRef] [PubMed]
- Bendlin, A.; Gemensky-Metzler, A.J.; Diaz-Campos, D.; Newbold, G.M.; Miller, E.J.; Chandler, H.L. Evaluation of a commercial NGS service for detection of bacterial and fungal pathogens in infectious ulcerative keratitis. Vet. Ophthalmol. 2023, 26, 500–513. [Google Scholar] [CrossRef]
- Derrick, T.; Roberts, C.H.; Last, A.R.; Burr, S.E.; Holland, M.J. Trachoma and ocular chlamydial infection in the era of genomics. Mediat. Inflamm. 2015, 2015, 791847. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, F.; Hutchinson, D.S.; Sun, W.; Ajami, N.J.; Lai, S.; Wong, M.C.; Petrosino, J.F.; Fang, J.; Jiang, J.; et al. Conjunctival microbiome changes associated with soft contact lens and orthokeratology lens wearing. Investig. Ophthalmol. Vis. Sci. 2017, 58, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lu, X.; Cheng, Z.; Zou, D.; Shi, W.; Wang, T. Alterations of conjunctival microbiota associated with orthokeratology lens wearing in myopic children. BMC Microbiol. 2023, 23, 397. [Google Scholar] [CrossRef] [PubMed]
- Curragh, D.S.; Bassiouni, A.; Macias-Valle, L.; Vreugde, S.; Wormald, P.-J.; Selva, D.; Psaltis, A.J. The microbiome of the nasolacrimal system and its role in nasolacrimal duct obstruction. Ophthal. Plast. Reconstr. Surg. 2020, 36, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Grondona, P.; Bucher, P.; Schulze-Osthoff, K.; Hailfinger, S.; Schmitt, A. NF-κB activation in lymphoid malignancies: Genetics, signaling, and targeted therapy. Biomedicines 2018, 6, 38. [Google Scholar] [CrossRef] [PubMed]
- Hotta, F.; Eguchi, H.; Kuwahara, T.; Nakayama-Imaohji, H.; Shimomura, Y.; Kusaka, S. Disturbances in the ocular surface microbiome by perioperative antimicrobial eye drops. Front. Cell. Infect. Microbiol. 2023, 13, 1172345. [Google Scholar] [CrossRef]
- Patangia, D.V.; Ryan, C.A.; Dempsey, E.; Ross, R.P.; Stanton, C. Impact of antibiotics on the human microbiome and consequences for host health. MicrobiologyOpen 2022, 11, e1260. [Google Scholar] [CrossRef]
- Zhao, A.; Sun, J.; Liu, Y. Understanding bacterial biofilms: From definition to treatment strategies. Front. Cell. Infect. Microbiol. 2023, 13, 1137947. [Google Scholar] [CrossRef]
- Lai, J.; Rigas, Y.; Kantor, N.; Cohen, N.; Tomlinson, A.; St Leger, A.J.; Galor, A. Living with your biome: How the bacterial microbiome impacts ocular surface health and disease. Expert Rev. Ophthalmol. 2024, 19, 89–103. [Google Scholar] [CrossRef]
- Chiang, M.-C.; Chern, E. More than antibiotics: Latest therapeutics in the treatment and prevention of ocular surface infections. J. Clin. Med. 2022, 11, 4195. [Google Scholar] [CrossRef]
- Chiang, M.C.; Chern, E. Ocular surface microbiota: Ophthalmic infectious disease and probiotics. Front. Microbiol. 2022, 13, 952473. [Google Scholar] [CrossRef]
- Pietrzak, B.; Tomela, K.; Olejnik-Schmidt, A.; Mackiewicz, A.; Schmidt, M. Secretory IgA in intestinal mucosal secretions as an adaptive barrier against microbial cells. Int. J. Mol. Sci. 2020, 21, 9254. [Google Scholar] [CrossRef] [PubMed]
- Coles, J.A.; Myburgh, E.; Brewer, J.M.; McMenamin, P.G. Where are we? The anatomy of the murine cortical meninges revisited for intravital imaging, immunology, and clearance of waste from the brain. Prog. Neurobiol. 2017, 156, 107–148. [Google Scholar] [CrossRef] [PubMed]
- Forrester, J.V.; McMenamin, P.G.; Dando, S.J. CNS infection and immune privilege. Nat. Rev. Neurosci. 2018, 19, 655–671. [Google Scholar] [CrossRef]
- Forrester, J.V.; McMenamin, P.G. Evolution of the ocular immune system. Eye 2025, 39, 468–477. [Google Scholar] [CrossRef] [PubMed]
- Boers, S.A.; Jansen, R.; Hays, J.P. Understanding and overcoming the pitfalls and biases of next-generation sequencing (NGS) methods for use in the routine clinical microbiological diagnostic laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1059–1070. [Google Scholar] [CrossRef]
- Kitazawa, K.; Sotozono, C.; Koizumi, N.; Nagata, K.; Inatomi, T.; Sasaki, H.; Kinoshita, S. Safety of anterior chamber paracentesis using a 30-gauge needle integrated with a specially designed disposable pipette. Br. J. Ophthalmol. 2017, 101, 548–550. [Google Scholar] [CrossRef]
- Tang, P.H.; Karkhur, S.; Nguyen, Q.D. Obtaining undiluted vitreous sample using small gauge pars plana vitrectomy and air infusion. Am. J. Ophthalmol. Case Rep. 2020, 19, 100768. [Google Scholar] [CrossRef]
- Asghar, M.A.; Tang, S.; Wong, L.P.; Yang, P.; Zhao, Q. Infectious uveitis: A comprehensive systematic review of emerging trends and molecular pathogenesis using network analysis. J. Ophthal. Inflamm. Infect. 2024, 14, 60. [Google Scholar] [CrossRef]
- Rosenbaum, J.T.; Asquith, M. The microbiome and HLA-B27-Associated acute anterior uveitis. Nat. Rev. Rheumatol. 2018, 14, 704–713. [Google Scholar] [CrossRef]
- Song, Z.-Y.; Yuan, D.; Zhang, S.-X. Role of the microbiome and its metabolites in ankylosing spondylitis. Front. Immunol. 2022, 13, 1010572. [Google Scholar] [CrossRef]
- Sharma, S.M.; Jackson, D. Uveitis and spondyloarthropathies. Best Pract. Res. Clin. Rheumatol. 2017, 31, 846–862. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.B.; Chen, Y.X.; Su, Z.Y.; Chen, X.Y.; Lai, Y.N.; Yang, J.H. Causal association of juvenile idiopathic arthritis or JIA-associated uveitis and gut microbiota: A bidirectional two-sample Mendelian randomisation study. Front. Immunol. 2024, 15, 1356414. [Google Scholar] [CrossRef] [PubMed]
- Mohan, K.V.; Mishra, A.; Muniyasamy, A.; Sinha, P.; Sahu, P.; Kesarwani, A.; Jain, K.; Nagarajan, P.; Scaria, V.; Agarwal, M.; et al. Immunological consequences of compromised ocular immune privilege accelerate retinal degeneration in retinitis pigmentosa. Orphanet J. Rare Dis. 2022, 17, 378. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Zhang, M.; Jiang, D.; Su, Q.; Shi, J. The role of inflammation in autoimmune disease: A therapeutic target. Front. Immunol. 2023, 14, 1267091. [Google Scholar] [CrossRef] [PubMed]
- Bhutta, N.K.; Xu, X.; Jian, C.; Wang, Y.; Liu, Y.; Sun, J.; Han, B.; Wu, S.; Javeed, A. Gut microbiota mediated T cells regulation and autoimmune diseases. Front. Microbiol. 2024, 15, 1477187. [Google Scholar] [CrossRef] [PubMed]
- Schlunck, G.; Maier, P.; Maier, B.; Maier, W.; Strempel, S.; Reinhard, T.; Heinzelmann, S. Sequencing of the human aqueous humour microbiome. Int. J. Mol. Sci. 2024, 25, 6128. [Google Scholar] [CrossRef]
- Deng, Y.; Ge, X.; Li, Y.; Zou, B.; Wen, X.; Chen, W.; Lu, L.; Zhang, M.; Zhang, X.; Li, C.; et al. Identification of an intraocular microbiota. Cell Discov. 2021, 7, 13. [Google Scholar] [CrossRef]
- Das, T.; Padakandla, S.R.; Shivaji, S.; Jayasudha, R.; Takkar, B. Intraocular microbiome in diabetes and diabetic retinopathy: A pilot study. Ophthalmol. Ther. 2023, 12, 1109–1126. [Google Scholar] [CrossRef] [PubMed]
- Das, T.; Takkar, B.; Padakandala, S.R.; Shivaji, S. Gut and intraocular fluid dysbiosis in people with type 2 diabetes-related retinopathy in India: A case for further research. Indian J. Ophthalmol. 2025, 73 (Suppl. 1), S144–S150. [Google Scholar] [CrossRef] [PubMed]
- Harper, D.R. A comparative study of the microbiological contamination of postmortem blood and vitreous humour samples taken for ethanol determination. Forensic Sci. Int. 1989, 43, 37–44. [Google Scholar] [CrossRef]
- Egger, S.F.; Buxbaum, A.; Georgopoulos, M.; Scholda, C.; Vecsei, V.P.; Huber-Spitzy, V.; Georgopoulos, A. Bacterial growth in human vitreous humour. Exp. Eye Res. 1997, 65, 791–795. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Chandran, K.; Fernandes, M. Practical tips and common mistakes in ocular microbiology sampling and processing. Indian J. Ophthalmol. 2023, 71, 1698–1705. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Xia, H.; Tang, R.; Ng, T.K.; Yao, F.; Liao, X.; Zhang, Q.; Ke, X.; Shi, T.; Chen, H. Metagenomic next-generation sequencing detects pathogens in endophthalmitis patients. Retina 2022, 42, 992–1000. [Google Scholar] [CrossRef]
- Bhende, M.; Raman, R.; Jain, M.; Shah, P.K.; Sharma, T.; Gopal, L.; Bhende, P.S.; Srinivasan, S.; Jambulingam, M.; Sankara Nethralaya Vitreoretinal Study Group (SNVR-Study Group). Incidence, microbiology, and outcomes of endophthalmitis after 111,876 pars plana vitrectomies at a single, tertiary eye care hospital. PLoS ONE 2018, 13, e0191173. [Google Scholar] [CrossRef] [PubMed]
- Levin, H.J.; Mehta, M.S.; Storey, P.P.; Patel, S.N.; Kuley, B.; Wibbelsman, T.D.; Obeid, A.; Garg, S.; Vander, J.; Dunn, J.P.; et al. Endophthalmitis following cataract surgery: Visual outcomes, microbial spectrum and complications. Curr. Opin. Ophthalmol. 2023, 34, 237–242. [Google Scholar] [CrossRef]
- Yap, A.; Kaur, D.; Muttaiyah, S.; Welch, S.; Lightman, S.; Tomkins-Netzer, O.; Niederer, R.L. Impact of microorganism virulence on endophthalmitis outcomes. Br. J. Ophthalmol. 2024, 109, 347–352. [Google Scholar] [CrossRef]
- Durand, M.L. Bacterial and fungal endophthalmitis. Clin. Microbiol. Rev. 2017, 30, 597–613. [Google Scholar] [CrossRef]
- Kessel, L.; Flesner, P.; Andresen, J.; Erngaard, D.; Tendal, B.; Hjortdal, J. Antibiotic prevention of postcataract endophthalmitis: A systematic review and meta-analysis. Acta Ophthalmol. 2015, 93, 303–317. [Google Scholar] [CrossRef]
- Xu, K.; Chin, E.K.; Bennett, S.R.; Williams, D.F.; Ryan, E.H.; Dev, S.; Mittra, R.A.; Quiram, P.A.; Davies, J.B.; Parke, D.W.; et al. Endophthalmitis after intravitreal injection of vascular endothelial growth factor inhibitors: Management and visual outcomes. Ophthalmology 2018, 125, 1279–1286. [Google Scholar] [CrossRef]
- Callegan, M.C.; Engelbert, M.; Parke, D.W.; Jett, B.D.; Gilmore, M.S. Bacterial endophthalmitis: Epidemiology, therapeutics, and bacterium-host interactions. Clin. Microbiol. Rev. 2002, 15, 111–124. [Google Scholar] [CrossRef]
- Endophthalmitis Vitrectomy Study Group. Results of the endophthalmitis vitrectomy study. A randomized trial of immediate vitrectomy and of intravenous antibiotics for the treatment of postoperative bacterial endophthalmitis. Arch. Ophthalmol. 1995, 113, 1479–1496. [Google Scholar] [CrossRef]
- Welch, S.; Bhikoo, R.; Wang, N.; Siemerink, M.J.; Shew, W.; Polkinghorne, P.J.; Niederer, R.L. Better visual outcome associated with early vitrectomy in the management of endophthalmitis. Br. J. Ophthalmol. 2022, 106, 1145–1149. [Google Scholar] [CrossRef]
- Michael, E.; Welch, S.; Niederer, R.L. Rapid treatment of endophthalmitis with intravitreal antibiotics is associated with better vision outcomes. Clin. Exp. Ophthalmol. 2023, 51, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Hashida, N.; Nishida, K. Recent advances and future prospects: Current status and challenges of the intraocular injection of drugs for vitreoretinal diseases. Adv. Drug Deliv. Rev. 2023, 198, 114870. [Google Scholar] [CrossRef]
- Jian, H.J.; Chiou, Y.R.; Anand, A.; Chen, C.F.; Ma, D.H.; Lai, J.Y.; Huang, C.C.; Chang, H.T. Carbon-in-carbon: Hybrid carbonized nanomaterials with multifunctional activities for the treatment of endophthalmitis. Chem. Eng. J. 2024, 491, 151997. [Google Scholar] [CrossRef]
- Deshmukh, D.; Joseph, J.; Chakrabarti, M.; Sharma, S.; Jayasudha, R.; Sama, K.C.; Sontam, B.; Tyagi, M.; Narayanan, R.; Shivaji, S. New insights into culture negative endophthalmitis by unbiased next generation sequencing. Sci. Rep. 2019, 9, 844. [Google Scholar] [CrossRef]
- Qian, Z.; Zhang, Y.; Wang, L.; Li, Z.; Wang, H.; Kang, H.; Feng, J.; Hu, X.; Tao, Y. Application of metagenomic next-generation sequencing in suspected intraocular infections. Eur. J. Ophthalmol. 2023, 33, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Naik, P.; Gandhi, J.; Joseph, J. Recent advances and ongoing challenges in the diagnosis of culture negative endophthalmitis. Semin. Ophthalmol. 2023, 38, 92–98. [Google Scholar] [CrossRef]
- Low, L.; Nakamichi, K.; Akileswaran, L.; Lee, C.S.; Lee, A.Y.; Moussa, G.; Murray, P.I.; Wallace, G.R.; Van Gelder, R.N.; Rauz, S.; et al. Deep metagenomic sequencing for endophthalmitis pathogen detection using a nanopore platform. Am. J. Ophthalmol. 2022, 242, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Z.; Wang, M.; Fu, A.; Hao, X.; Guo, X.; Gu, J.; Jin, W.; Yang, A. The diagnostic utility of nanopore targeted sequencing in suspected endophthalmitis. Int. Ophthalmol. 2023, 43, 2653–2668. [Google Scholar] [CrossRef]
- Hao, X.; Wang, M.; Yuan, M.; Zhang, R.; Jin, W.; Yang, A. Identification of pathogens in the intraocular fluid samples of patients with endogenous endophthalmitis using rapid nanopore targeted sequencing. Retina 2023, 43, 606–615. [Google Scholar] [CrossRef] [PubMed]
- Jun, K.I.; Oh, B.L.; Kim, N.; Shin, J.Y.; Moon, J. Microbial diagnosis of endophthalmitis using nanopore amplicon sequencing. Int. J. Med. Microbiol. 2021, 311, 151505. [Google Scholar] [CrossRef] [PubMed]
- Omi, M.; Matsuo, Y.; Araki-Sasaki, K.; Oba, S.; Yamada, H.; Hirota, K.; Takahashi, K. 16S rRNA nanopore sequencing for the diagnosis of ocular infection: A feasibility study. BMJ Open Ophthalmol. 2022, 7, e000910. [Google Scholar] [CrossRef] [PubMed]
- Ciuffreda, L.; Rodríguez-Pérez, H.; Flores, C. Nanopore sequencing and its application to the study of microbial communities. Comput. Struct. Biotechnol. J. 2021, 19, 1497–1511. [Google Scholar] [CrossRef]
- Huang, Q.; Fu, A.; Wang, Y.; Zhang, J.; Zhao, W.; Cheng, Y. Microbiological diagnosis of endophthalmitis using nanopore targeted sequencing. Clin. Exp. Ophthalmol. 2021, 49, 1060–1068. [Google Scholar] [CrossRef]
- Eguchi, H.; Hotta, F.; Kusaka, S. Applying metagenomic analysis using nanopore sequencer (MinION) for precision medicine in bacterial keratoconjunctivitis: Comprehensive validation of molecular biological and conventional examinations. Int. J. Mol. Sci. 2023, 24, 2611. [Google Scholar] [CrossRef]
- Song, J.; Dong, H.; Wang, T.; Yu, H.; Yu, J.; Ma, S.; Song, X.; Sun, Q.; Xu, Y.; Liu, M. What is the impact of microbiota on dry eye: A literature review of the gut-eye axis. BMC Ophthalmol. 2024, 24, 262. [Google Scholar] [CrossRef]
- Segal, E.; Romano, A.; Eylan, E.; Stein, R. Fungal flora of the normal conjunctival sac. Mykosen 1977, 20, 9–14. [Google Scholar] [CrossRef]
- Ozkan, J.; Nielsen, S.; Diez-Vives, C.; Coroneo, M.; Thomas, T.; Willcox, M. Temporal stability and composition of the ocular surface microbiome. Sci. Rep. 2017, 7, 9880. [Google Scholar] [CrossRef]
- Shivaji, S.; Jayasudha, R.; Sai Prashanthi, G.; Kalyana Chakravarthy, S.; Sharma, S. The human ocular surface fungal microbiome. Investig. Ophthalmol. Vis. Sci. 2019, 60, 451–459. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, H.; Xia, T.; Huang, Y. Characterization of fungal microbiota on normal ocular surface of humans. Clin. Microbiol. Infect. 2020, 26, 123.e9–123.e13. [Google Scholar] [CrossRef] [PubMed]
- Prashanthi, G.S.; Jayasudha, R.; Chakravarthy, S.K.; Padakandla, S.R.; SaiAbhilash, C.R.; Sharma, S.; Bagga, B.; Murthy, S.I.; Garg, P.; Shivaji, S. Alterations in the ocular surface fungal microbiome in fungal keratitis patients. Microorganisms 2019, 7, 309. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Ogulur, I.; Mitamura, Y.; Yazici, D.; Pat, Y.; Bu, X.; Li, M.; Zhu, X.; Babayev, H.; Ardicli, S.; et al. The epithelial barrier theory and its associated diseases. Allergy 2024, 79, 3192–3237. [Google Scholar] [CrossRef] [PubMed]
- Jo, J.H.; Kennedy, E.A.; Kong, H.H. Topographical and physiological differences of the skin mycobiome in health and disease. Virulence 2017, 8, 324–333. [Google Scholar] [CrossRef]
- Zhang, S.D.; He, J.N.; Niu, T.T.; Chan, C.Y.; Ren, C.Y.; Liu, S.S.; Qu, Y.; Chong, K.L.; Wang, H.L.; Tao, J.; et al. Bacteriological profile of ocular surface flora in Meibomian gland dysfunction. Ocul. Surf. 2017, 15, 242–247. [Google Scholar] [CrossRef]
- Denoyer, A.; Godefroy, D.; Célérier, I.; Frugier, J.; Riancho, L.; Baudouin, F.; Rostène, W.; Baudouin, C. CX3CL1 expression in the conjunctiva is involved in immune cell trafficking during toxic ocular surface inflammation. Mucosal Immunol. 2012, 5, 702–711. [Google Scholar] [CrossRef]
- Corthésy, B. Multi-faceted functions of secretory IgA at mucosal surfaces. Front. Immunol. 2013, 4, 185. [Google Scholar] [CrossRef]
- Tajbakhsh, Z.; Golebiowski, B.; Stapleton, F.; Alghamdi, A.; Gray, P.E.; Altavilla, B.; Briggs, N.; Jalbert, I. Increased dendritic cell density and altered morphology in allergic conjunctivitis. Eye 2023, 37, 2896–2904. [Google Scholar] [CrossRef]
- Kubaisi, B.; Samra, K.; Syeda, S.; Schmidt, A.; Foster, S. Ocular allergy: An updated review. J. Allergy Immunol. 2017, 1, 002. [Google Scholar]
- Humeniuk, P.; Dubiela, P.; Hoffmann-Sommergruber, K. Dendritic cells and their role in allergy: Uptake, proteolytic processing and presentation of allergens. Int. J. Mol. Sci. 2017, 18, 1491. [Google Scholar] [CrossRef]
- Chigbu, D.G.I.; Karbach, N.J.; Abu, S.L.; Hehar, N.K. Cytokines in Allergic Conjunctivitis: Unraveling Their Pathophysiological Roles. Life 2024, 14, 350. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, H.; Takatsu, K. Role of cytokines in allergic airway inflammation. Int. Arch. Allergy Immunol. 2007, 142, 265–273. [Google Scholar] [CrossRef]
- Chen, J.; Bielory, L. Comparison of cytokine mediators in type 2 inflammatory conditions on the skin and ocular surface. Curr. Opin. Allergy Clin. Immunol. 2022, 22, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Triviño, F.J.; Ayén-Rodríguez, Á. Study of hypersensitivity to Malassezia furfur in patients with atopic dermatitis with head and neck pattern: Is it useful as a biomarker and therapeutic indicator in these patients? Life 2022, 12, 299. [Google Scholar] [CrossRef]
- Alofi, R.M.; Alrohaily, L.S.; Alharthi, N.N.; Almouteri, M.M. Ocular manifestations in seborrheic dermatitis epidemiology, clinical features, and management: A comprehensive review. Cureus 2024, 16, e70335. [Google Scholar] [CrossRef]
- Sugita, S.; Takase, H.; Nakano, S. Role of recent PCR tests for infectious ocular diseases: From laboratory-based studies to the clinic. Int. J. Mol. Sci. 2023, 24, 8146. [Google Scholar] [CrossRef] [PubMed]
- Santos, H.N.V.D.; Ferracioli-Oda, E.; Barbosa, T.S.; Otani, C.S.V.; Tanaka, T.; Silva, L.C.S.D.; Lopes, G.O.; Doi, A.; Hirata, C.E.; Yamamoto, J.H. Usefulness of aqueous and vitreous humour analysis in infectious uveitis. Clinics 2020, 75, e1498. [Google Scholar] [CrossRef]
- Nafea, A.M.; Wang, Y.; Wang, D.; Salama, A.M.; Aziz, M.A.; Xu, S.; Tong, Y. Application of next-generation sequencing to identify different pathogens. Front. Microbiol. 2023, 14, 1329330. [Google Scholar] [CrossRef] [PubMed]
- Kishore, K.; McGowan, D.S.; Chatterjee, T.; Hassanzadeh, B. A case of bilateral endogenous Candida dubliniensis endophthalmitis treated with aggressive local and systemic therapy. Case Rep. Ophthalmol. 2020, 11, 561–573. [Google Scholar] [CrossRef]
- Haseeb, A.A.; Elhusseiny, A.M.; Siddiqui, M.Z.; Ahmad, K.T.; Sallam, A.B. Fungal endophthalmitis: A comprehensive review. J. Fungi 2021, 7, 996. [Google Scholar] [CrossRef]
- Peng, K.L.; Kung, Y.H.; Tsai, H.S.; Wu, T.T. Treatment outcomes of acute postoperative infectious endophthalmitis. BMC Ophthalmol. 2021, 21, 384. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Soto, M.C.; Bonifaz, A. Ocular fungal infections. J. Fungi. 2022, 8, 1078. [Google Scholar] [CrossRef]
- Jiang, S.; Chen, Y.; Han, S.; Lv, L.; Li, L. Next-generation sequencing applications for the study of fungal pathogens. Microorganisms 2022, 10, 1882. [Google Scholar] [CrossRef] [PubMed]
- Lema, N.K.; Gemeda, M.T.; Woldesemayat, A.A. Recent advances in metagenomic approaches, applications, and challenges. Curr. Microbiol. 2023, 80, 347. [Google Scholar] [CrossRef]
- Petrillo, F.; Petrillo, A.; Sasso, F.P.; Schettino, A.; Maione, A.; Galdiero, M. Viral infection and antiviral treatments in ocular pathologies. Microorganisms 2022, 10, 2224. [Google Scholar] [CrossRef]
- Koganti, R.; Yadavalli, T.; Naqvi, R.A.; Shukla, D.; Naqvi, A.R. Pathobiology and treatment of viral keratitis. Exp. Eye Res. 2021, 205, 108483. [Google Scholar] [CrossRef]
- Hoffman, J.; Foster, A. Viral diseases of the eye. Community Eye Health 2020, 33, 65–67. [Google Scholar] [PubMed]
- Zimmerman, K.; Kearns, F.; Tzekov, R. Natural protection of ocular surface from viral infections—A hypothesis. Med. Hypotheses 2020, 143, 110082. [Google Scholar] [CrossRef]
- Gershon, A.A.; Breuer, J.; Cohen, J.I.; Cohrs, R.J.; Gershon, M.D.; Gilden, D.; Grose, C.; Hambleton, S.; Kennedy, P.G.E.; Oxman, M.N.; et al. Varicella zoster virus infection. Nat. Rev. Dis. Primers 2015, 1, 15016. [Google Scholar] [CrossRef]
- Bialasiewicz, A. Adenoviral keratoconjunctivitis. Sultan Qaboos Univ. Med. J. 2007, 7, 15–23. [Google Scholar] [CrossRef]
- Nichols, W.G.; Peck Campbell, A.J.; Boeckh, M. Respiratory viruses other than influenza virus: Impact and therapeutic advances. Clin. Microbiol. Rev. 2008, 21, 274–290. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Shimomura, Y.; Kinoshita, S.; Nishida, K.; Yamamoto, R.; Tano, Y. Detection of herpes simplex virus DNA in human tear film by the polymerase chain reaction. Am. J. Ophthalmol. 1994, 117, 160–163. [Google Scholar] [CrossRef]
- Rajalakshmy, A.R.; Malathi, J.; Madhavan, H.N.; Bhaskar, S.; Iyer, G.K. Patients with dry eye without hepatitis C virus infection possess the viral RNA in their tears. Cornea 2015, 34, 28–31. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Iyer, G.; Srinivasan, B.; Agarwal, S.; Kuila, J. Severe acute onset dry eye following presumed Epstein-Barr viral infection. Indian J. Ophthalmol. 2020, 68, 642–644. [Google Scholar] [CrossRef]
- Blyden, K.; Thomas, J.; Emami-Naeini, P.; Fashina, T.; Conrady, C.D.; Albini, T.A.; Carag, J.; Yeh, S. Emerging infectious diseases and the eye: Ophthalmic manifestations, pathogenesis, and one health perspectives. Int. Ophthalmol. Clin. 2024, 64, 39–54. [Google Scholar] [CrossRef]
- Letafati, A.; Jazayeri, S.M.; Atwan, H.; Mahmoudi, M.K.; Sarrafzadeh, S.; Ardekani, O.S.; Norouzi, M.; Ghaziasadi, A. Utilization of multiplex polymerase chain reaction for simultaneous and rapid detection of viral infections from different ocular structures. Sci. Rep. 2024, 14, 17997. [Google Scholar] [CrossRef] [PubMed]
- Siegal, N.; Gutowski, M.; Akileswaran, L.; Beauchamp, N.J.; Ding, L.-C.; Chambers, C.B.; Van Gelder, R.N. Elevated levels of Merkel cell polyoma virus in the anophthalmic conjunctiva. Sci. Rep. 2021, 11, 15366. [Google Scholar] [CrossRef]
- Doan, T.; Hinterwirth, A.; Worden, L.; Arzika, A.M.; Maliki, R.; Chen, C.; Zhong, L.; Zhou, Z.; Acharya, N.R.; Porco, T.C.; et al. Post-antibiotic ocular surface microbiome in children: A cluster-randomized trial. Ophthalmology 2020, 127, 1127–1130. [Google Scholar] [CrossRef]
- Nakano, S.; Sugita, S.; Tomaru, Y.; Hono, A.; Nakamuro, T.; Kubota, T.; Takase, H.; Mochizuki, M.; Takahashi, M.; Shimizu, N. Establishment of multiplex solid-phase strip PCR test for detection of 24 ocular infectious disease pathogens. Investig. Ophthalmol. Vis. Sci. 2017, 58, 1553–1559. [Google Scholar] [CrossRef]
- Nakano, S.; Tomaru, Y.; Kubota, T.; Takase, H.; Mochizuki, M.; Shimizu, N.; Sugita, S.; Direct Strip Polymerase Chain Reaction Project Groups. Multiplex solid-phase real-time polymerase chain reaction without DNA extraction: A rapid intraoperative diagnosis using microvolumes. Ophthalmology 2021, 128, 729–739. [Google Scholar] [CrossRef]
- Quentin, C.D.; Reiber, H. Fuchs heterochromic cyclitis: Rubella virus antibodies and genome in aqueous humour. Am. J. Ophthalmol. 2004, 138, 46–54. [Google Scholar] [CrossRef]
- Gallon, P.; Parekh, M.; Ferrari, S.; Fasolo, A.; Ponzin, D.; Borroni, D. Metagenomics in ophthalmology: Hypothesis or real prospective? Biotechnol. Rep. 2019, 23, e00355. [Google Scholar] [CrossRef] [PubMed]
- Arunasri, K.; Mahesh, M.; Sai Prashanthi, G.; Jayasudha, R.; Kalyana Chakravarthy, S.; Tyagi, M.; Pappuru, R.R.; Shivaji, S. Comparison of the vitreous fluid bacterial microbiomes between individuals with post fever retinitis and healthy controls. Microorganisms 2020, 8, 751. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.Y.; Akileswaran, L.; Tibbetts, M.D.; Garg, S.J.; Van Gelder, R.N. Identification of torque Teno virus in culture-negative endophthalmitis by representational deep-DNA sequencing. Ophthalmology 2014, 122, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Hong, B.; Kasi, S.K.; Aderman, C.; Talcott, K.E.; Adam, M.K.; Yue, B.; Akileswaran, L.; Nakamichi, K.; Wu, Y.; et al. Prognostic utility of whole-genome sequencing and polymerase chain reaction tests of ocular fluids in postprocedural endophthalmitis. Am. J. Ophthalmol. 2020, 217, 325–334. [Google Scholar] [CrossRef]
- Naik, P.; Dave, V.P.; Joseph, J. Detection of Torque Teno Virus (TTV) and TTV-Like Minivirus in patients with presumed infectious endophthalmitis in India. PLoS ONE 2020, 15, e0227121. [Google Scholar] [CrossRef]
- Komatsu, H.; Usui, Y.; Sukeda, A.; Yamamoto, Y.; Ohno, S.-I.; Goto, K.; Kuroda, M.; Nagao, T.; Goto, H. Prevalence of Merkel cell polyomavirus in primary eyelid Merkel cell carcinomas and association with clinicopathological features. Am. J. Ophthalmol. 2023, 249, 49–56. [Google Scholar] [CrossRef]
- Roux, S.; Adriaenssens, E.M.; Dutilh, B.E.; Koonin, E.V.; Kropinski, A.M.; Krupovic, M.; Kuhn, J.H.; Lavigne, R.; Brister, J.R.; Varsani, A.; et al. Minimum information about an uncultivated virus genome (MIUViG). Nat. Biotechnol. 2019, 37, 29–37. [Google Scholar] [CrossRef]
- Nayfach, S.; Camargo, A.P.; Schulz, F.; Eloe-Fadrosh, E.; Roux, S.; Kyrpides, N.C. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat. Biotechnol. 2021, 39, 578–585. [Google Scholar] [CrossRef]
- Wishart, D.S.; Han, S.; Saha, S.; Oler, E.; Peters, H.; Grant, J.R.; Stothard, P.; Gautam, V. PHASTEST: Faster than PHASTER, better than PHAST. Nucleic Acids Res. 2023, 51, W443–W450. [Google Scholar] [CrossRef]
- Camargo, A.P.; Roux, S.; Schulz, F.; Babinski, M.; Xu, Y.; Hu, B.; Chain, P.S.G.; Nayfach, S.; Kyrpides, N.C. Identification of mobile genetic elements with geNomad. bioRxiv 2023, 42, 1303–1312. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. Blast+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Camargo, A.P.; Nayfach, S.; Chen, I.A.; Palaniappan, K.; Ratner, A.; Chu, K.; Ritter, S.J.; Reddy, T.B.K.; Mukherjee, S.; Schulz, F.; et al. IMG/VR v4: An expanded database of uncultivated virus genomes within a framework of extensive functional, taxonomic, and ecological metadata. Nucleic Acids Res. 2023, 51, D733–D743. [Google Scholar] [CrossRef]
| Generation | First-Generation Sequencing | Second-Generation Sequencing | Third-Generation Sequencing |
|---|---|---|---|
| Detection methods | Sanger sequencing capillary electrophoresis | Bridge PCR Sequencing by synthesis | DNA elongation in a microwell Nanopore sequencing |
| Since | Late 1990s | ~2005 | ~2017 |
| PCR | Yes | Yes | No |
| Read lengths | ~1000 bp | 100–200 bp | >10 Kbp |
| Characteristics | Low reading error rate and relatively long read length approach | Short read length approach Large-scale parallel sequencing Reduced run costs Improved analysis speed | Long read length approach; Fastest sequencer; Whole-genome scan within 15 min |
| Disadvantages | High run costs Low analysis speed | Identify microorganisms at the genus level Laboratory-based study | Relatively high error rates |
| Sequencing accuracy (Q score) | >Q20 | >Q31 | >Q21 |
| Representative equipment | SeqStudio Genetic Analyzer (Thermo Fisher Scientific) | 454 sequencing (454 Life Sciences) NovaSeq (Illumina) Ion Torrent Sequencing (Thermo Fisher) | Sequel II (Pacific Biosciences) MinION platform (Oxford Nanopore Technologies) |
| Authors | Participants | Locations | Major Findings | References | Category | Generation |
|---|---|---|---|---|---|---|
| Zhou Y et al., 2014 | 105 healthy volunteers 115 patients with suspected trachoma | Conjunctiva | In trachomatous disease, changes in the conjunctival microbiome could occur | [140] | Bacterial Microbiome | Second |
| Doan T et al., 2016 | 107 healthy volunteers | Conjunctiva | On the healthy ocular surface, Corynebacteria, Propionibacteria, and coagulase-negative Staphylococci were the predominant organisms. TTV were also detected. | [164] | Bacterial Microbiome Virome | Second |
| Deng Y et al., 2021 | 41 patients with cataract, glaucoma and AMD | Conjunctiva Aqueous Humor Plasma Skin | Cutibacterium acnes was the most abundant. Complex community of bacteria might be present inside the eyes. | [195] | Bacterial Microbiome | Second |
| Deshmukh D et al., 2019 | 34 patients with endophthalmitis 30 participates with non-infectious retinal disorders as controls | Vitreous Fluid | Culture-based diagnosis was achieved in 44% of cases. NGS diagnosed the presence of microbes in 88% of cases. | [214] | Bacterial Microbiome Mycobiome | Second |
| Low L et al., 2022 | 23 patients with suspected endophthalmitis | Aqueous Humor Vitreous Fluid | At genus level, the coincidence between culture and 16S Nanopore, Nanopore WGS, and Illumina WGS were 75%, 100%, and 78%, respectively. | [217] | Bacterial Microbiome | Second and Third |
| Hao X et al., 2023 | 34 eyes with endogenous endophthalmitis | Aqueous Humor Vitreous Fluid | NTS and culture detected pathogens in 89.28% and 35.71% of cases. The average detection time of NTS (1.11 days) was shorter than that of culture (2.50 days). | [219] | Bacterial Microbiome Mycobiome | Third |
| Eguchi H et al., 2023 | 8 patients with clinically diagnosed bacterial keratoconjunctivitis | Ocular Surface Specimens | In 66% of culture-negative cases, the smear positivity closely resembled the MinION results. In 80% of culture-positive cases, culture and sequencing results were consistent. | [224] | Bacterial Microbiome | Third |
| Shivaji S et al., 2019 | 34 healthy volunteers | Conjunctiva | The genera Aspergillus, Setosphaeria, Malassezia, and Haematonectria were present. | [228] | Mycobiome | Second |
| Wang Y et al., 2020 | 90 healthy volunteers | Conjunctiva | Two phyla, Basidiomycota and Ascomycota, and five genera, Malassezia, Rhodotorula, Davidiella, Aspergillus, and Alternaria, were identified, accounting for >80% of the fungal microbiome. | [229] | Mycobiome | Second |
| Prashanthi GS et al., 2019 | 25 healthy controls 35 patients with fungal keratitis patients | Conjunctiva Corneal Epithelium | Alteration in the fungal microbiota was observed both at the phylum and genera levels. The ocular microbiome analysis identified 11 genera. | [230] | Mycobiome | Second |
| Siegal N et al., 2021 | 20 anophthalmic and 20 fellow-eye | Conjunctiva | TTV and MCPyV were detected frequently in healthy and anophthalmic conjunctiva. | [265] | Bacterial Microbiome Virome | Second |
| Doan T et al., 2020 | Not available | Conjunctiva | The structure of the ocular surface virome was not altered after azithromycin treatment | [266] | Bacterial Microbiome Virome | Second |
| Arunasri K et al., 2020 | 19 healthy volunteers 9 patients with post fever retinitis | Vitreous Fluid | An increase in abundance of anti-inflammatory and antimicrobial genera and decrease in proinflammatory genera were detected compared with that in healthy controls. | [271] | Bacterial Microbiome | Second |
| Lee et al., 2014 | 21 patients with presumed infectious endophthalmitis 7 patients with noninfectious retinal disorders or culture-negative endophthalmitis | Aqueous Humor Vitreous Fluid | 57.1% of culture-positive and 100% of culture-negative samples demonstrated the presence of TTV. | [272] | Bacterial Microbiome Virome | Second |
| Lee CS et al., 2020 | 50 patients with postprocedural endophthalmitis | Aqueous Humor Vitreous Fluid | In post-procedure endophthalmitis, a higher load of bacteria other than that of S. epidermidis and the presence of TTV DNA are associated with worse outcomes. | [273] | Bacterial Microbiome Virome | Second |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asao, K.; Hashida, N. Overview of Microorganisms: Bacterial Microbiome, Mycobiome, Virome Identified Using Next-Generation Sequencing, and Their Application to Ophthalmic Diseases. Microorganisms 2025, 13, 1300. https://doi.org/10.3390/microorganisms13061300
Asao K, Hashida N. Overview of Microorganisms: Bacterial Microbiome, Mycobiome, Virome Identified Using Next-Generation Sequencing, and Their Application to Ophthalmic Diseases. Microorganisms. 2025; 13(6):1300. https://doi.org/10.3390/microorganisms13061300
Chicago/Turabian StyleAsao, Kazunobu, and Noriyasu Hashida. 2025. "Overview of Microorganisms: Bacterial Microbiome, Mycobiome, Virome Identified Using Next-Generation Sequencing, and Their Application to Ophthalmic Diseases" Microorganisms 13, no. 6: 1300. https://doi.org/10.3390/microorganisms13061300
APA StyleAsao, K., & Hashida, N. (2025). Overview of Microorganisms: Bacterial Microbiome, Mycobiome, Virome Identified Using Next-Generation Sequencing, and Their Application to Ophthalmic Diseases. Microorganisms, 13(6), 1300. https://doi.org/10.3390/microorganisms13061300





