Additions to Pleosporalean Taxa Associated with Xanthoceras sorbifolium from Jilin and Hebei, China
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, Isolation, and Morphological Observation
2.2. DNA Extraction, PCR Amplification, and Sequencing
2.3. Molecular Phylogeny
| Species | Strain/Isolate | GenBank Accession Numbers | |||
|---|---|---|---|---|---|
| ITS | LSU | SSU | tub2 | ||
| Alloleptosphaeria italica | MFLUCC 14-0934 | KT454722 | KT454714 | _ | _ |
| A. clematidis | MFLUCC 17-2071 | MT310604 | MT214557 | MT226674 | _ |
| A. iridicola | CBS 143395 | MH107919 | MH107965 | _ | NA |
| A. shangrilana | HKAS:112210 | MW431059 | MW431315 | MW431058 | NA |
| A. xanthoceratis | CCMJ 13066 | PP151694 | PP153449 | PV569760 | PV670045 |
| Didymella exigua | CBS 183.55 | GU237794 | EU754155 | EU754056 | GU237525 |
| D. rumicicola | CBS 683.79 | KT389503 | KT389721 | _ | KT389800 |
| Heterospora chenopodii | CBS 448.68 | FJ427023 | EU754187 | EU754088 | _ |
| H. chenopodii | CBS 115.96 | JF740227 | EU754188 | EU754089 | _ |
| H. dimorphospora | CBS 165.78 | JF740204 | JF740281 | JF740098 | _ |
| H. dimorphospora | CBS 345.78 | JF740203 | GU238069 | GU238213 | _ |
| Lep. conoidea | CBS 616.75 | JF740201 | JF740279 | _ | KT389804 |
| Lep. doliolum | CBS 155.94 | JF740207 | JF740282 | _ | JF740146 |
| Lep. doliolum | CBS 505.75 | JF740205 | GQ387576 | GQ387515 | JF740144 |
| Lep. ebuli | MFLUCC 14-0828 | KP744446 | KP744488 | KP753954 | _ |
| Lep. irregularis | MFLUCC 15-1118 | KX856056 | KX856055 | _ | _ |
| Lep. slovacica | CBS 389.80 | JF740247 | JF740315 | JF740101 | _ |
| Lep. urticae | MFLU 18-0591 | MK123333 | MK123332 | MK123329 | _ |
| Neoleptosphaeria jonesii | MFLUCC 16-1442 | KY211869 | KY211870 | KY211871 | _ |
| Neol. rubefaciens | CBS 223.77 | JF740243 | JF740312 | _ | _ |
| Neol. rubefaciens | CBS 387.80 | JF740242 | JF740311 | _ | _ |
| Pseudoleptosphaeria etheridgei | CBS 125980 | JF740221 | JF740291 | _ | _ |
| Querciphoma carteri | CBS 105.91 | KF251209 | GQ387594 | GQ387533 | KF252700 |
| Q. carteri | CBS 101633 | KF251210 | GQ387593 | GQ387532 | KF252701 |
| Sclerenchymomyces clematidis | MFLUCC 17-2180 | MT310605 | MT214558 | MT226675 | _ |
| Species | Strain/Isolate | GenBank Accession Numbers | ||||
|---|---|---|---|---|---|---|
| ITS | LSU | SSU | tef1-α | rpb2 | ||
| Crassiclypeus aquaticus | KH 104 | LC312499 | LC312528 | LC312470 | LC312557 | LC312586 |
| C. aquaticus | KT 970 | LC312501 | LC312530 | LC312472 | LC312559 | LC312588 |
| Dimorphiopsis brachystegiae | CPC 22679 | KF777160 | KF777213 | _ | _ | _ |
| Flabellascoma aquaticum | KUMCC 15-0258 | MN304827 | MN274564 | MN304832 | MN328898 | MN328895 |
| F. cycadicola | KT 2034 | LC312502 | LC312531 | LC312473 | LC312560 | LC312589 |
| F. fusiforme | MFLUCC 18-1584 | MN304830 | MN274567 | _ | MN328902 | _ |
| F. minimum | KT 2013 | LC312503 | LC312532 | LC312474 | LC312561 | LC312590 |
| F. minimum | KT 2040 | LC312504 | LC312533 | LC312475 | LC312562 | LC312591 |
| Lentistoma bipolare | KT 3056 | LC312513 | LC312542 | LC312484 | LC312571 | LC312600 |
| Len. bipolare | CBS 115375 | LC312506 | LC312535 | LC312477 | LC312564 | LC312593 |
| Leptoparies palmarum | KT 1653 | LC312514 | LC312543 | LC312485 | LC312572 | LC312601 |
| Lophiostoma arundinis | KT 606 | JN942964 | AB618998 | AB618679 | LC001737 | JN993482 |
| Lop. arundinis | KT 651 | JN942965 | AB618999 | AB618680 | LC001738 | JN993486 |
| Lop. biappendiculatum | KT 975 | _ | GU205228 | GU205254 | _ | _ |
| Lop. biappendiculatum | KT 1124 | _ | GU205227 | GU205256 | _ | _ |
| Lop. caespitosum | CBS 147391 | MW759252 | MW750387 | _ | MW752404 | MW752383 |
| Lop. caespitosum | MFLUCC 13-0442 | KP899134 | KP888639 | KP899125 | KR075161 | _ |
| Lop. caespitosum | MFLUCC 14-0993 | KP899135 | KP888640 | KP899126 | KR075162 | _ |
| Lop. carabassense | CBS 149324 | MT679671 | OL544969 | OL544968 | OL554876 | _ |
| Lop. carpini | CBS 147279 | MW759258 | MW750386 | _ | MW752405 | MW752384 |
| Lop. caryophyllacearum | MFLUCC 17-0749 | MG828964 | MG829076 | MG829176 | MG829238 | _ |
| Lop. caudatum | KT 530 | LC001723 | AB619000 | AB618681 | LC001739 | _ |
| Lop. caulium | KT 603 | LC001724 | AB619001 | AB618682 | LC001740 | _ |
| Lop. caulium | KT 633 | LC001725 | AB619002 | AB618683 | LC001741 | _ |
| Lop. clavatum | CBS 147278 | MW759259 | MW750385 | _ | MW752406 | MW752385 |
| Lop. clavatum | MFLUCC 18-1316 | _ | MN274566 | MN304835 | MN328901 | _ |
| Lop. clematidicola | MFLUCC 16-0446 | MT310609 | MT214563 | MT226680 | MT394742 | _ |
| Lop. clematidis | MFLUCC 17-2081 | MN393004 | MT214562 | MT226679 | MT394741 | MT394689 |
| Lop. clematidis-subumbellatae | MFLUCC 17-2063 | MT310607 | MT214560 | MT226677 | MT394739 | MT394687 |
| Lop. clematidis-vitalbae | MFLUCC 16-1368 | MT310610 | MT214564 | MT226681 | MT394743 | _ |
| Lop. compressum | CBS 147276 | MW759272 | MW750382 | _ | MW752408 | MW752381 |
| Lop. compressum | IFRD 2014 | _ | FJ795437 | FJ795480 | _ | FJ795457 |
| Lop. cornisporum | KH 322 | LC312515 | LC312544 | LC312486 | LC312573 | LC312602 |
| Lop. coronillae | MFLUCC 14-0941 | KT026120 | KT026112 | KT026116 | _ | _ |
| Lop. crenatum | AFTOL-ID 1581 | _ | DQ678069 | DQ678017 | DQ677912 | DQ677965 |
| Lop. dictyosporum | CBS 147389 | MW759251 | MW750379 | _ | MW752411 | MW752388 |
| Lop. erumpens | CBS 147275 | MW759262 | MW750381 | _ | MW752409 | MW752386 |
| Lop. fusisporum | CBS 147891 | MW759253 | _ | _ | MW752401 | MW752382 |
| Lop. helichrysi | IT-1296 | KT333435 | KT333436 | KT333437 | KT427535 | _ |
| Lop. heterosporum | AFTOL-ID 1036 | GQ203795 | AY016369 | _ | DQ497609 | DQ497615 |
| Lop. japonicum | KT 686-1 | LC001729 | AB619006 | AB618687 | LC001745 | _ |
| Lop. japonicum | KT 573 | LC001728 | AB619005 | AB618686 | LC001744 | _ |
| Lop. jonesii | GAAZ 54-1 | KX687757 | KX687753 | KX687755 | KX687759 | _ |
| Lop. jonesii | GAAZ 54-2 | KX687758 | KX687754 | KX687756 | KX687760 | _ |
| Lop. jotunheimenense | CBS 147522 | MW759261 | MW750394 | _ | MW752392 | _ |
| Lop. junci | MFLUCC 14-0938 | MG828966 | MG829078 | MG829178 | NA | _ |
| Lop. khanzada-kirgizbaeva | TASM 6158 | MZ966265 | OK017520 | OK017525 | MZ997338 | _ |
| Lop. khanzada-kirgizbaeva | TASM 6164 | MZ966266 | OK017521 | OK017526 | MZ997339 | _ |
| Lop. longiappendiculatum | MFLUCC 17-1452 | MT214368 | MT214462 | MT214415 | MT235783 | _ |
| Lop. longiappendiculatum | MFLUCC 17-1457 | MT214369 | MT214463 | MT214416 | MT235784 | MT235821 |
| Lop. macrostomoides | CBS 147523 | MW759256 | MW750389 | _ | _ | _ |
| Lop. macrostomoides | CBS 147277 | MW759257 | MW750384 | _ | MW752407 | MW752380 |
| Lop. macrostomoides | CBS 123097 | _ | FJ795439 | FJ795482 | GU456277 | FJ795458 |
| Lop. macrostomum | KT 508 | JN942961 | AB619010 | _ | LC001751 | JN993491 |
| Lop. macrostomum | KT 709/HHUF 27293 | AB433276 | AB433274 | AB521732 | LC001753 | JN993493 |
| Lop. macrostomum | KT 635/HHUF 27290 | AB433275 | AB433273 | AB521731 | LC001752 | JN993484 |
| Lop. mangiferae | MFLUCC 17-2651 | MG931031 | MG931025 | MG931028 | _ | _ |
| Lop. mangiferae | MFLUCC 17-2653 | MG931032 | MG931026 | MG931029 | _ | _ |
| Lop. medicaginicola | MFLUCC 17-0681 | MG828967 | MG829079 | MG829179 | _ | _ |
| Lop. montanae | MFLUCC 16-0999 | MT310611 | MT214565 | MT226682 | MT394744 | _ |
| Lop. montanae | UESTCC 23.0038 | OR253137 | OR253296 | OR253209 | OR263570 | OR253750 |
| Lop. montanae | UESTCC 23.0039 | OR253138 | OR253297 | OR253210 | OR251148 | OR253751 |
| Lop. montanae | CCMJ 13067 | PV569791 | PV569911 | PV569764 | PV670048 | PV670046 |
| Lop. montanae | CCMJ 13068 | PV569792 | PV569912 | PV569765 | PV670049 | PV670047 |
| Lop. multiforme | CCMJ 13069 | PP151705 | PP153460 | PV569761 | PV670043 | PV670041 |
| Lop. multiforme | CCMJ 13070 | PP151706 | PP153461 | PV569762 | PV670044 | PV670042 |
| Lop. multiseptatum | CBS 623.86 | _ | GU301833 | GU296163 | _ | GU371791 |
| Lop. multiseptatum | KT 604 | LC001726 | AB619003 | AB618684 | LC001742 | _ |
| Lop. neomuriforme | MFLUCC 13-0744 | KY496740 | KY496719 | KY501110 | _ | _ |
| Lop. obtusisporum | KT 3098 | LC312519 | LC312548 | LC312490 | LC312577 | LC312606 |
| Lop. obtusisporum | KT 2838 | LC312518 | LC312547 | LC312489 | LC312576 | LC312605 |
| Lop. oleae | CGMCC 3.24426 | OR253081 | OR253233 | OR253172 | OR262141 | OR262130 |
| Lop. oleae | UESTCC 23.0036 | OR253079 | OR253231 | OR253171 | OR262139 | OR262129 |
| Lop. ononidis | MFLUCC 14-0613 | KU243128 | KU243125 | KU243126 | KU243127 | _ |
| Lop. paramacrostomum | MFLUCC 11-0463 | _ | KP888636 | KP899122 | _ | _ |
| Lop. plantaginis | CBS 147527 | MW759250 | MW750378 | _ | _ | MW752375 |
| Lop. pseudodictyosporium | MFLUCC 13-0451 | KR025858 | KR025862 | _ | _ | _ |
| Lop. pseudomacrostomum | CBS 147525 | MW759255 | MW750391 | _ | MW752395 | _ |
| Lop. pseudomacrostomum | CBS 147526 | MW759254 | MW750392 | _ | MW752394 | _ |
| Lop. ravennicum | MFLUCC 14-0005 | KP698413 | KP698414 | KP698415 | _ | _ |
| Lop. rosae-ecae | MFLUCC 17-0807 | MG828924 | MG829033 | MG829139 | MG829217 | _ |
| Lop. rosicola | MFLU 15-1888 | MG828968 | MG829080 | MG829180 | MG829240 | _ |
| Lop. sagittiforme | KT 1934 | AB369268 | AB369267 | AB618693 | LC001756 | _ |
| Lop. scabridisporum | BCC 22835 | _ | GQ925844 | GQ925831 | GU479857 | GU479830 |
| Lop. scabridisporum | BCC 22836 | _ | GQ925845 | GQ925832 | GU479856 | GU479829 |
| Lop. scrophulariicola | MFLUCC 17-0689 | MG828969 | MG829081 | _ | _ | _ |
| Lop. semiliberum | KT 622 | JN942966 | AB619012 | AB618694 | LC001757 | JN993483 |
| Lop. semiliberum | KT 652 | JN942967 | AB619013 | AB618695 | LC001758 | JN993485 |
| Lop. semiliberum | KT 828 | JN942970 | AB619014 | AB618696 | LC001759 | JN993489 |
| Lop. spartii-juncei | MFLUCC 13-0351 | KP899136 | KP888641 | KP899127 | KR075163 | _ |
| Lop. submuriforme | CBS 147274 | MW759260 | MW750380 | _ | MW752410 | MW752387 |
| Lop. terricola | SC-12 | JN662930 | JX985750 | JX985749 | _ | _ |
| Lop. thymi | MFLU 15-2131 | MG828970 | MG829082 | MG829182 | MG829241 | _ |
| Lop. tropicum | KH 352 | LC312521 | LC312550 | LC312492 | LC312579 | LC312608 |
| Lop. tropicum | KT 3134 | LC312522 | LC312551 | LC312493 | LC312580 | LC312609 |
| Lop. vitigenum | HH 26930 | LC001735 | AB619015 | AB618697 | LC001761 | _ |
| Lop. vitigenum | HH 26931 | LC001736 | AB619016 | AB618698 | LC001762 | _ |
| Lop. winteri | KT 740 | JN942969 | AB619017 | AB618699 | LC001763 | JN993487 |
| Lop. winteri | KT 764 | JN942968 | AB619018 | AB618700 | LC001764 | JN993488 |
| Neovaginatispora clematidis | MFLUCC 17-2149 | MT310606 | MT214559 | MT226676 | MT394738 | _ |
| Neov. fuckelii | MFLUCC 17-1334 | MN304828 | MN274565 | MN304833 | MN328899 | MN328896 |
| Neov. fuckelii | KT 634 | LC001732 | AB619009 | AB618690 | LC001750 | _ |
| Parapaucispora pseudoarmatispora | KT 2237 | LC100021 | LC100026 | LC100018 | LC100030 | _ |
| Paucispora kunmingense | MFLUCC 17-0932 | MF173432 | MF173428 | MF173430 | MF173434 | MF173436 |
| Pa. quadrispora | KH 448 | LC001733 | LC001722 | LC001720 | LC001754 | _ |
| Pa. quadrispora | KT 843 | LC001734 | AB619011 | AB618692 | LC001755 | _ |
| Pa. versicolor | KH 110 | AB918731 | AB918732 | LC001721 | LC001760 | _ |
| Pa. xishanensis | HKAS 115905 | MZ966267 | OK017522 | OK017527 | MZ997340 | _ |
| Pa. xishanensis | HKAS 115906 | MZ966268 | OK017523 | OK017528 | MZ997341 | _ |
| Platystomum actinidiae | KT 521 | JN942963 | JN941380 | JN941375 | LC001747 | JN993490 |
| Pl. actinidiae | KT 534 | JN942962 | JN941379 | JN941376 | LC001748 | JN993492 |
| Pl. crataegi | MFLUCC 14-0925 | KT026117 | KT026109 | KT026113 | KT026121 | _ |
| Pl. rosae | MFLUCC 15-0633 | KT026119 | KT026111 | KT026115 | _ | _ |
| Pl. salicicola | MFLUCC 15-0632 | KT026118 | KT026110 | KT026114 | _ | _ |
| Pseudopaucispora brunneospora | KH 227 | LC312523 | LC312552 | LC312494 | LC312581 | LC312610 |
| Vaginatispora amygdali | KT 2248 | LC312524 | LC312553 | LC312495 | LC312582 | LC312611 |
| V. amygdali | MFLUCC 18-1526 | MK085055 | MK085059 | MK085057 | MK087657 | _ |
| V. appendiculata | MFLUCC 16-0314 | KU743217 | KU743218 | KU743219 | KU743220 | _ |
| V. appendiculata | MFLUCC 13-0835 | _ | KY264745 | KY264749 | _ | _ |
| V. aquatica | MFLUCC 11-0083 | KJ591577 | KJ591576 | KJ591575 | _ | _ |
| V. armatispora | MFLUCC 18-0247 | MK085056 | MK085060 | MK085058 | MK087658 | MK087669 |
| V. armatispora | MFLUCC 18-0213 | MN304826 | MN274563 | MN304831 | MN328897 | MN328894 |
| V. microarmatispora | MTCC 12733 | MF142592 | MF142593 | MF142594 | MF142595 | MF142596 |
| V. scabrispora | KT 2443 | LC312525 | LC312554 | LC312496 | LC312583 | LC312612 |
| Teichospora rubriostiolata | TR7 | KU601590 | _ | _ | KU601609 | KU601599 |
| T. trabicola | C134 | KU601591 | _ | _ | KU601601 | KU601600 |
3. Results
3.1. Phylogenetic Analyses
3.1.1. Phylogenetic Analyses of Alloleptosphaeria

3.1.2. Phylogenetic Analyses of Lophiostoma
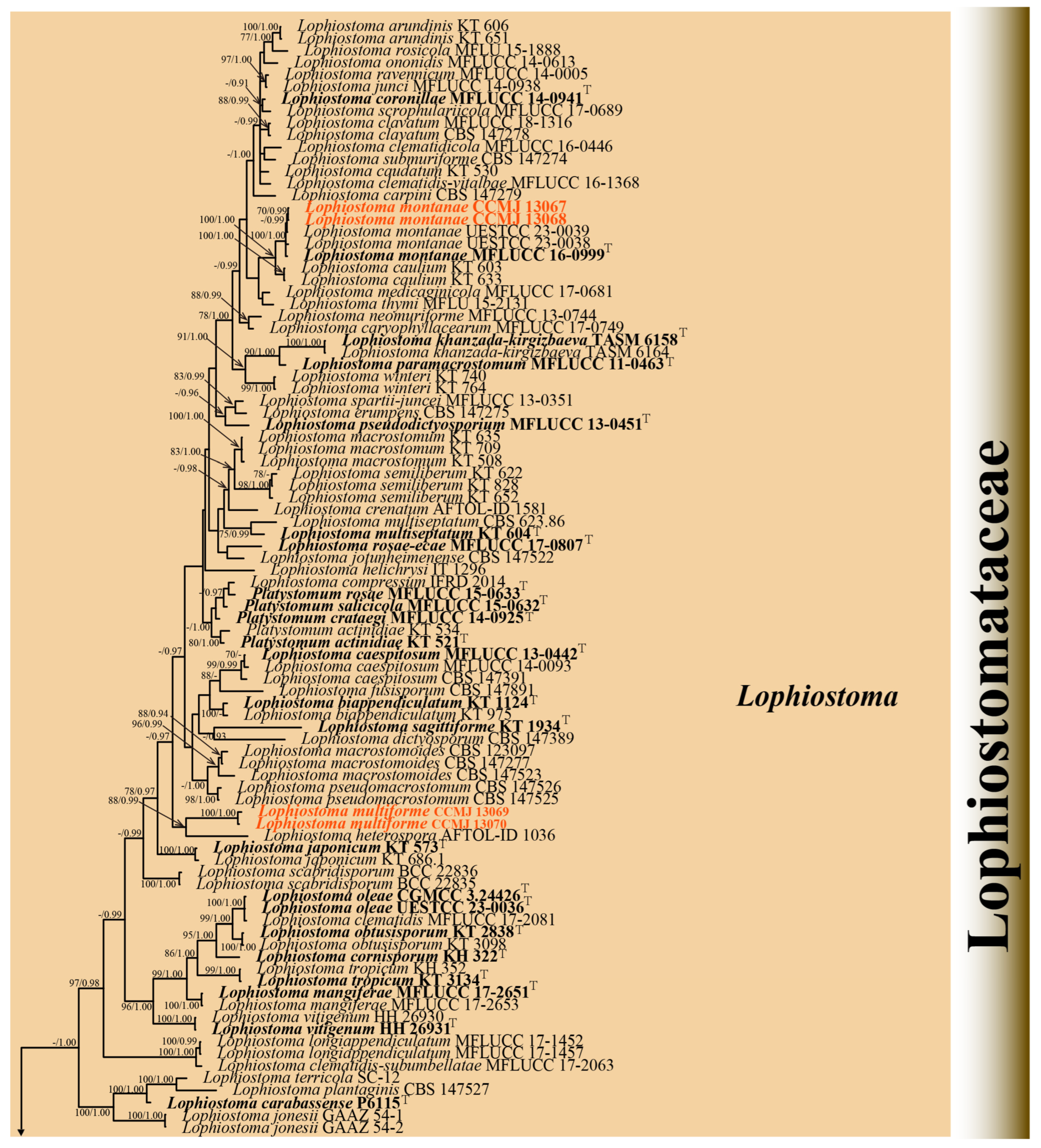
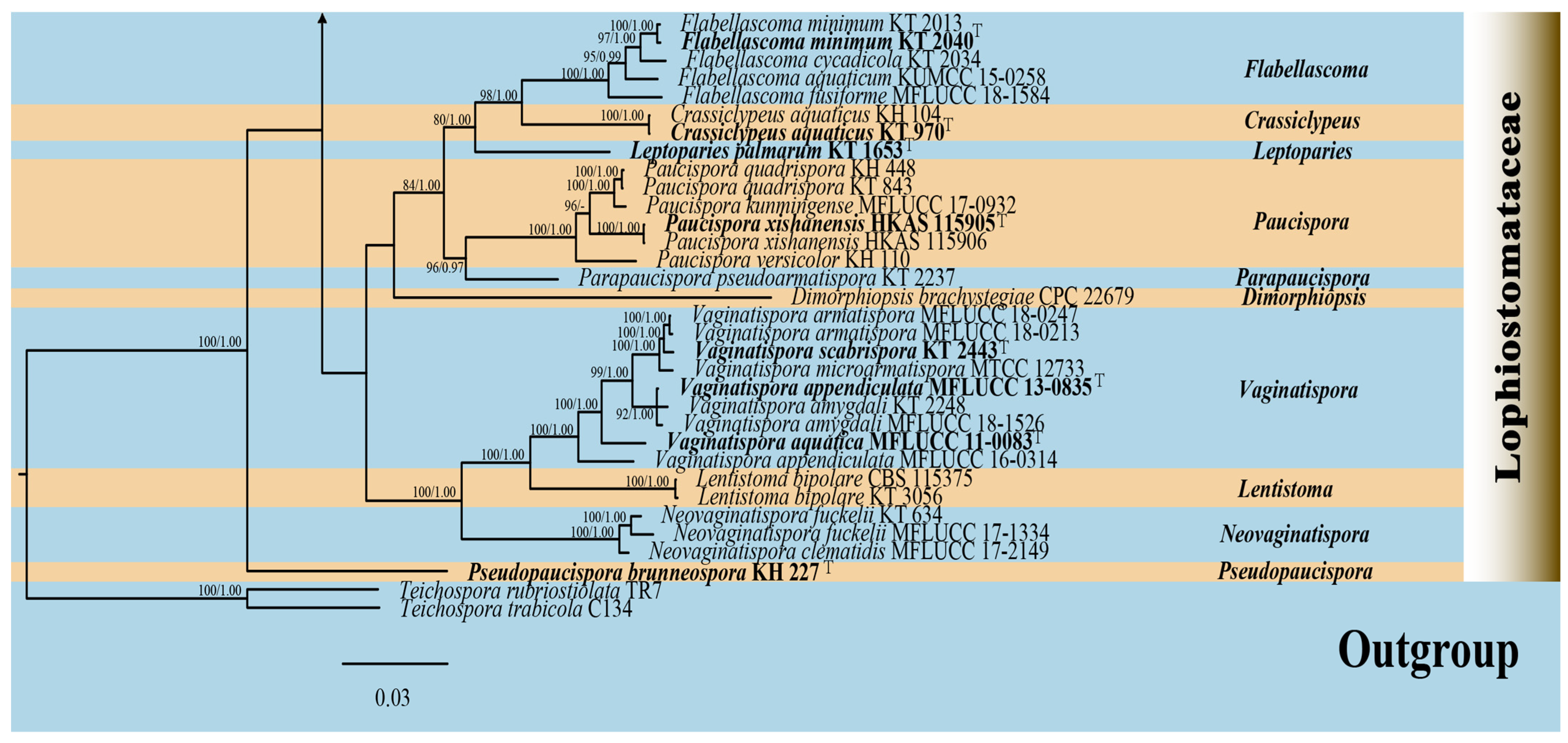
3.2. Taxonomy
3.2.1. Alloleptosphaeria xanthoceratis R. Xu and Y. Li, sp. nov. (Figure 3)

3.2.2. Lophiostoma montanae (Phukhams., Sue, and K.D. Hyde) Andreasen, Jaklitsch, and Voglmayr, Persoonia 46: 259 (2021) [41] = Sigarispora montanae Phukhams. Sue et al., Fungal Diversity 102: 55 [42], (Figure 4)

3.2.3. Lophiostoma multiforme R. Xu and Y. Li, sp. nov. (Figure 5)
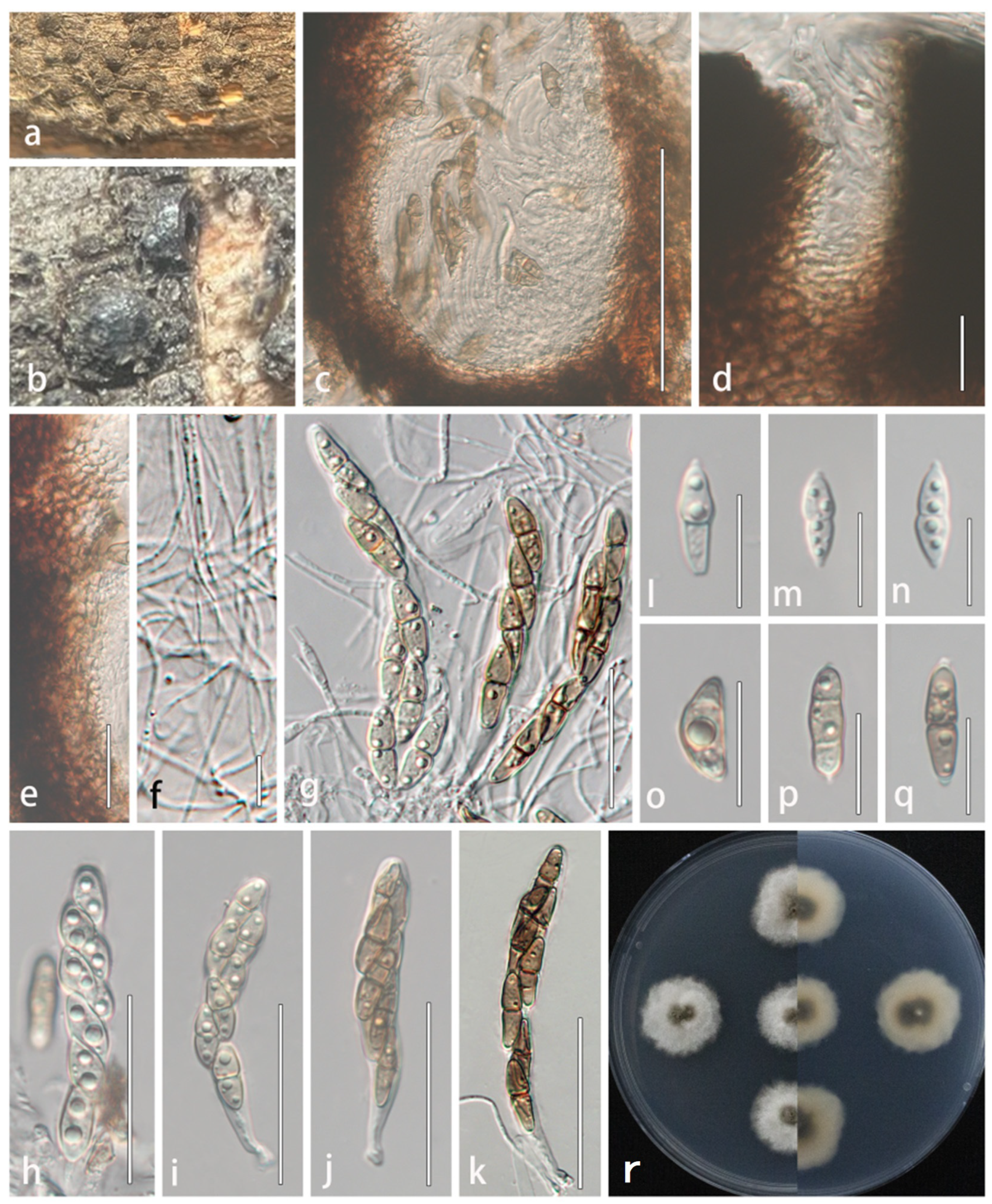
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bánki, O.; Roskov, Y.; Döring, M.; Ower, G.; Vandepitte, L.; Hobern, D.; Remsen, D.; Schalk, P.; DeWalt, R.E.; Keping, M.; et al. Catalogue of Life Checklist (Version 2022-01-14); Catalogue of Life: Leiden, NL, USA, 2022. [Google Scholar]
- Nagy, L.G.; Kovács, G.M.; Krizsán, K. Repeated Evolution of Complex Multicellularity in Fungi. Curr. Biol. 2018, 24, 2384–2394. [Google Scholar]
- Schoch, C.L.; Sung, G.H.; López-Giráldez, F.; Townsend, J.P.; Miadlikowska, J.; Hofstetter, V.; Robbertse, B.; Brandon Matheny, P.; Kauff, F.; Wang, Z.; et al. The Ascomycota tree of life: A phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst. Biol. 2009, 58, 224–239. [Google Scholar] [CrossRef] [PubMed]
- Esser, K. The Mycota: A Comprehensive Treatise on Fungi as Experimental Systems for Basic and Applied Research; Springer: Berlin/Heidelberg, Germany, 2001. [Google Scholar]
- Hyde, K.D.; Xu, J.; Rapior, S.; Jeewon, R.; Lumyong, S.; Niego, A.G.T.; Abeywickrama, P.D.; Aluthmuhandiram, J.V.; Brahamanage, R.S.; Brooks, S.; et al. The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Divers. 2019, 97, 1–136. [Google Scholar]
- Hyde, K.D.; Noordeloos, M.E.; Savchenko, K.G.; Jeewon, R.; Lumyong, S.; Niego, A.G.T.; Abeywickrama, P.D.; Aluthmuhandiram, J.V.S.; Brahamanage, R.S.; Brooks, S.; et al. The 2024 Outline of Fungi and fungus-like taxa. Mycosphere 2024, 15, 5146–6239. [Google Scholar] [CrossRef]
- Hongsanan, S.; Hyde, K.D.; Phookamsak, R.; Wanasinghe, D.N.; McKenzie, E.H.C.; Sarma, V.V.; Boonmee, S.; Lücking, R.; Pem, D.; Bhat, J.D.; et al. Refned families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere 2020, 11, 1553–2107. [Google Scholar] [CrossRef]
- Hyde, K.D.; Jones, E.B.G.; Liu, J.K.; Ariyawansa, H.; Boehm, E.; Boonmee, S.; Braun, U.; Chomnunti, P.; Crous, P.W.; Dai, D.Q.; et al. Families of Dothideomycetes. Fungal Divers. 2013, 63, 1–313. [Google Scholar]
- Zhang, Y.; Crous, P.W.; Schoch, C.L.; Hyde, K.D. Pleosporales. Fungal Divers. 2012, 53, 1–221. [Google Scholar] [CrossRef]
- Ariyawansa, H.A.; Thambugala, K.M.; Manamgoda, D.S.; Jayawardena, R.; Camporesi, E.; Boonmee, S.; Wanasinghe, D.N.; Phookamsak, R.; Hongsanan, S.; Singtripop, C.; et al. Towards a natural classifcation and backbone tree for Pleosporaceae. Fungal Divers. 2015, 71, 85–139. [Google Scholar] [CrossRef]
- Zhang, Y.; Schoch, C.L.; Fournier, J.; Crous, P.W.; de Gruyter, J.; Woudenberg, J.H.; Hirayama, K.; Tanaka, K.; Pointing, S.B.; Spatafora, J.W.; et al. Multi-locus phylogeny of Pleosporales: A taxonomic, ecological and evolutionary re-evaluation. Stud. Mycol. 2009, 64, 85–102. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Dai, D.Q.; Sánchez-García, M.; Goto, B.T.; Saxena, R.K.; Erdoğdu, M.; Selçuk, F.; Rajeshkumar, K.C.; Aptroot, A.; et al. Outline of Fungi and fungus-like taxa-2021. Mycosphere 2022, 13, 53–453. [Google Scholar] [CrossRef]
- Li, W.L.; Liang, R.R.; Dissanayake, A.J.; Liu, J.K. Botryosphaerialean fungi associated with woody oil plants cultivated in Sichuan Province, China. MycoKeys 2023, 97, 71–116. [Google Scholar] [CrossRef] [PubMed]
- Li, W.L.; Liang, R.R.; Dissanayake, A.J.; Liu, J.K. Mycosphere Notes 413–448: Dothideomycetes associated with woody oil plants in China. Mycosphere 2023, 14, 1436–1529. [Google Scholar] [CrossRef]
- Li, W.L.; Dissanayake, A.J.; Zhang, T.; Maharachchikumbura, S.S.N.; Liu, J.K. Identification and pathogenicity of Pestalotiod fungi associated with woody oil plants in Sichuan Province, China. J. Fungi 2022, 8, 1175. [Google Scholar] [CrossRef]
- Liang, Q.; Li, H.Y.; Li, S.K.; Yuan, F.L.; Sun, J.F.; Duan, Q.C.; Li, Q.Y.; Zhang, R.; Sang, Y.L.; Wang, N.; et al. The genome assembly and annotation of yellowhorn (Xanthoceras sorbifolium Bunge). GigaScience 2019, 8, giz071. [Google Scholar] [CrossRef]
- Zhao, Z.; Liang, C.J.; Zhang, W.; Yang, Y.Y.; Bi, Q.X.; Yu, H.Y.; Wang, L.B. Genome-wide association analysis identifies a candidate gene controlling seed size and yield in Xanthoceras sorbifolium Bunge. Hortic. Res. 2024, 11, 243. [Google Scholar] [CrossRef]
- Zang, E.H.; Qiu, B.; Chen, N.H.; Li, C.F.; Liu, Q.; Zhang, M.; Liu, Y.C.; Li, M.H. Xanthoceras sorbifolium Bunge: A review on botany, phytochemistry, pharmacology, and applications. Front. Pharmacol. 2021, 12, 708549. [Google Scholar] [CrossRef]
- Xu, R.; Su, W.; Tian, S.; Bhunjun, C.S.; Tibpromma, S.; Hyde, K.D.; Li, Y.; Phukhamsakda, C. Synopsis of Leptosphaeriaceae and introduction of three new taxa and one new record from China. J. Fungi 2022, 8, 416. [Google Scholar] [CrossRef]
- Xu, R.; Su, W.; Wang, Y.; Tian, S.; Li, Y.; Phukhamsakda, C. Morphological characteristics and phylogenetic evidence reveal two new species and the first report of Comoclathris (Pleosporaceae, Pleosporales) on dicotyledonous plants from China. MycoKeys 2024, 101, 95–112. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Su, W.; Li, Y.; Li, X.; Li, C. Multiple evidence reveals a new species of Neocucurbitaria (Cucurbitariaceae, Pleosporales) from China. Phytotaxa 2024, 650, 249–261. [Google Scholar] [CrossRef]
- Yang, G. Study on Main Diseases and insect pests and control techniques in Xanthoceras sorbifolia Bunge. Hortic. Seed 2022, 42, 37–39. [Google Scholar]
- Senanayake, I.C.; Rathnayaka, A.R.; Marasinghe, D.S.; Calabon, M.S.; Gentekaki, E.; Lee, H.B.; Hurdeal, V.G.; Pem, D.; Dissanayake, L.S.; Wijesinghe, S.N.; et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere 2020, 11, 2678–2754. [Google Scholar] [CrossRef]
- Crous, P.W.; Gams, W.; Stalpers, J.A.; Robert, V.; Stegehuis, G. MycoBank: An online initiative to launch mycology into the 21st century. Stud. Mycol. 2004, 50, 19–22. [Google Scholar]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guid. Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- Rehner, S.A.; Buckley, E.A. Beauveria phylogeny inferred from nuclear ITS and EF-1α sequences: Evidence for cryptic diversifcation and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for PCR to amplify conserved genes from filamentous Ascomycetes. Appl. Environ. Microb. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- McGinnis, S.; Madden, T.L. BLAST: At the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004, 32, W20–W25. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Nylander, J.A.A. MrModeltest, 2.0; Program Distributed by the Author; Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree, v.1.4.4; UnSiversity of Edinburgh: Edinburgh, UK, 2018. [Google Scholar]
- Crous, P.W.; Schumacher, R.K.; Wingfield, M.J.; Akulov, A.; Denman, S.; Roux, J.; Braun, U.; Burgess, T.I.; Carnegie, A.J.; Váczy, K.Z.; et al. New and Interesting Fungi. 1. Fungal Syst. Evol. 2018, 1, 169–216. [Google Scholar] [CrossRef] [PubMed]
- Aiello, D.; Vitale, A.; Polizzi, G.; Voglmayr, H. Ochraceocephala foeniculi gen. et sp. nov., a new pathogen causing crown rot of fennel in Italy. MycoKeys 2020, 66, 1–22. [Google Scholar] [CrossRef]
- Andreasen, M.; Skrede, I.; Jaklitsch, W.M.; Voglmayr, H.; Nordén, B. Multi-locus phylogenetic analysis of lophiostomatoid fungi motivates a broad concept of Lophiostoma and reveals nine new species. Persoonia 2021, 46, 240–271. [Google Scholar] [CrossRef]
- Phukhamsakda, C.; McKenzie, E.H.C.; Phillips, A.J.L.; Jones, E.B.G.; Bhat, D.J.; Stadler, M.; Bhunjun, C.S.; Wanasinghe, D.N.; Thongbai, B.; Camporesi, E.; et al. Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Divers. 2020, 102, 1–203. [Google Scholar] [CrossRef]
- Saccardo, P.A. Sylloge Fungorum. II; Biodiversity Heritage Library: Washington, DC, USA, 1883. [Google Scholar]
- Liu, F.; Ma, Z.Y.; Hou, L.W.; Diao, Y.Z.; Wu, W.P.; Damm, U.; Song, S.; Cai, L. Updating species diveristy of Colletotrichum, with a phylogenomic overview. Stud. Mycol. 2022, 101, 1–56. [Google Scholar] [CrossRef]
- Han, S.L.; Wang, M.M.; Ma, Z.Y.; Raza, M.; Zhao, P.; Liang, J.M.; Gao, M.; Li, Y.J.; Wang, J.W.; Hu, D.M.; et al. Fusarium diversity associated with diseased cereals in China, with an updated phylogenomic assessment of the genus. Stud. Mycol. 2023, 104, 87–148. [Google Scholar] [CrossRef]
- Razaghi, P.; Raza, M.; Han, L.S.; Ma, Z.Y.; Cai, L.; Zhao, P.; Chen, Q.; Phurbu, D.; Liu, F. Sporocadaceae revisited. Stud. Mycol. 2024, 109, 155–272. [Google Scholar] [CrossRef]
- Steenwyk, L.J.; Balamurugan, C.; Raja, A.H.; Gonçalves, C.; Li, N.X.; Martin, F.; Berman, J.; Oberlies, N.H.; Gibbons, J.G.; Goldman, G.H. Phylogenomics reveals extensive misidentification of fungal strains from the genus Aspergillus. Microbiol. Spectr. 2024, 12, e0398023. [Google Scholar] [CrossRef] [PubMed]
- Ariyawansa, H.A.; Phukhamsakda, C.; Thambugala, K.M.; Bulgakov, T.S.; Wanasinghe, D.N.; Perera, R.H.; Mapook, A.; Camporesi, E.; Kang, J.C.; Jones, E.B.G.; et al. Revision and phylogeny of Leptosphaeriaceae. Fungal Divers. 2015, 74, 19–51. [Google Scholar] [CrossRef]
- Thiyagaraja, V.; Wanasinghe, D.N.; Karunarathna, S.C.; Tennakoon, D.S.; Hyde, K.D.; To-anun, C.; Cheewangkoon, R. Alloleptosphaeria shangrilana sp. nov. and first report of the genus (Leptosphaeriaceae, Dothideomycetes) from China. Phytotaxa 2021, 491, 12–22. [Google Scholar] [CrossRef]
- Tanaka, K.; Harada, Y. Pleosporales in Japan (1): The genus Lophiostoma. Mycoscience 2003, 44, 85–96. [Google Scholar] [CrossRef]
- Hirayama, K.; Tanaka, K. Taxonomic revision of Lophiostoma and Lophiotrema based on reevaluation of morphological characters and molecular analyses. Mycoscience 2011, 52, 401–412. [Google Scholar] [CrossRef]
- Thambugala, K.M.; Hyde, K.D.; Tanaka, K.; Tian, Q.; Wanasinghe, D.N.; Ariyawansa, H.A.; Jayasiri, S.C. Towards a natural classification and backbone tree for Lophiostomataceae, Floricolaceae, and Amorosiaceae fam. nov. Fungal Divers. 2015, 74, 199–266. [Google Scholar] [CrossRef]
- Aluthmuhandiram, J.V.; Wanasinghe, D.N.; Thilini Chethana, K.W.; Gafforov, Y.; Saichana, N.; Li, X.; Yan, J.; Mamarakhimov, O.M. Lophiostomataceae (Dothideomycetes): Introducing Lophiostoma khanzada-kirgizbaeva sp. nov. and Paucispora xishanensis sp. nov. Phytotaxa 2022, 559, 247–262. [Google Scholar] [CrossRef]
- Hawksworth, D.L.; Lücking, R. Fungal Diversity Revisited: 2.2 to 3.8 Million Species. Microbiol. Spectr. 2017, 5, 1–17. [Google Scholar] [CrossRef]
- Wanasinghe, D.N.; Mortimer, P.E.; Bezerra, J. Editorial: Fungal Systematics and Biogeography. Front. Microbiol. 2022, 12, 827725. [Google Scholar] [CrossRef]
| Loci | Primer Pair Forward/Reverse | Sequence (5′–3′) | Reference |
|---|---|---|---|
| ITS | ITS5/ITS4 | GGAAGTAAAAGTCGTAACAAGG TCCTCCGCTTATTGATATGC | [26] |
| LSU | LR0R/LR5 | ACCCGCTGAACTTAAGC ATCCTGAGGGAAACTTC | [27] |
| SSU | NS1/NS4 | GTAGTCATATGCTTGTCTC CTTCCGTCAATTCCTTTAAG | [26] |
| tef1-α | 2218F/983R | GCYCCYGGHCAYCGTGAYTTYAT ATGACACCRACRGCRACACRGTYTG | [28] |
| rpb2 | fRPB2-5F/fRPB2-7cR | GAYGAYMGWGATCAYTTYGG CCCATRGCTTGYTTRCCCAT | [29] |
| tub2 | T1/Bt2b | AACATGCGTGAGATTGTAAGT ACCCTCAGTGTAGTGACCCTTGGC | [30] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, R.; Li, Y. Additions to Pleosporalean Taxa Associated with Xanthoceras sorbifolium from Jilin and Hebei, China. Microorganisms 2025, 13, 1296. https://doi.org/10.3390/microorganisms13061296
Xu R, Li Y. Additions to Pleosporalean Taxa Associated with Xanthoceras sorbifolium from Jilin and Hebei, China. Microorganisms. 2025; 13(6):1296. https://doi.org/10.3390/microorganisms13061296
Chicago/Turabian StyleXu, Rong, and Yu Li. 2025. "Additions to Pleosporalean Taxa Associated with Xanthoceras sorbifolium from Jilin and Hebei, China" Microorganisms 13, no. 6: 1296. https://doi.org/10.3390/microorganisms13061296
APA StyleXu, R., & Li, Y. (2025). Additions to Pleosporalean Taxa Associated with Xanthoceras sorbifolium from Jilin and Hebei, China. Microorganisms, 13(6), 1296. https://doi.org/10.3390/microorganisms13061296




