Isolation, Characterization, and Preliminary Application of Staphylococcal Bacteriophages in Sichuan Paocai Fermentation
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Sichuan Paocai
2.2. Amplification and Concentration of Phages in the Sichuan Paocai
2.3. Isolation of Specific Phages and Host Bacteria
2.4. Identification of Specific Host Bacteria
2.5. Characterization of Phages
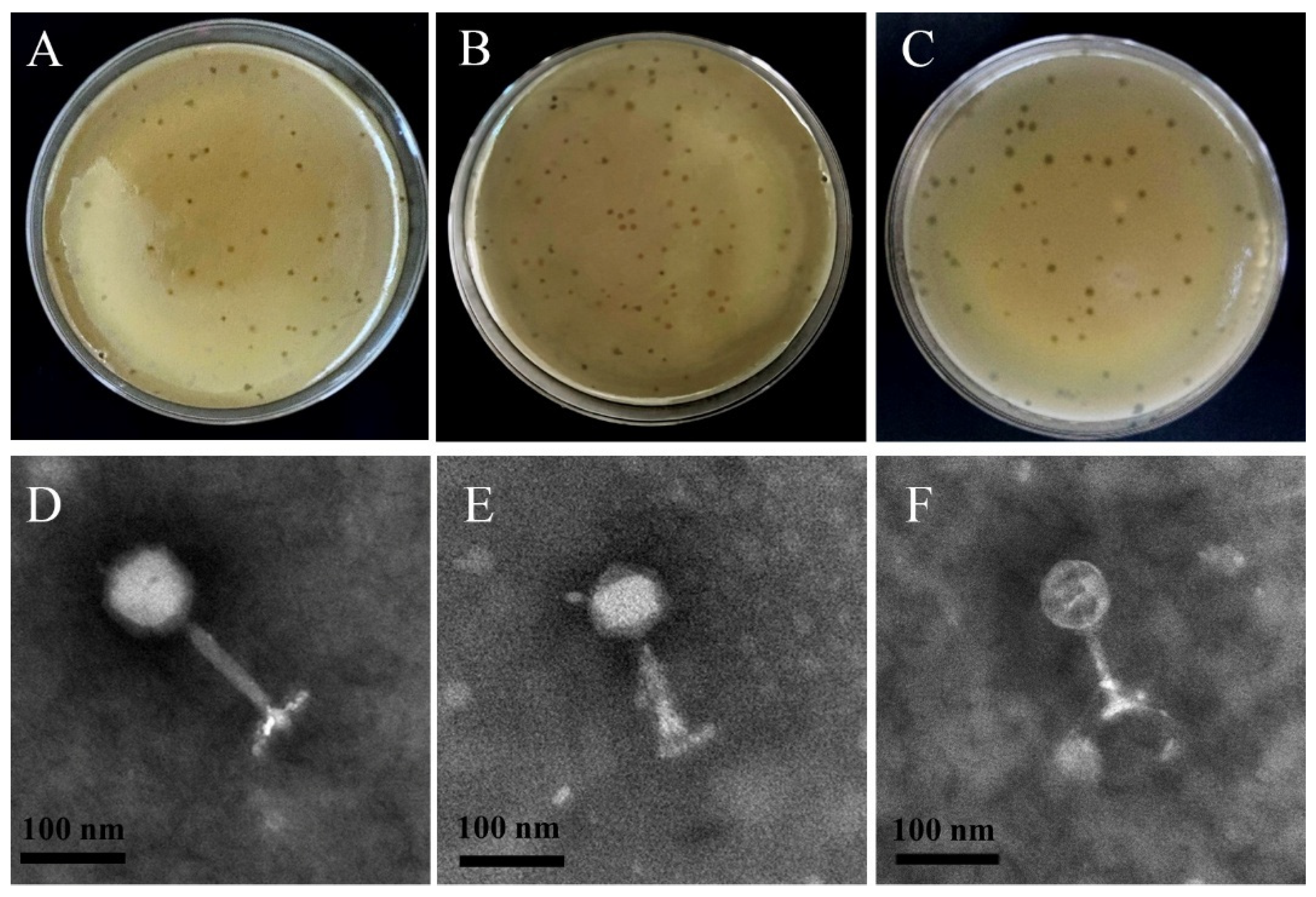
2.5.1. Transmission Electron Microscopy
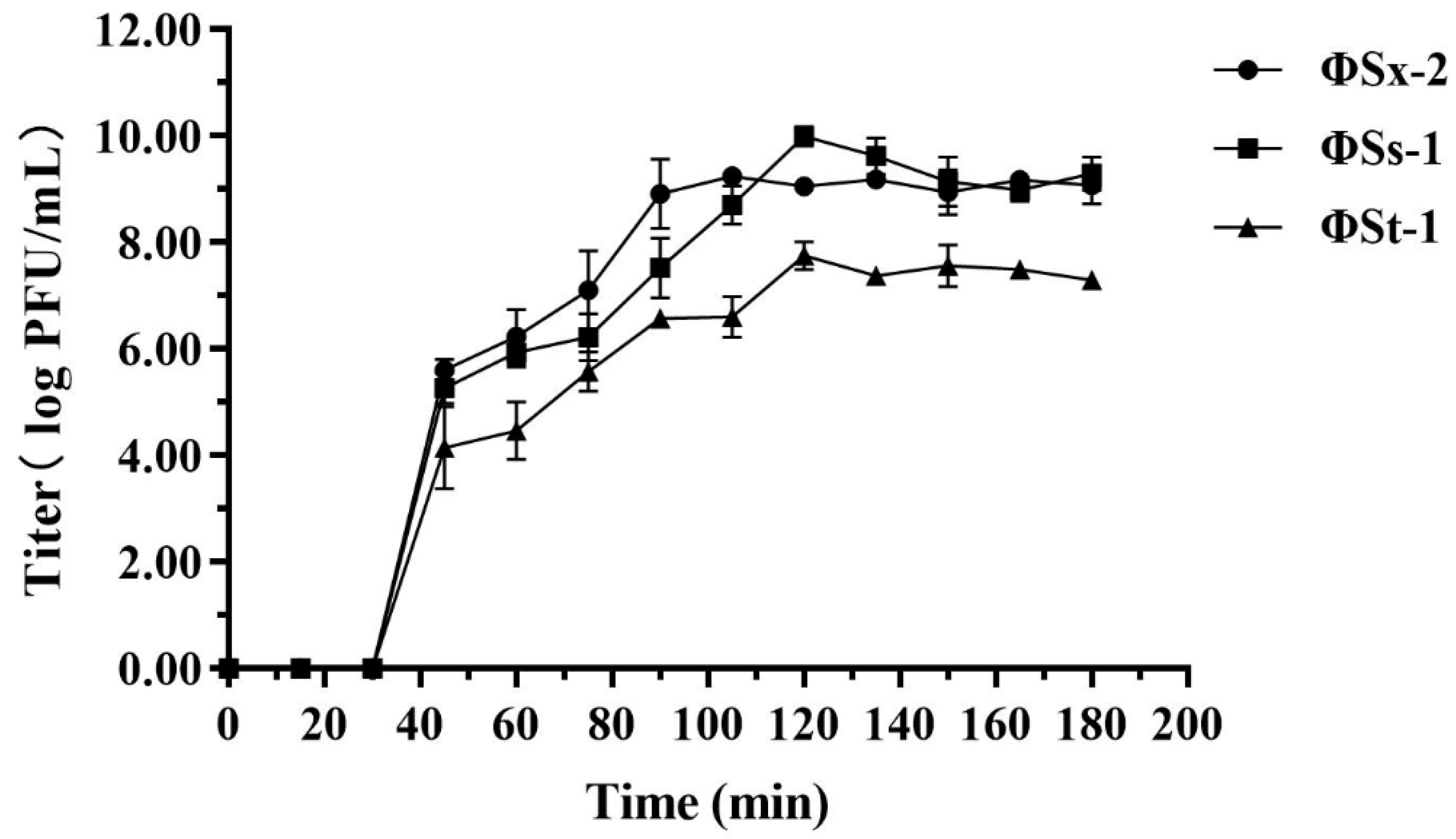
2.5.2. One-Step Growth Curve
2.5.3. Phage Tolerance Test
2.6. The Impact of Phages ΦSx-2, ΦSs-1, and ΦSs-2 on the Fermentation of Sichuan Paocai
2.6.1. Inoculating Phages into Sichuan Paocai
2.6.2. Determination of pH Value and Microbial Count of Sichuan Paocai
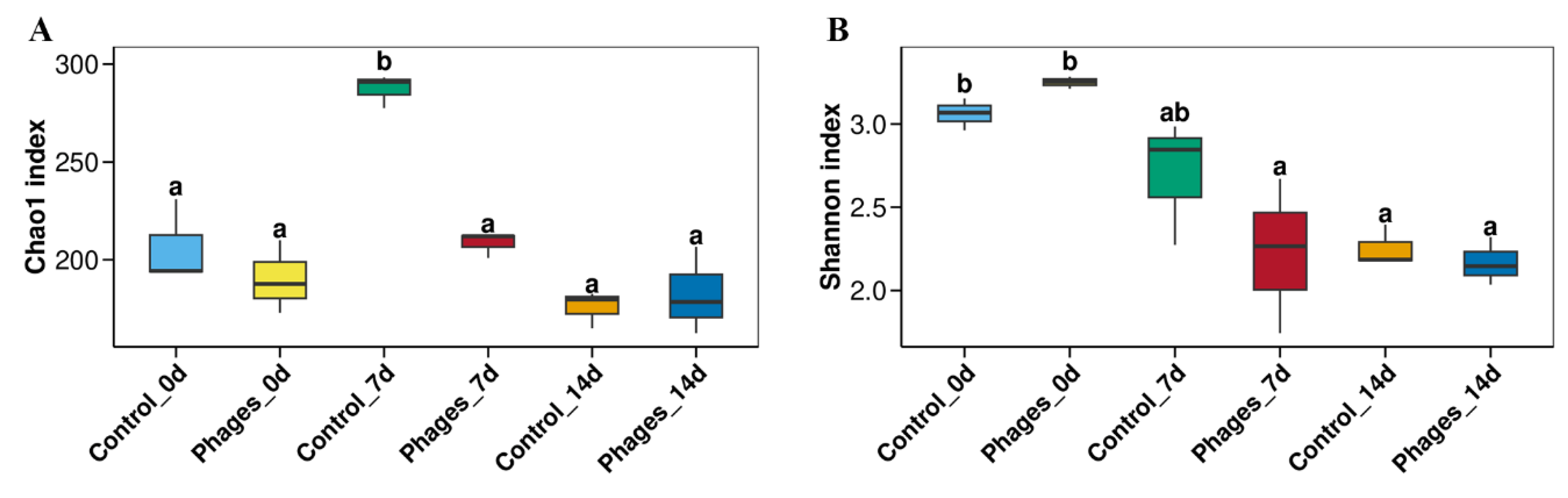
2.6.3. Analysis of the Microbial Diversity of Sichuan Paocai
2.6.4. Determination of Physicochemical Indexes of Sichuan Paocai
2.6.5. Flavor Determination and Color Observation of Sichuan Paocai
2.7. Data Analysis
3. Results
3.1. Identification of Specific Hosts for Phages
3.2. Phage Characterization
3.2.1. Phage Morphology
3.2.2. One-Step Growth Curve of Phages
3.2.3. Analysis of Phage Tolerance
3.3. The Impact of Phages ΦSx-2, ΦSs-1, and ΦSs-2 on Sichuan Paocai Fermentation
3.3.1. Changes in pH Value, Total Bacterial Count, and Lactic Acid Bacteria Count
3.3.2. The Impact of Phages ΦSx-2, ΦSs-1, and ΦSs-2 on Microbial Community Structure
Alpha Diversity Indices of Different Paocai Samples
Analysis of Microbial Community Structure During Paocai Fermentation
Microecological Correlation Analysis
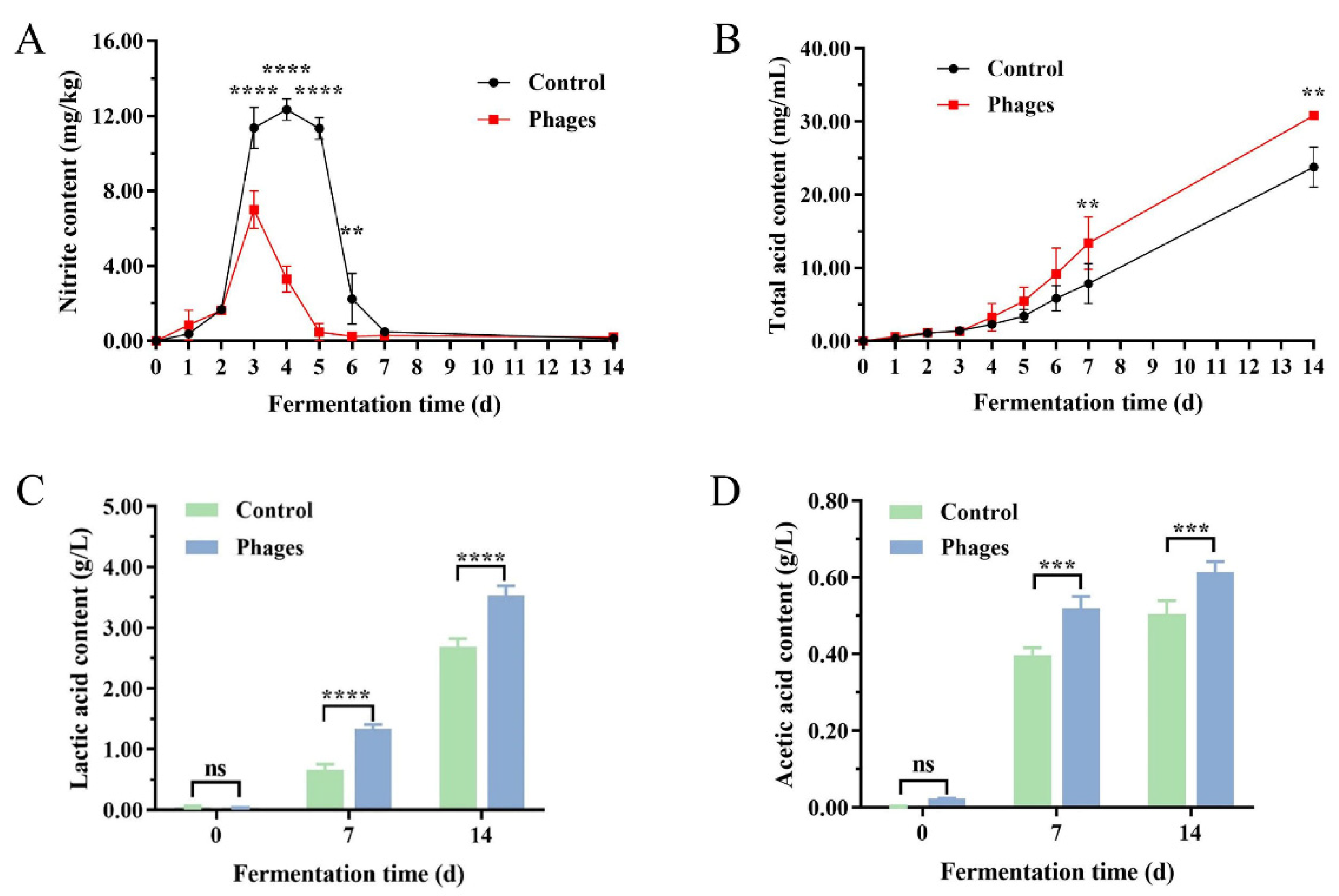
3.3.3. Changes in Nitrite, Total Acid, and Organic Acid Contents
3.3.4. Color Changes of Sichuan Paocai During Fermentation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dion, M.B.; Oechslin, F.; Moineau, S. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 2020, 18, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, L.L.; Lacorte, G.A.; Isidorio, W.R.; Landgraf, M.; de Melo Franco, B.D.G.; Pinto, U.M.; Hoffmann, C.; Gambino, M. High Level of Interaction between Phages and Bacteria in an Artisanal Raw Milk Cheese Microbial Community. mSystems 2023, 8, e00564-22. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, H.; Du, H.; Yu, X.; Xu, Y. The insights into the phage communities of fermented foods in the age of viral metagenomics. Crit. Rev. Food Sci. Nutr. 2024, 65, 1656–1668. [Google Scholar] [CrossRef]
- Agyirifo, D.S.; Wamalwa, M.; Otwe, E.P.; Galyuon, I.; Runo, S.; Takrama, J.; Ngeranwa, J. Metagenomics analysis of cocoa bean fermentation microbiome identifying species diversity and putative functional capabilities. Heliyon 2019, 5, e02170. [Google Scholar] [CrossRef]
- Alexeeva, S.; Liu, Y.; Zhu, J.; Kaczorowska, J.; Kouwen, T.R.H.M.; Abee, T.; Smid, E.J. Genomics of tailless bacteriophages in a complex lactic acid bacteria starter culture. Int. Dairy J. 2021, 114, 104900. [Google Scholar] [CrossRef]
- Park, E.-J.; Kim, K.-H.; Abell, G.C.J.; Kim, M.-S.; Roh, S.W.; Bae, J.-W. Metagenomic Analysis of the Viral Communities in Fermented Foods. Appl. Environ. Microbiol. 2011, 77, 1284–1291. [Google Scholar] [CrossRef]
- Zheng, X.-F.; Yang, Z.-Q.; Zhang, H.; Jin, W.-X.; Xu, C.-W.; Gao, L.; Rao, S.-Q.; Jiao, X.-A. Isolation of virulent phages infecting dominant mesophilic aerobic bacteria in cucumber pickle fermentation. Food Microbiol. 2020, 86, 103330. [Google Scholar] [CrossRef]
- Behera, S.S.; El Sheikha, A.F.; Hammami, R.; Kumar, A. Traditionally fermented pickles: How the microbial diversity associated with their nutritional and health benefits? J. Funct. Foods 2020, 70, 103971. [Google Scholar] [CrossRef]
- Jiang, L.; Xian, S.; Liu, X.; Shen, G.; Zhang, Z.; Hou, X.; Chen, A. Metagenomic Study on Chinese Homemade Paocai: The Effects of Raw Materials and Fermentation Periods on the Microbial Ecology and Volatile Components. Foods 2022, 11, 62. [Google Scholar] [CrossRef]
- Rao, Y.; Tao, Y.; Chen, X.; She, X.; Qian, Y.; Li, Y.; Du, Y.; Xiang, W.; Li, H.; Liu, L. The characteristics and correlation of the microbial communities and flavors in traditionally pickled radishes. LWT 2020, 118, 108804. [Google Scholar] [CrossRef]
- Rao, Y.; Qian, Y.; Tao, Y.; She, X.; Li, Y.; Chen, X.; Guo, S.; Xiang, W.; Liu, L.; Du, H.; et al. Characterization of the microbial communities and their correlations with chemical profiles in assorted vegetable Sichuan pickles. Food Control 2020, 113, 107174. [Google Scholar] [CrossRef]
- Tan, G.; Qi, S.; Wang, Y.; Li, X.; Li, X.; Li, M.; Li, L.; Zhao, L.; Hu, M. Uncovering differences in the composition and function of phage communities and phage-bacterium interactions in raw soy sauce. Front. Microbiol. 2023, 14, 1328158. [Google Scholar] [CrossRef] [PubMed]
- He, G.-Q.; Liu, T.-J.; Sadiq, F.A.; Gu, J.-S.; Zhang, G.-H. Insights into the microbial diversity and community dynamics of Chinese traditional fermented foods from using high-throughput sequencing approaches. J. Zhejiang Univ. Sci. B 2017, 18, 289–302. [Google Scholar] [CrossRef]
- Ávila, M.; Sánchez, C.; Calzada, J.; Mayer, M.J.; Berruga, M.I.; López-Díaz, T.M.; Narbad, A.; Garde, S. Isolation and characterization of new bacteriophages active against Clostridium tyrobutyricum and their role in preventing the late blowing defect of cheese. Food Res. Int. 2023, 163, 112222. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Pérez-Díaz, I.M.; Hayes, J.S.; Breidt, F. Bacteriophage Ecology in a Commercial Cucumber Fermentation. Appl. Environ. Microbiol. 2012, 78, 8571–8578. [Google Scholar] [CrossRef]
- Garcia, E.F.; Luciano, W.A.; Xavier, D.E.; da Costa, W.C.A.; de Sousa Oliveira, K.; Franco, O.L.; de Morais Júnior, M.A.; Lucena, B.T.L.; Picão, R.C.; Magnani, M.; et al. Identification of Lactic Acid Bacteria in Fruit Pulp Processing Byproducts and Potential Probiotic Properties of Selected Lactobacillus Strains. Front. Microbiol. 2016, 7, 1371. [Google Scholar] [CrossRef]
- Oh, H.; Seo, D.J.; Jeon, S.B.; Park, H.; Jeong, S.; Chun, H.S.; Oh, M.; Choi, C. Isolation and Characterization of Bacillus cereus Bacteriophages from Foods and Soil. Food Environ. Virol. 2017, 9, 260–269. [Google Scholar] [CrossRef]
- Lin, X.; Bakyrbay, S.; Liu, L.; Tang, X.; Liu, Y. Microbiota Succession and Chemical Composition Involved in Lactic Acid Bacteria-Fermented Pickles. Fermentation 2023, 9, 330. [Google Scholar] [CrossRef]
- Liu, L.; She, X.; Qian, Y.; Li, Y.; Tao, Y.; Che, Z.; Liu, G.; Rao, Y. Effect of different fermenting containers on the deterioration of Sichuan pickle. LWT 2019, 111, 829–836. [Google Scholar] [CrossRef]
- Leng, L. Statistical Computing with R. J. Am. Stat. Assoc. 2020, 115, 1557–1558. [Google Scholar] [CrossRef]
- Mahony, J.; van Sinderen, D. Current taxonomy of phages infecting lactic acid bacteria. Front. Microbiol. 2014, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Wu, W.; Du, X.; Yu, Y.; Wu, J.; Xu, Y.; Li, L. Regulation of the nitrite, biogenic amine and flavor quality of Cantonese pickle by selected lactic acid bacteria. Food Biosci. 2023, 53, 102554. [Google Scholar] [CrossRef]
- Fan, Y.; Yang, X.; Hu, C.; Wei, B.; Xu, F.; Guo, Q. Fermentation Performance Evaluation of Lactic Acid Bacteria Strains for Sichuan Radish Paocai Production. Foods 2024, 13, 1813. [Google Scholar] [CrossRef]
- Jurado, A.; Fernández, L.; Rodríguez, A.; García, P. Understanding the Mechanisms That Drive Phage Resistance in Staphylococci to Prevent Phage Therapy Failure. Viruses 2022, 14, 1061. [Google Scholar] [CrossRef]
- Ali Gharieb, R.M.; Saad, M.F.; Mohamed, A.S.; Tartor, Y.H. Characterization of two novel lytic bacteriophages for reducing biofilms of zoonotic multidrug-resistant Staphylococcus aureus and controlling their growth in milk. LWT 2020, 124, 109145. [Google Scholar] [CrossRef]
- Hatoum-Aslan, A. The phages of staphylococci: Critical catalysts in health and disease. Trends Microbiol. 2021, 29, 1117–1129. [Google Scholar] [CrossRef] [PubMed]
- Deghorain, M.; Van Melderen, L. The Staphylococci Phages Family: An Overview. Viruses 2012, 4, 3316–3335. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Ma, Y.; Guan, Y.; Zhu, Y.; Wang, K.; Wang, Y.; Liu, P.; Chen, J.; Yu, Y. Metagenomics of Virus Diversities in Solid-State Brewing Process of Traditional Chinese Vinegar. Foods 2022, 11, 3296. [Google Scholar] [CrossRef]
- Wang, D.; Chen, G.; Tang, Y.; Ming, J.; Huang, R.; Li, J.; Ye, M.; Fan, Z.; Yin, L.; Zhang, Q.; et al. Effect of non-core microbes on the key odorants of paocai. LWT 2022, 172, 114211. [Google Scholar] [CrossRef]
- White, K.; Yu, J.-H.; Eraclio, G.; Dal Bello, F.; Nauta, A.; Mahony, J.; van Sinderen, D. Bacteriophage-host interactions as a platform to establish the role of phages in modulating the microbial composition of fermented foods. Microbiome Res. Rep. 2022, 1, 3. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, C.; Liu, F.; Jin, Z.; Xia, X. Ecological succession and functional characteristics of lactic acid bacteria in traditional fermented foods. Crit. Rev. Food Sci. Nutr. 2023, 63, 5841–5855. [Google Scholar] [CrossRef]
- Zapaśnik, A.; Sokołowska, B.; Bryła, M. Role of Lactic Acid Bacteria in Food Preservation and Safety. Foods 2022, 11, 1283. [Google Scholar] [CrossRef]
- Huang, Y.Y.; Huang, F.; Huang, J.Y.; Liu, Y.; Li, M.N.; Yao, Q.B.; Zeng, X.A.; Chen, B.R. Repercussions of Lactiplantibacillus plantarum HYY-DB9 and Levilactobacillus brevis GIM 1.773 on the degradation of nitrite, volatile compounds, and sensory assessment of traditional Chinese paocai. Int. J. Food Sci. Technol. 2024, 59, 6522–6532. [Google Scholar] [CrossRef]
- Hang, S.; Zeng, L.; Han, J.; Zhang, Z.; Zhou, Q.; Meng, X.; Gu, Q.; Li, P. Lactobacillus plantarum ZJ316 improves the quality of Stachys sieboldii Miq. pickle by inhibiting harmful bacteria growth, degrading nitrite and promoting the gut microbiota health in vitro. Food Funct. 2022, 13, 1551–1562. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, Y.; Zhao, Y.; Guan, H.; Jin, C.; Gong, H.; Sun, X.; Wang, P.; Li, H.; Liu, W. Effect of Levilactobacillus brevis as a starter on the flavor quality of radish paocai. Food Res. Int. 2023, 168, 112780. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.Y.; Lee, M.; Song, J.H.; Choi, E.J.; Mun, S.Y.; Kim, D.; Lim, S.K.; Kim, N.; Park, B.Y.; Chang, J.Y. Organic acid type in kimchi is a key factor for determining kimchi starters for kimchi fermentation control. Heliyon 2024, 10, e36860. [Google Scholar] [CrossRef]
- Liang, H.; He, Z.; Wang, X.; Song, G.; Chen, H.; Lin, X.; Ji, C.; Li, S. Effects of salt concentration on microbial diversity and volatile compounds during suancai fermentation. Food Microbiol. 2020, 91, 103537. [Google Scholar] [CrossRef]
- Xiao, M.; Xiong, T.; Peng, Z.; Liu, C.; Huang, T.; Yu, H.; Xie, M. Correlation between microbiota and flavours in fermentation of Chinese Sichuan Paocai. Food Res. Int. 2018, 114, 123–132. [Google Scholar] [CrossRef]
- Zhao, H.; Li, Y.; Lv, P.; Huang, J.; Tai, R.; Jin, X.; Wang, J.; Wang, X. Salmonella Phages Affect the Intestinal Barrier in Chicks by Altering the Composition of Early Intestinal Flora: Association with Time of Phage Use. Front. Microbiol. 2022, 13, 947640. [Google Scholar] [CrossRef]
- Blaser, M.J.; Kirschner, D. The equilibria that allow bacterial persistence in human hosts. Nature 2007, 449, 843–849. [Google Scholar] [CrossRef]
- Hsu, B.B.; Gibson, T.E.; Yeliseyev, V.; Liu, Q.; Lyon, L.; Bry, L.; Silver, P.A.; Gerber, G.K. Dynamic Modulation of the Gut Microbiota and Metabolome by Bacteriophages in a Mouse Model. Cell Host Microbe 2019, 25, 803–814.e805. [Google Scholar] [CrossRef] [PubMed]
- Koskella, B.; Brockhurst, M.A. Bacteria–phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol. Rev. 2014, 38, 916–931. [Google Scholar] [CrossRef] [PubMed]




| Phage | Host | ||
|---|---|---|---|
| ID | ID | ID by 16S rRNA Sequencing | % Identity/Coverage |
| ΦSx-2 | Sx-2 | Staphylococcus xylosus | 99.36/99 |
| ΦSs-1 | Ss-1 | Staphylococcus sciuri | 99.23/99 |
| ΦSs-2 | Ss-2 | Staphylococcus sciuri | 98.25/99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, X.; Deng, C.; Wang, L.; Shu, Y.; Li, S.; Song, Y.; Kong, H.; Liang, Z.; Liu, L.; Rao, Y. Isolation, Characterization, and Preliminary Application of Staphylococcal Bacteriophages in Sichuan Paocai Fermentation. Microorganisms 2025, 13, 1273. https://doi.org/10.3390/microorganisms13061273
Lin X, Deng C, Wang L, Shu Y, Li S, Song Y, Kong H, Liang Z, Liu L, Rao Y. Isolation, Characterization, and Preliminary Application of Staphylococcal Bacteriophages in Sichuan Paocai Fermentation. Microorganisms. 2025; 13(6):1273. https://doi.org/10.3390/microorganisms13061273
Chicago/Turabian StyleLin, Xia, Chunhui Deng, Luya Wang, Yue Shu, Shengshuai Li, Yunlong Song, Hong Kong, Ziwei Liang, Lei Liu, and Yu Rao. 2025. "Isolation, Characterization, and Preliminary Application of Staphylococcal Bacteriophages in Sichuan Paocai Fermentation" Microorganisms 13, no. 6: 1273. https://doi.org/10.3390/microorganisms13061273
APA StyleLin, X., Deng, C., Wang, L., Shu, Y., Li, S., Song, Y., Kong, H., Liang, Z., Liu, L., & Rao, Y. (2025). Isolation, Characterization, and Preliminary Application of Staphylococcal Bacteriophages in Sichuan Paocai Fermentation. Microorganisms, 13(6), 1273. https://doi.org/10.3390/microorganisms13061273





