Functional Outlook of Penicillium digitatum PdMFS6 Transporter to Elucidate Its Role in Fungicide Resistance and Virulence
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Media
2.2. Nucleic Acids Isolations
2.3. Cloning and Characterization of PdMFS6
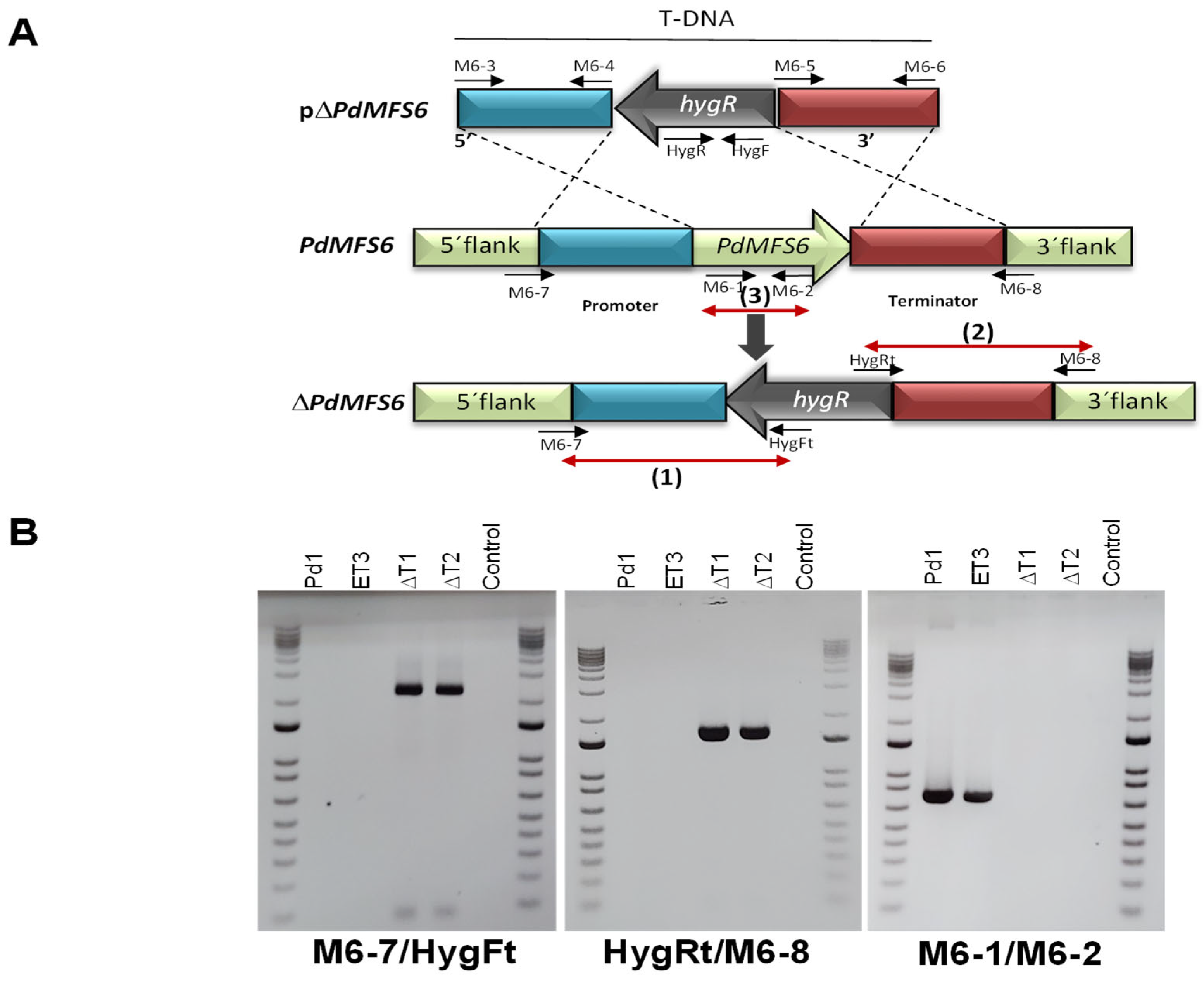
2.4. PdMFS6 Gene Elimination and Gene Over-Expression
2.5. Infection Analyses
2.6. Chemical Sensitivity
2.7. Quantitative Real-Time PCR
2.8. Statistical Analysis
3. Results
3.1. Investigation of PdMFS6
3.2. Characterization of Deletion and Over-Expression Mutants
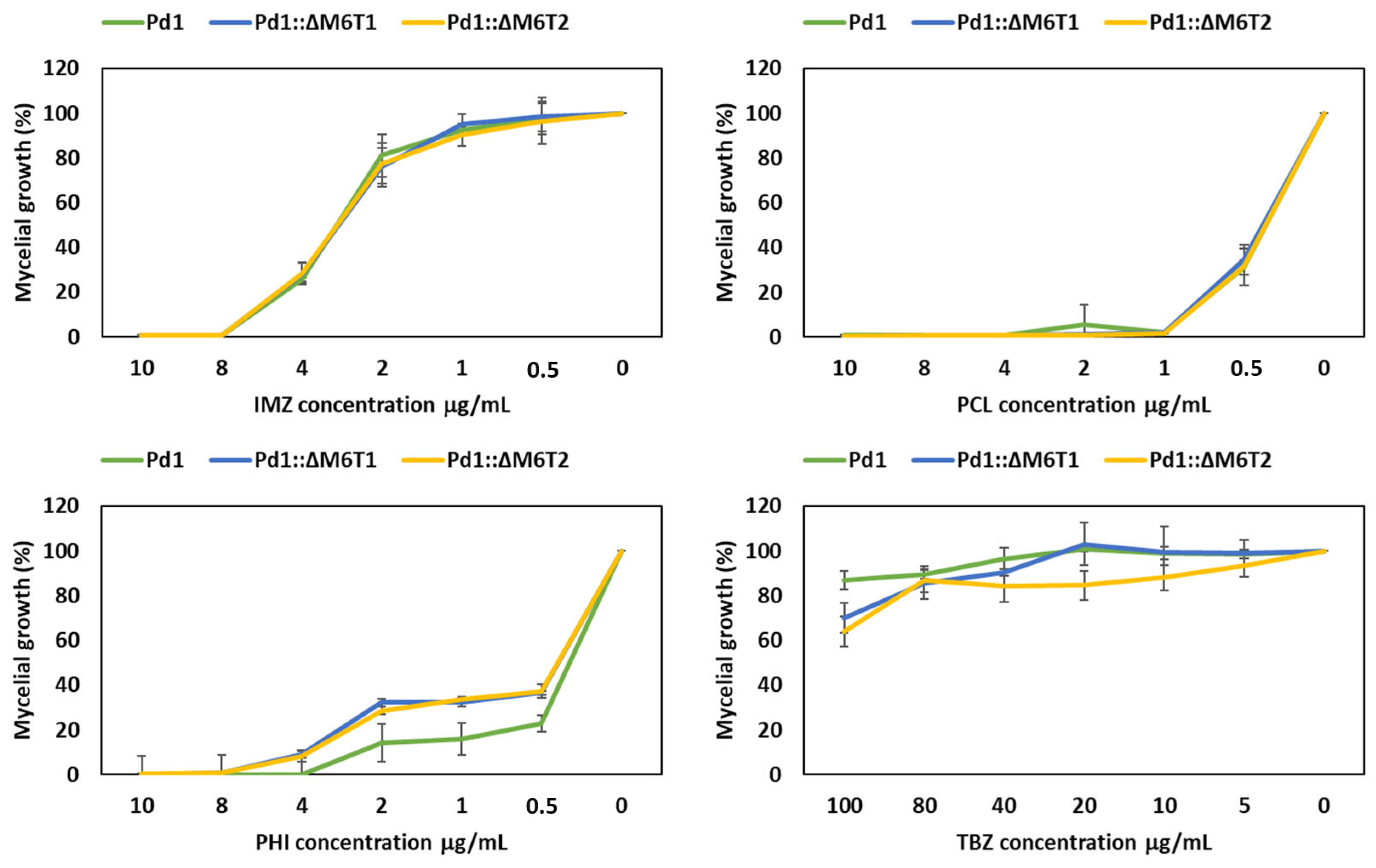
3.3. Fungicide Evaluation
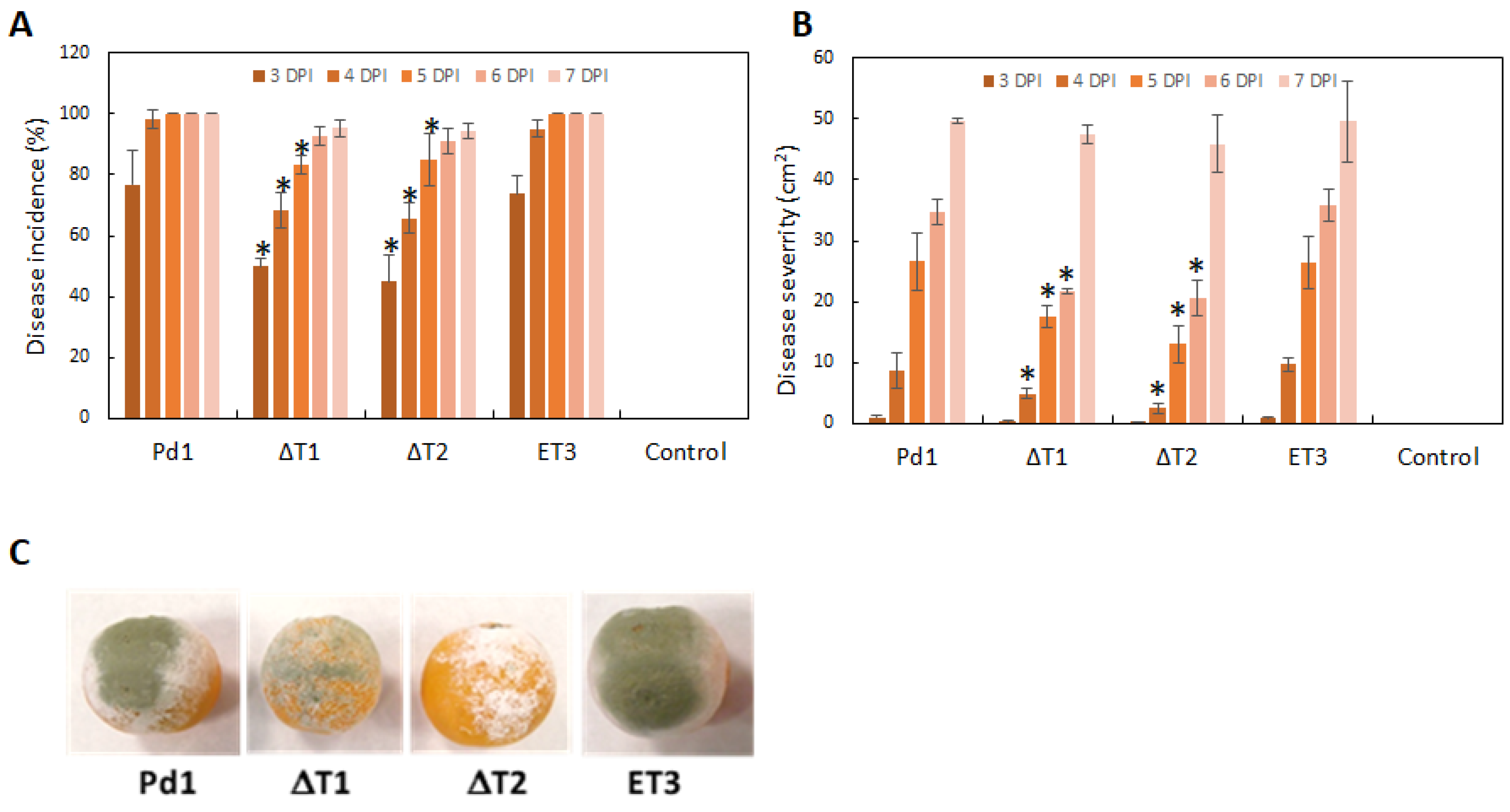
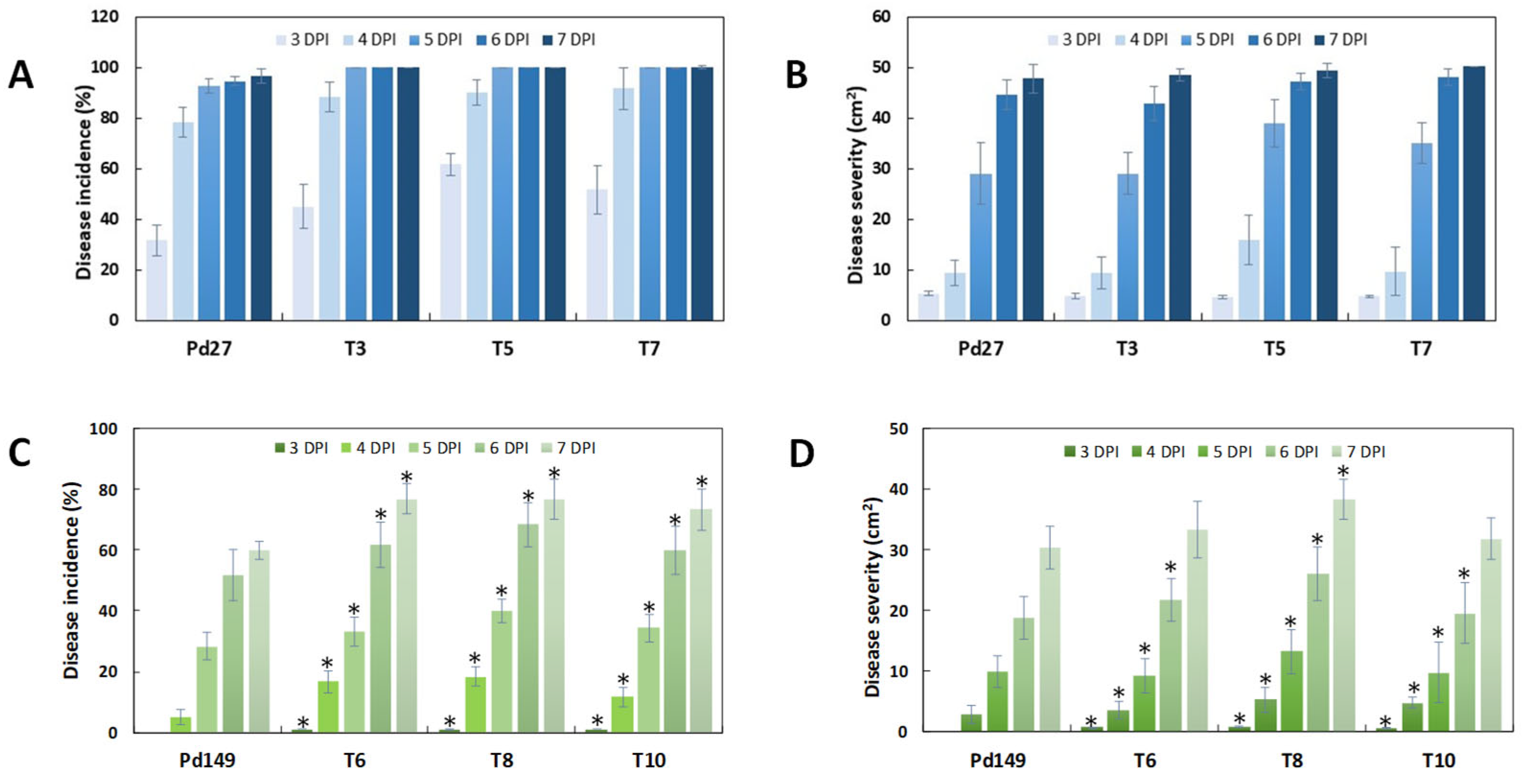
3.4. Infection Evaluation
3.5. Transcription Profile of PdMFS6
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ATMT | Agrobacterium tumefaciens mediated transformation |
| MFS | Major Facilitator Superfamily |
| MDR | Multi Drug Resistance |
| DMI | Demethylation inhibitor |
| TF | Transcription Factor |
| MAPK | mitogen-activated protein kinase |
| IMZ | Imazalil |
| TBZ | Thiabendazole |
| PHI | Philabuster |
| PCL | Prochloraz |
| DPI | Days Post Infection |
References
- Palou, L.; Smilanick, J.L.; Usall, J.; Viñas, I. Control of postharvest blue and green molds of oranges by hot water, sodium carbonate, and sodium bicarbonate. Plant Dis. 2001, 85, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Lin, Y.; Cao, H.; Li, Z. Citrus Postharvest Green Mold: Recent Advances in Fungal Pathogenicity and Fruit Resistance. Microorganisms 2020, 8, 449. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Chen, S. Non-Target Site Mechanisms of Fungicide Resistance in Crop Pathogens: A Review. Microorganisms 2021, 9, 502. [Google Scholar] [CrossRef]
- Perlin, M.H.; Andrews, J.; San Toh, S. Essential letters in the fungal alphabet: ABC and MFS transporters and their roles in survival and pathogenicity. In Advances in Genetics; Elsevier: Amsterdam, The Netherlands, 2014; Volume 85, pp. 201–253. [Google Scholar]
- Rybenkov, V.V.; Zgurskaya, H.I.; Ganguly, C.; Leus, I.V.; Zhang, Z.; Moniruzzaman, M. The Whole Is Bigger than the Sum of Its Parts: Drug Transport in the Context of Two Membranes with Active Efflux. Chem. Rev. 2021, 121, 5597–5631. [Google Scholar] [CrossRef]
- Del Sorbo, G.; Schoonbeek, H.-J.; de Waard, M.A. Fungal transporters involved in efflux of natural toxic compounds and fungicides. Fungal Gen. Biol. 2000, 30, 1–15. [Google Scholar] [CrossRef]
- Nakaune, R.; Hamamoto, H.; Imada, J.; Akutsu, K.; Hibi, T. A novel ABC transporter gene, PMR5, is involved in multidrug resistance in the phytopathogenic fungus Penicillium digitatum. Mol. Genet. Genom. 2002, 267, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Kretschmer, M.; Leroch, M.; Mosbach, A.; Walker, A.-S.; Fillinger, S.; Mernke, D.; Schoonbeek, H.-J.; Pradier, J.-M.; Leroux, P.; De Waard, M.A.; et al. Fungicide-driven evolution and molecular basis of multidrug resistance in field populations of the grey mould fungus Botrytis cinerea. PLoS Pathog. 2009, 5, e1000696. [Google Scholar] [CrossRef]
- Omrane, S.; Sghyer, H.; Audéon, C.; Lanen, C.; Duplaix, C.; Walker, A.-S.; Fillinger, S. Fungicide efflux and the MgMFS 1 transporter contribute to the multidrug resistance phenotype in Zymoseptoria tritici Field Isolates. Environ. Microbiol. 2015, 17, 2805–2823. [Google Scholar] [CrossRef]
- Dos Santos, S.C.; Teixeira, M.C.; Dias, P.J.; Sá-Correia, I. MFS transporter required for multidrug/multixenobiotic (MD/MX) resistance in the model yeast: Understanding their physiological function through postgenomic approaches. Front. Physiol. 2014, 5, 180. [Google Scholar] [CrossRef]
- Beseli, A.; Amnuaykanjanasin, A.; Herrero, S.; Thomas, E.; Daub, M.E. Membrane transporters in self-resistance of Cercospora nicotianae to the photoactivated toxin cercosporin. Curr. Genet. 2015, 61, 601–620. [Google Scholar] [CrossRef]
- Thomas, E.; Herrero, S.; Eng, H.; Gomaa, N.; Gillikin, J.; Noar, R.; Beseli, A.; Daub, M.E. Engineering Cercospora disease resistance via expression of Cercospora nicotianae cercosporin-resistance genes and silencing of cercosporin production in tobacco. PLoS ONE 2020, 15, e0230362. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M. The rising threat of fungicide resistance in plant pathogenic fungi: Botrytis as a case study. J. Chem. Biol. 2014, 7, 133–141. [Google Scholar] [CrossRef]
- Liu, M.; Peng, J.; Wang, X.; Zhang, W.; Zhou, Y.; Wang, H.; Li, X.; Yan, J.; Duan, L. Transcriptomic Analysis of Resistant and Wild-Type Botrytis cinerea Isolates Revealed Fludioxonil-Resistance Mechanisms. Int. J. Mol. Sci. 2023, 24, 988. [Google Scholar] [CrossRef]
- Leroux, P.; Walker, A.S. Activity of fungicides and modulators of membrane drug transporters in field strains of Botrytis cinerea displaying multidrug resistance. Eur. J. Plant Pathol. 2013, 135, 683–693. [Google Scholar] [CrossRef]
- Roohparvar, R.; De Waard, M.A.; Kema, G.H.; Zwiers, L.H. MgMfs1, a major facilitator superfamily transporter from the fungal wheat pathogen Mycosphaerella graminicola, is a strong protectant against natural toxic compounds and fungicides. Fungal Genet. Biol. 2007, 44, 378–388. [Google Scholar] [CrossRef]
- Roohparvar, R.; Mehrabi, R.; Van Nistelrooy, J.G.M.; Zwiers, L.; De Waard, M.A. The drug transporter MgMfs1 can modulate sensitivity of field strains of the fungal wheat pathogen Mycosphaerella graminicola to the strobilurin fungicide trifloxystrobin. Pest. Manag. Sci. 2008, 64, 685–693. [Google Scholar] [CrossRef]
- Crespo-Sempere, A.; Martínez-Culebras, P.V.; González-Candelas, L. The loss of the inducible Aspergillus carbonarius MFS transporter MfsA leads to ochratoxin A overproduction. Int. J. Food Microbiol. 2014, 181, 1–9. [Google Scholar] [CrossRef]
- Omrane, S.; Audéon, C.; Ignace, A.; Duplaix, C.; Aouini, L.; Kema, G.; Walker, A.; Fillinger, S. Plasticity of the MFS1 Promoter Leads to Multidrug Resistance in the Wheat Pathogen Zymoseptoria tritici. mSphere 2017, 2, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.H.; Tsai, H.C.; Chung, K.R. A Major Facilitator Superfamily Transporter-Mediated Resistance to Oxidative Stress and Fungicides Requires Yap1, Skn7, and MAP Kinases in the Citrus Fungal Pathogen Alternaria alternata. PLoS ONE 2017, 12, e0169103. [Google Scholar] [CrossRef]
- Samaras, A.; Ntasiou, P.; Myresiotis, C.; Karaoglanidis, G. Multidrug resistance of Penicillium expansum to fungicides: Whole transcriptome analysis of MDR strains reveals overexpression of efflux transporter genes. Int. J. Food Microbiol. 2020, 335, 108896. [Google Scholar] [CrossRef]
- Marcet-Houben, M.; Ballester, A.R.; de la Fuente, B.; Harries, E.; Marcos, J.F.; González-Candelas, L.; Gabaldón, T. Genome sequence of the necrotrophic fungus Penicillium digitatum, the main postharvest pathogen of citrus. BMC Genom. 2012, 13, 646. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Sun, X.P.; Lin, L.Y.; Zhang, T.Y.; Ma, Z.H.; Li, H.Y. PdMfs1, a major facilitator superfamily transporter from Penicillium digitatum, is partially involved in the imazalil-resistance and pathogenicity. Afr. J. Microbiol. Res. 2012, 6, 95–105. [Google Scholar]
- Wu, Z.; Wang, S.Q.; Yuan, Y.Z.; Zhang, T.F.; Liu, J.; Liu, D.L. A novel major facilitator superfamily transporter in Penicillium digitatum (PdMFS2) is required for prochloraz resistance, conidiation and full virulence. Biotechnol. Lett. 2016, 38, 1349–1357. [Google Scholar] [CrossRef]
- De Ramón-Carbonell, M.; López-Pérez, M.; González-Candelas, L.; Sánchez-Torres, P. PdMFS1 transporter contributes to Penicilliun digitatum fungicide resistance and fungal virulence during citrus fruit infection. J. Fungi 2019, 5, 100. [Google Scholar] [CrossRef]
- De Ramón-Carbonell, M.; Sánchez-Torres, P. Penicillium digitatum MFS transporters can display different roles during pathogen-fruit interaction. Int. J. Food Microbiol. 2021, 337, 108918. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Wang-Kan, X.; Neuberger, A.; van Veen, H.W.; Pos, K.M.; Piddock, L.J.V.; Luisi, B.F. Multidrug efflux pumps: Structure, function and regulation. Nat. Rev. Microbiol. 2018, 16, 523–539. [Google Scholar] [CrossRef]
- Prober, J.M.; Trainor, G.L.; Dam, R.J.; Hobbs, F.W.; Robertson, C.W.; Zagursky, R.J.; Cocuzza, A.J.; Jensen, M.A.; Baumeister, K. A system for rapid DNA sequencing with fluorescent chain terminating dideoxynucleotides. Science 1987, 238, 336–341. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Frandsen, R.J.N.; Andersson, J.A.; Kristensen, M.B.; Giese, H. Efficient four fragment cloning for the construction of vectors for targeted gene replacement in filamentous fungi. BMC Mol. Biol. 2008, 9, 70. [Google Scholar] [CrossRef]
- Sánchez-Torres, P. Molecular Mechanisms Underlying Fungicide Resistance in Citrus Postharvest Green Mold. J. Fungi 2021, 7, 783. [Google Scholar] [CrossRef] [PubMed]
- Vela-Corcía, D.; Aditya Srivastava, D.; Dafa-Berger, A.; Rotem, N.; Barda, O.; Levy, M. MFS transporter from Botrytis cinerea provides tolerance to glucosinolate-breakdown products and is required for pathogenicity. Nat. Commun. 2019, 10, 2886. [Google Scholar] [CrossRef] [PubMed]
- Bulasag, A.S.; Camagna, M.; Kuroyanagi, T.; Ashida, A.; Ito, K.; Tanaka, A.; Sato, I.; Chiba, S.; Ojika, M.; Takemoto, D. Botrytis cinerea tolerates phytoalexins produced by Solanaceae and Fabaceae plants through an efflux transporter BcatrB and metabolizing enzymes. Front. Plant Sci. 2023, 14, 1177060. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, G.; Liu, J. A major facilitator superfamily transporter in Colletotrichum fructicola (CfMfs1) is required for sugar transport, appressorial turgor pressure, conidiation and pathogenicity. For. Pathol. 2019, 49, e12558. [Google Scholar] [CrossRef]
- Liu, N.; Wang, Q.; He, C.; An, B. CgMFS1, a Major Facilitator Superfamily Transporter, Is Required for Sugar Transport, Oxidative Stress Resistance, and Pathogenicity of Colletotrichum gloeosporioides from Hevea brasiliensis. Curr. Issues Mol. Biol. 2021, 43, 1548–1557. [Google Scholar] [CrossRef]
- De Waard, M.A.; Andrade, A.C.; Hayashi, K.; Schoonbeek, H.; Stergiopoulos, I.; Zwiers, L.H. Impact of fungal drug transporters on fungicide sensitivity, multidrug resistance and virulence. Pest Manag. Sci. 2006, 62, 195–207. [Google Scholar] [CrossRef]
- Madej, M.G.; Sun, L.; Yan, N.; Kaback, H.R. Functional architecture of MFS D-glucose transporters. Proc. Natl. Acad. Sci. USA 2014, 111, E719–E727. [Google Scholar] [CrossRef]
- Drew, D.; North, R.A.; Nagarathinam, K.; Tanabe, M. Structures and General Transport Mechanisms by the Major Facilitator Superfamily (MFS). Chem. Rev. 2021, 21, 5289–5335. [Google Scholar] [CrossRef]
- De Ramón-Carbonell, M.; Sánchez-Torres, P. Involvement of Penicillium digitatum PdSUT1 in fungicide sensitivity and virulence during citrus fruit infection. Microbiol. Res. 2017, 203, 57–67. [Google Scholar] [CrossRef]
- Quistgaard, E.M.; Low, C.; Guettou, F.; Nordlund, P. Understanding transport by the major facilitator superfamily (MFS): Structures pave the way. Nat. Rev. Mol. Cell Biol. 2016, 17, 123–132. [Google Scholar] [CrossRef]
- Chen, Q.; Lei, L.; Liu, C.; Zhang, Y.; Xu, Q.; Zhu, J.; Guo, Z.; Wang, Y.; Li, Q.; Li, Y.; et al. Major Facilitator Superfamily Transporter Gene FgMFS1 Is Essential for Fusarium graminearum to Deal with Salicylic Acid Stress and for Its Pathogenicity Towards Wheat. Int. J. Mol. Sci. 2021, 22, 8497. [Google Scholar] [CrossRef] [PubMed]
- Choquer, M.; Lee, M.; Bau, H.; Chung, K. Deletion of a MFS transporter like gene in Cercospora nicotianae reduces cercosporin toxin accumulation and fungal virulence. FEMS Lett. 2007, 581, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Schoonbeek, H.J.; De Waard, M.A. Modulators of membrane drug transporters potentiate the activity of the DMI fungicide oxpoconazole against Botrytis cinerea. Pest. Manag. Sci. 2003, 59, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Stergiopoulos, I.; Gielkens, M.M.; Goodall, S.D.; Venema, K.; De Waard, M.A. Molecular cloning and characterisation of three new ATP-binding cassette transporter genes from the wheat pathogen Mycosphaerella graminicola. Gene 2002, 289, 141–149. [Google Scholar] [CrossRef]
- Sang, H.; Hulvey, J.P.; Green, R.; Xu, H.; Im, J.; Chang, T.; Jung, G. A xenobiotic detoxification pathway through transcriptional regulation in filamentous fungi. mBio 2018, 9, e00457-18. [Google Scholar] [CrossRef]
- Vilanova, L.; Teixidó, N.; Torres, R.; Usall, J.; Viñas, I.; Sánchez-Torres, P. Relevance of the transcription factor PdSte12 in Penicillium digitatum conidiation and virulence during citrus fruit infection. Int. J. Food Microbiol. 2016, 235, 93–102. [Google Scholar] [CrossRef]
- De Ramón-Carbonell, M.; Sánchez-Torres, P. PdSlt2 Penicillium digitatum mitogen-activated-protein kinase controls sporulation and virulence during citrus fruit infection. Fungal Biol. 2017, 121, 1063–1074. [Google Scholar] [CrossRef]
- Banerjee, A.; Rahman, H.; Prasad, R.; Golin, J. How fungal multidrug transporters mediate hyper resistance through DNA amplification and mutation. Mol. Microbiol. 2022, 118, 3–15. [Google Scholar] [CrossRef]
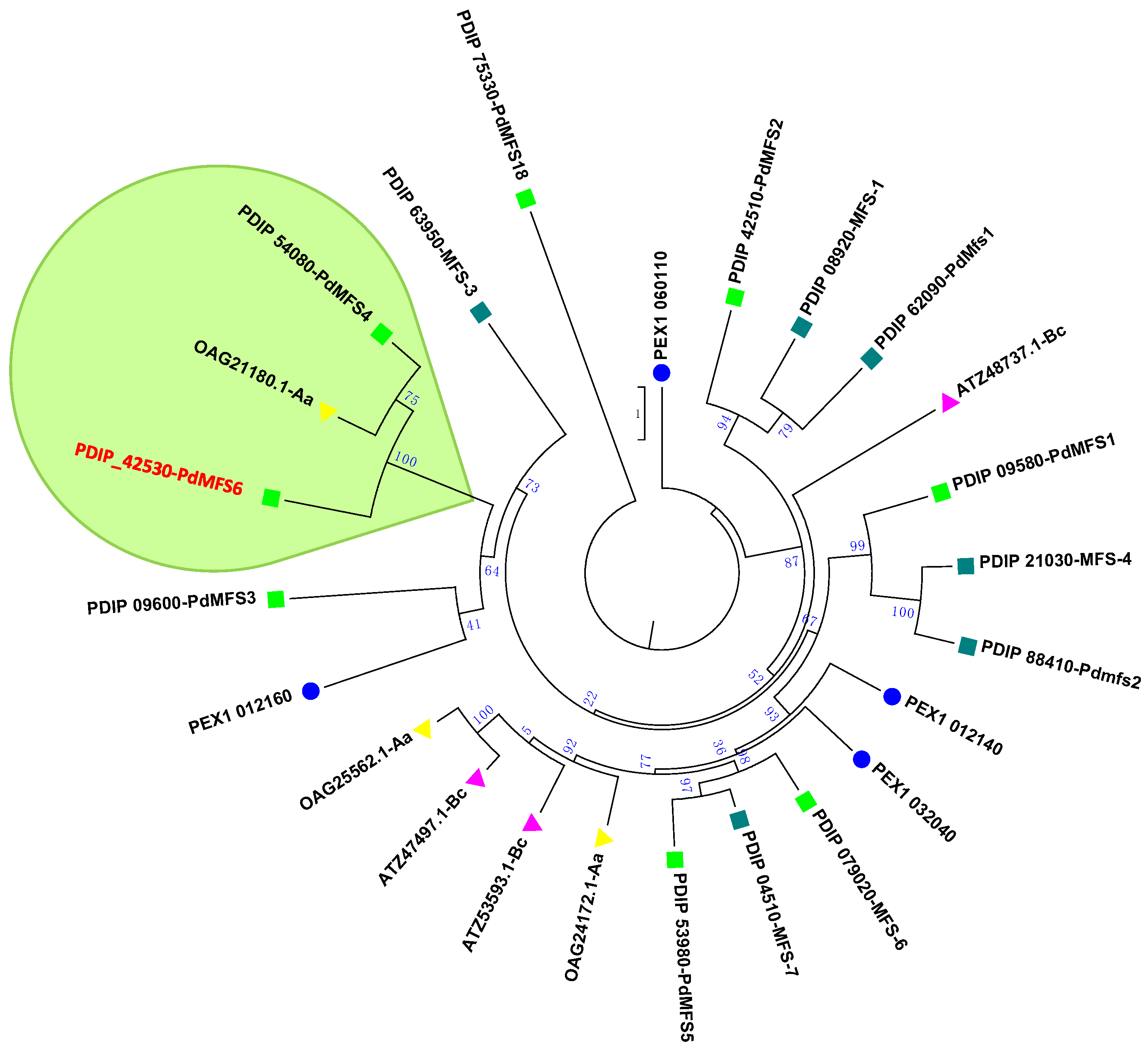


| Gene Identification | PDIP_42530 |
|---|---|
| Gene size (bp) | 1727 |
| N° Introns | 5 |
| Transmembrane helices | 12 |
| Protein Size (aa) | 502 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Torres, P. Functional Outlook of Penicillium digitatum PdMFS6 Transporter to Elucidate Its Role in Fungicide Resistance and Virulence. Microorganisms 2025, 13, 1213. https://doi.org/10.3390/microorganisms13061213
Sánchez-Torres P. Functional Outlook of Penicillium digitatum PdMFS6 Transporter to Elucidate Its Role in Fungicide Resistance and Virulence. Microorganisms. 2025; 13(6):1213. https://doi.org/10.3390/microorganisms13061213
Chicago/Turabian StyleSánchez-Torres, Paloma. 2025. "Functional Outlook of Penicillium digitatum PdMFS6 Transporter to Elucidate Its Role in Fungicide Resistance and Virulence" Microorganisms 13, no. 6: 1213. https://doi.org/10.3390/microorganisms13061213
APA StyleSánchez-Torres, P. (2025). Functional Outlook of Penicillium digitatum PdMFS6 Transporter to Elucidate Its Role in Fungicide Resistance and Virulence. Microorganisms, 13(6), 1213. https://doi.org/10.3390/microorganisms13061213






