Familial Reclassification Within Order Lysobacterales and Proposal of Four Novel Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain and Culture Conditions
2.2. Genomic Data
2.3. Phylogenetic Analyses
2.4. Genome-Based Metrics Analyses
2.5. Chemotaxonomy and Physiology
3. Results
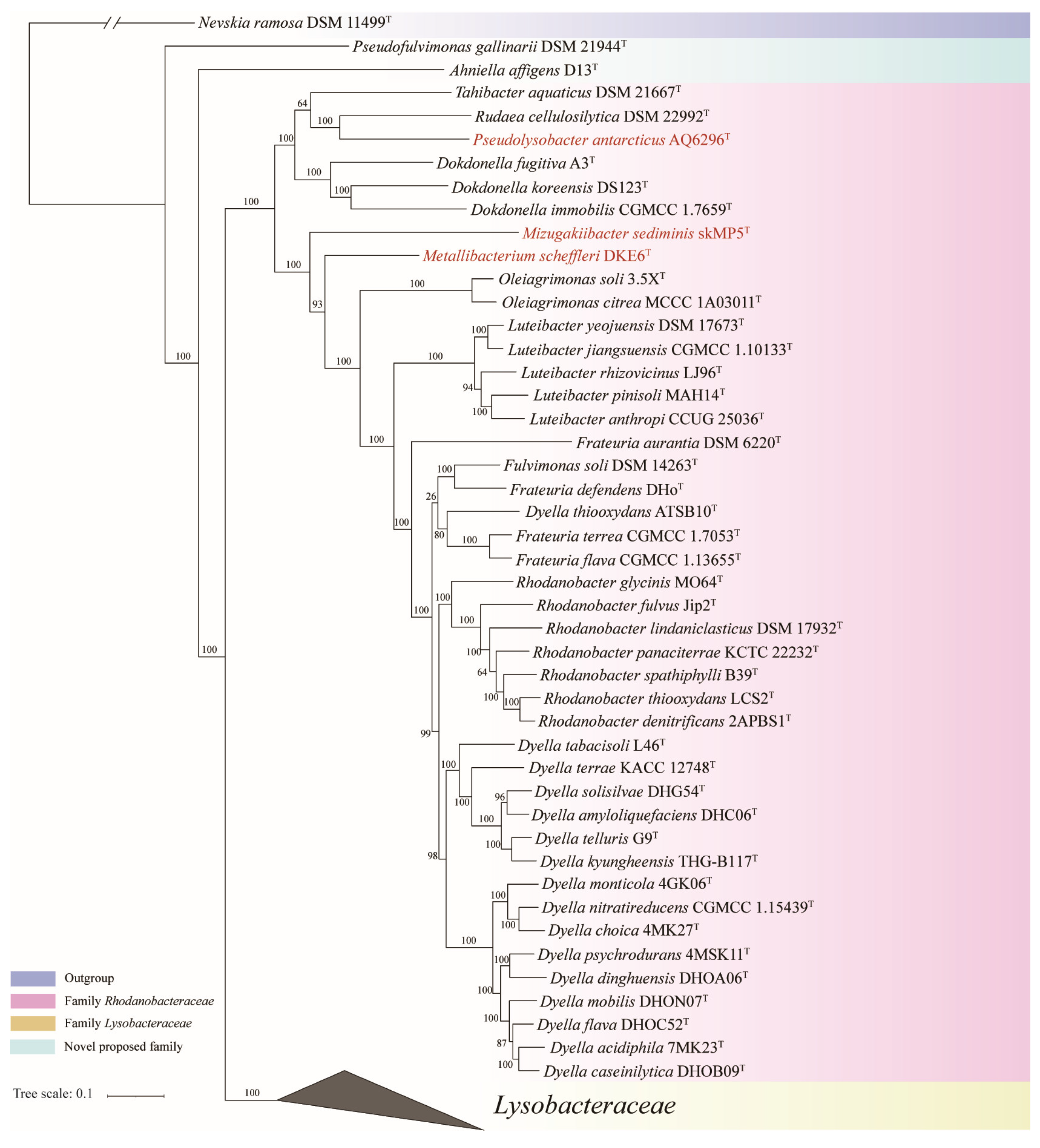
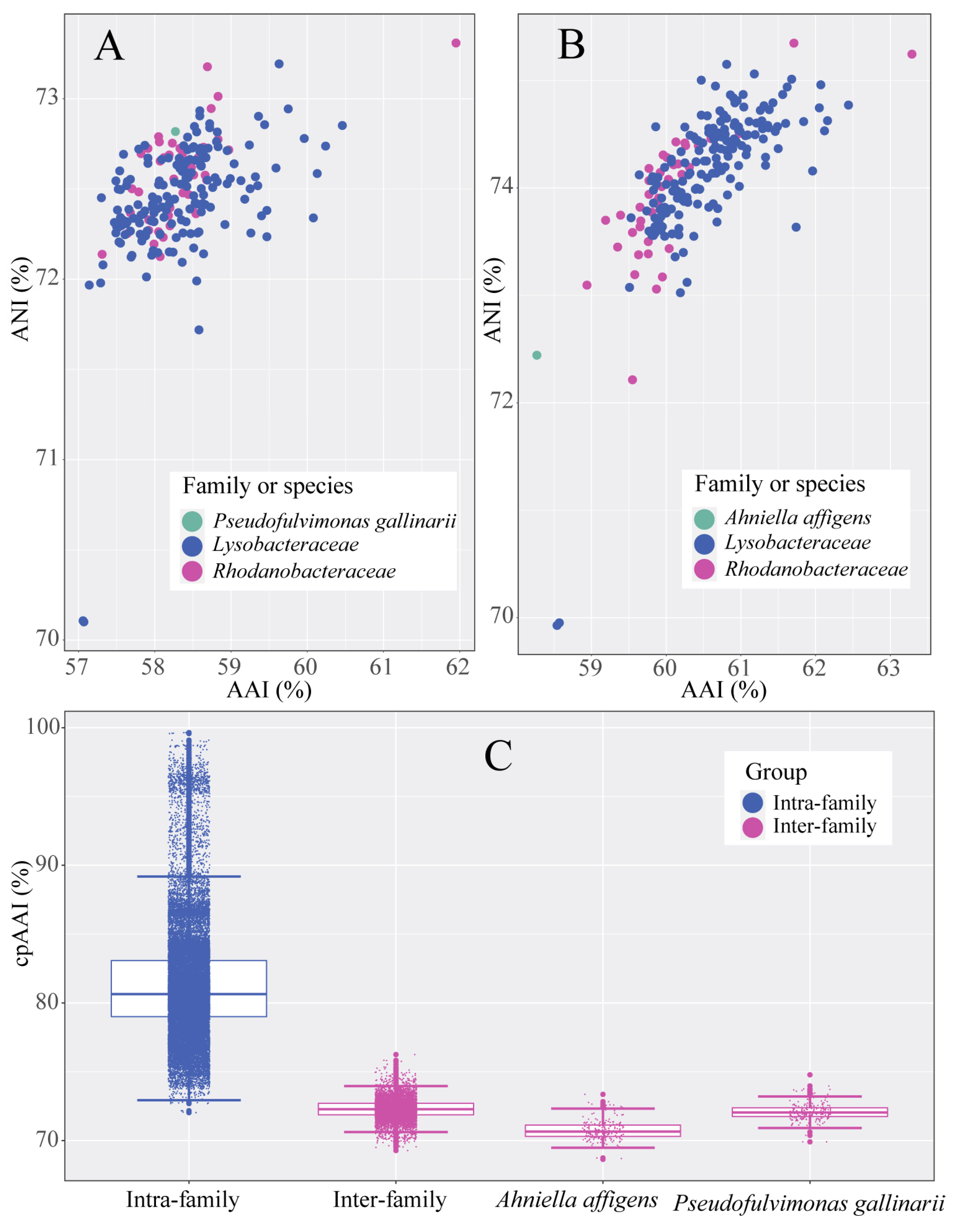
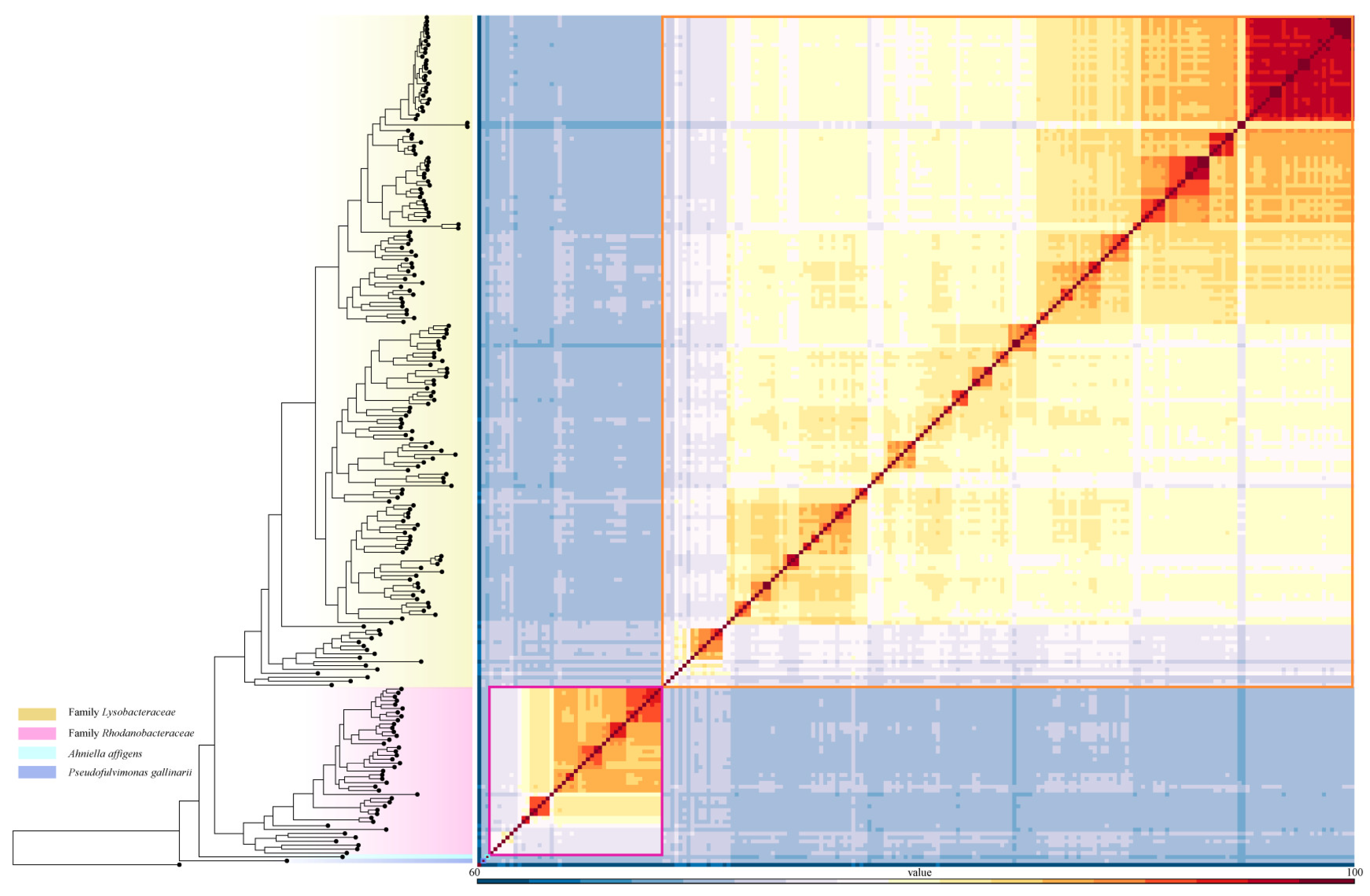
3.1. Proposal for New Family
3.2. Proposal for the Transfer of Genera Between Families Lysobacteraceae and Rhodanobacteraceae
3.3. Proposal for Four Novel Species
3.3.1. Genome-Based Phylogenetic and Metrics Analyses
3.3.2. Chemotaxonomic and Physiological Analysis
4. Discussion and Conclusions
4.1. Taxonomic Levels: New Family
Description of Pseudofulvimonadaceae fam. nov.
4.2. The Transfer for the Member of Family Rhodanobacteraceae and Lysobacteraceae
4.3. Taxonomic Levels: New Species
4.3.1. Description of Alterluteimonas quercicellularis sp. nov.
4.3.2. Description of Alterluteimonas muca sp. nov.
4.3.3. Description of Proluteimonas luteida sp. nov.
4.3.4. Description of Proluteimonas flavola sp. nov.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AAI | average amino acid identity |
| ANI | average nucleotide identity |
| cpAAI | core-proteome average amino acid identity |
| dDDH | digital DNA–DNA hybridization |
References
- Kumar, S.; Bansal, K.; Patil, P.P.; Patil, P.B. Phylogenomics insights into order and families of Lysobacterales. Access Microbiol. 2019, 1, e000015. [Google Scholar] [CrossRef] [PubMed]
- Naushad, S.; Adeolu, M.; Wong, S.; Sohail, M.; Schellhorn, H.E.; Gupta, R.S. A phylogenomic and molecular marker based taxonomic framework for the order Xanthomonadales: Proposal to transfer the families Algiphilaceae and Solimonadaceae to the order Nevskiales ord. nov. and to create a new family within the order Xanthomonadales, the family Rhodanobacteraceae fam. nov., containing the genus Rhodanobacter and its closest relatives. Antonie Van Leeuwenhoek 2015, 107, 467–485. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Chen, L.; Wang, Y.; Xing, P.; Wu, Q. Luteimonas flava sp. nov. and Aquilutibacter rugosus gen. nov., sp. nov., isolated from freshwater environments in China and re-examining the taxonomic status of genera Luteimonas and Lysobacter. Int. J. Syst. Evol. Microbiol. 2024, 74, 006585. [Google Scholar] [CrossRef] [PubMed]
- Romanenko, L.; Otstavnykh, N.; Tanaka, N.; Kurilenko, V.; Svetashev, V.; Tekutyeva, L.; Mikhailov, V.; Isaeva, M. Characterization and genomic analysis of Fererhizobium litorale gen. nov., sp. nov., isolated from the sandy sediments of the sea of Japan seashore. Microorganisms 2023, 11, 2385. [Google Scholar] [CrossRef]
- Yin, J.; He, M.; Liu, X.X.; Ren, C.B.; Liu, H.H.; Luo, H.; Chen, G.; Wang, Z.F.; Debnath, S.C.; Wang, P.M.; et al. Peteryoungia algae sp. nov. isolated from seaweeds of Gouqi Island, China, and its unique genetic features among Peteryoungia strains. Antonie Van Leeuwenhoek 2024, 117, 112. [Google Scholar] [CrossRef]
- De Lajudie, P.M.; Andrews, M.; Ardley, J.; Eardly, B.; Jumas-Bilak, E.; Kuzmanović, N.; Lassalle, F.; Lindström, K.; Mhamdi, R.; Martínez-Romero, E. Minimal standards for the description of new genera and species of rhizobia and agrobacteria. Int. J. Syst. Evol. Microbiol. 2019, 69, 1852–1863. [Google Scholar] [CrossRef]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.-A.; Hugenholtz, P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018, 36, 996–1004. [Google Scholar] [CrossRef]
- Ma, T.; Xue, H.; Piao, C.; Liu, C.; Yang, M.; Bian, D.; Li, Y. Reclassification of 11 members of the family Rhodobacteraceae at genus and species levels and proposal of Pseudogemmobacter hezensis sp. nov. Front. Microbiol. 2022, 13, 849695. [Google Scholar] [CrossRef]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef]
- Li, R.; Zhu, H.; Ruan, J.; Qian, W.; Fang, X.; Shi, Z.; Li, Y.; Li, S.; Shan, G.; Kristiansen, K. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010, 20, 265–272. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.M.; Birol, I. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009, 19, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.H.; Liao, Y.C. CISA: Contig integrator for sequence assembly of bacterial genomes. PLoS ONE 2013, 8, e60843. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Kim, J.; Na, S.I.; Kim, D.; Chun, J. UBCG2: Up-to-date bacterial core genes and pipeline for phylogenomic analysis. J. Microbiol. 2021, 59, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Na, S.I.; Kim, Y.O.; Yoon, S.H.; Ha, S.M.; Baek, I.; Chun, J. UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 2018, 56, 280–285. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Pritchard, L.; Cock, P.; Esen, Ö. Pyani, version 0.2.8. Average Nucleotide Identity (ANI) and Related Measures for Whole Genome Comparisons. Zenodo: San Francisco, CA, USA, 2019. [CrossRef]
- Ma, T.; Xue, H.; Piao, C.; Jiang, N.; Li, Y. Genome-based analyses of family Oxalobacteraceae reveal the taxonomic classification. Res. Microbiol. 2023, 7, 104076. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Paradis, E.; Schliep, K. ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef] [PubMed]
- Minnikin, D.E.; O’Donnell, A.G.; Goodfellow, M.; Alderson, G.; Athalye, M.; Schaal, A.; Parlett, J.H. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Meth. 1984, 2, 233–241. [Google Scholar] [CrossRef]
- Collins, M.D.; Pirouz, T.; Goodfellow, M.; Minnikin, D.E. Distribution of menaquinones in actinomycetes and corynebacteria. Microbiology 1977, 100, 221–230. [Google Scholar] [CrossRef]
- Groth, I.; Schumann, P.; Rainey, F.A.; Martin, K.; Schuetze, B.; Augsten, K. Demetria terragena gen. nov., sp. nov., a new genus of Actinomycetes isolated from compost soil. Int. J. Syst. Evol. Microbiol. 1997, 47, 1129–1133. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Du, H.J.; Zhang, Y.Q.; Liu, H.Y.; Su, J.; Wei, Y.Z.; Ma, B.P.; Guo, B.L.; Yu, L.Y. Allonocardiopsis opalescens gen. nov., sp. nov., a new member of the suborder Streptosporangineae, from the surface-sterilized fruit of a medicinal plant. Int. J. Syst. Evol. Microbiol. 2013, 63, 900–904. [Google Scholar] [CrossRef]
- Kuykendall, L.D.; Roy, M.A.; O’Neill, J.J.; Devine, T.E. Fatty acids, antibiotic resistance, and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int. J. Syst. Evol. Microbiol. 1988, 38, 358–361. [Google Scholar] [CrossRef]
- Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids; MIDI Inc.: Newark, NJ, USA, 1990; pp. 1–6. [Google Scholar]
- Li, Y.; Song, L.M.; Guo, M.W.; Wang, L.F.; Liang, W.X. Sphingobacterium populi sp. nov., isolated from bark of Populus × euramericana. Int. J. Syst. Evol. Microbiol. 2016, 66, 3456–3462. [Google Scholar] [CrossRef] [PubMed]
- Delory, G.E.; King, E.J. A sodium carbonate-bicarbonate buffer for alkaline phosphatases. Biochem. J. 1945, 39, 245. [Google Scholar] [CrossRef]
- Gomori, G. Preparation of buffers for use in enzyme studies. In Methods in Enzymology; Academic Press: New York, NY, USA, 1955; pp. 138–146. [Google Scholar] [CrossRef]
- Jenkins, D.; Richard, M.G.; Daigger, G.T. Manual on the Causes and Control of Activated Sludge Bulking, Foaming, and Other Solids Separation Problems; CRC Press: Boca Raton, FL, USA, 2003; pp. 9–56. [Google Scholar] [CrossRef]
- Smibert, R.M. Phenotypic characterization. In Methods for General and Molecular Bacteriology, 1st ed.; Gerhardt, P., Murray, R.G.E., Eds.; American Society for Microbiology: Washington, DC, USA, 1993; pp. 607–654. [Google Scholar]
- Zayulina, K.S.; Prokofeva, M.I.; Elcheninov, A.G.; Voytova, M.P.; Novikov, A.A.; Kochetkova, T.V.; Kublanov, I.V. Arenimonas fontis sp. nov., a bacterium isolated from Chukotka hot spring, Arctic region, Russia. Int. J. Syst. Evol. Microbiol. 2020, 70, 2726–2731. [Google Scholar] [CrossRef]
- Liang, K.Y.H.; Orata, F.D.; Boucher, Y.F.; Case, R.J. Roseobacters in a sea of poly-and paraphyly: Whole genome-based taxonomy of the family Rhodobacteraceae and the proposal for the split of the “Roseobacter clade” into a novel family, Roseobacteraceae fam. nov. Front. Microbiol. 2021, 12, 1635. [Google Scholar] [CrossRef]
- Xu, Z.; Masuda, Y.; Wang, X.; Ushijima, N.; Shiratori, Y.; Senoo, K.; Itoh, H. Genome-Based taxonomic rearrangement of the order Geobacterales including the description of Geomonas azotofigens sp. nov. and Geomonas diazotrophica sp. nov. Front. Microbiol. 2021, 12, 737531. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Lo, B.; Son, J. Phylogenomics and comparative genomic studies robustly support division of the genus Mycobacterium into an emended genus Mycobacterium and four novel genera. Front. Microbiol. 2018, 9, 67. [Google Scholar] [CrossRef] [PubMed]
- Wirth, J.S.; Whitman, W.B. Phylogenomic analyses of a clade within the roseobacter group suggest taxonomic reassignments of species of the genera Aestuariivita, Citreicella, Loktanella, Nautella, Pelagibaca, Ruegeria, Thalassobius, Thiobacimonas and Tropicibacter, and the proposal of six novel genera. Int. J. Syst. Evol. Microbiol. 2018, 68, 2393–2411. [Google Scholar] [CrossRef]
- Kuzmanović, N.; Fagorzi, C.; Mengoni, A.; Lassalle, F. Taxonomy of Rhizobiaceae revisited: Proposal of a new framework for genus delimitation. Int. J. Syst. Evol. Microbiol. 2022, 72, 005243. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613. [Google Scholar] [CrossRef]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.H.; Whitman, W.B.; Euzéby, J.; Amann, R.; Rosselló-Móra, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef]
- Hackmann, T.J. Setting new boundaries of 16S rRNA gene identity for prokaryotic taxonomy. Int. J. Syst. Evol. Microbiol. 2025, 75, 006747. [Google Scholar] [CrossRef]
- Chuvochina, M.; Mussig, A.J.; Chaumeil, P.; Skarshewski, A.; Rinke, C.; Parks, D.H.; Hugenholtz, P. Proposal of names for 329 higher rank taxa defined in the Genome Taxonomy Database under two prokaryotic codes. FEMS Microbiol. Lett. 2023, 370, fnad071. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Liu, Y.; Li, Q.; Du, H.; You, J.; Li, H.; Ke, C.; Zhang, X.; Yu, J.; Zhao, T. Luteimonas huabeiensis sp. nov., isolated from stratum water. Int. J. Syst. Evol. Microbiol. 2013, 63, 3352–3357. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, X.; Yang, J.; Lu, S.; Lai, X.H.; Jin, D.; Pu, J.; Li, J.; Huang, Y.; Zhang, G. Luteimonas yindakuii sp. nov. isolated from the leaves of Dandelion (Taraxacum officinale) on the Qinghai-Tibetan Plateau. Int. J. Syst. Evol. Microbiol. 2020, 70, 1007–1014. [Google Scholar] [CrossRef]
- Zhang, G.; Lai, X.H.; Yang, J.; Jin, D.; Pu, J.; Xiong, Y.; Yang, C.; Dong, K.; Huang, Y.; Luo, X. Luteimonas chenhongjianii, a novel species isolated from rectal contents of Tibetan Plateau pika (Ochotona curzoniae). Int. J. Syst. Evol. Microbiol. 2020, 70, 3186–3193. [Google Scholar] [CrossRef]
- Ngo, H.T.T.; Yin, C.S. Luteimonas terrae sp. nov., isolated from rhizosphere soil of Radix ophiopogonis. Int. J. Syst. Evol. Microbiol. 2016, 66, 1920–1925. [Google Scholar] [CrossRef]
- Roh, S.W.; Kim, K.H.; Nam, Y.D.; Chang, H.W.; Kim, M.S.; Yoon, J.H.; Oh, H.M.; Bae, J.W. Luteimonas aestuarii sp. nov., isolated from tidal flat sediment. J. Microbiol. 2008, 46, 525–529. [Google Scholar] [CrossRef] [PubMed]
- Riesco, R.; Trujillo, M.E. Update on the proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2024, 74, 006300. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.L.; Xie, B.B.; Zhang, X.Y.; Chen, X.L.; Zhou, B.C.; Zhou, J.; Oren, A.; Zhang, Y.Z. A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 2014, 12, 2210–2215. [Google Scholar] [CrossRef]
- Ma, T.; Xue, H.; Piao, C.; Jiang, N.; Li, Y. Phylogenomic reappraisal of the family Rhizobiaceae at the genus and species levels, including the description of Ectorhizobium quercum gen. nov., sp. nov. Front. Microbiol. 2023, 14, 1207256. [Google Scholar] [CrossRef]
- Bansal, K.; Kumar, S.; Singh, A.; Chaudhary, A.; Patil, P.B. Redefining the taxonomic boundaries of genus Xanthomonas. Taxonomy 2023, 3, 452–465. [Google Scholar] [CrossRef]
- Kämpfer, P.; Martin, E.; Lodders, N.; Langer, S.; Schumann, P.; Jäckel, U.; Busse, H.J. Pseudofulvimonas gallinarii gen. nov., sp. nov., a new member of the family Xanthomonadaceae. Int. J. Syst. Evol. Microbiol. 2010, 60, 1427–1431. [Google Scholar] [CrossRef] [PubMed][Green Version]

| Strain | Y-2-3-4FT | MHLX1AT | BDR2-5T | XNQY3-4T | ||||
|---|---|---|---|---|---|---|---|---|
| ANI | dDDH | ANI | dDDH | ANI | dDDH | ANI | dDDH | |
| MHLX1AT | 81.52 | 24.5 | ||||||
| BDR2-5T | 79.43 | 22.9 | 77.84 | 21.6 | ||||
| XNQY3-4T | 78.07 | 21.6 | 77.92 | 21.3 | 82.20 | 25.7 | ||
| Luteimonas huabeiensis HB2T | 90.78 | 41.9 | 87.27 | 24.2 | 79.50 | 22.7 | 77.85 | 21.6 |
| Luteimonas yindakuii S-1072T | 81.74 | 24.5 | 87.73 | 34.1 | 77.94 | 21.8 | 77.24 | 21.2 |
| Luteimonas chenhongjianii 100111T | 77.42 | 21.2 | 76.97 | 20.8 | 81.17 | 24.2 | 84.25 | 28.0 |
| Luteimonas terrae THG-MD21T | 77.80 | 21.5 | 76.61 | 20.8 | 81.55 | 24.6 | 88.53 | 30.0 |
| Luteimonas aestuarii B9T | 77.92 | 21.8 | 77.32 | 21.4 | 78.20 | 22.0 | 77.30 | 21.2 |
| Characteristic | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|
| Cell size (µm) | 0.6–0.8 × 1.2–1.8 | 0.5–0.8 × 1.5–2 | 0.5–0.7 × 1.5–1.8 | 0.6–0.8 × 1.6–2.0 | 0.4–0.5 × 0.9–1.6 | 0.7–1 × 1.7–2.8 | 1.1–1.4 × 1.5–1.8 | 0.4–0.5 × 1.1–1.7 | 0.5 × 1.5–2.0 |
| pH (optimum pH) | 6–9 (7–8) | 6–8.5 (7–7.5) | 6–8.5 (7–7.5) | 6–9 (7–8) | 6–11 (7) | 6.5–9.5 (7) | 6–10 (7–8) | 6.5–8 (7–7.5) | 6.5–11 (8) |
| Temperature (Optimum temperature °C) | 10–41 (28–30) | 10–37 (28) | 10–37 (28) | 10–37 (25–28) | 20–45 (30) | 4–40 (28) | 22–40 (35–37) | 4–45 (25–30) | 15–40 (34–37) |
| NaCl range (%, w/v) | 0–5 | 0–4 | 0–4 | 0–3 | 0–5 | 0–2.5 | 0–3.5 | 0–5.5 | 0–4 |
| Reduction of nitrate | + | − | − | − | − | − | − | + | − |
| Utilization of: | |||||||||
| D-mannose | − | + | − | − | + | − | − | − | + |
| D-mannitol | − | − | − | − | + | − | − | − | + |
| Methyl α-D-mannopyranoside | − | − | − | − | − | − | − | + | − |
| D-maltose | + | − | − | + | + | − | + | + | + |
| Glycogen | + | − | − | − | − | − | − | + | W |
| Xylitol | − | − | − | − | − | − | − | + | − |
| Enzyme activities: | |||||||||
| Esterase (C4) | W | + | + | + | + | + | − | + | W |
| Lipase (C14) | W | W | − | − | + | − | − | − | − |
| Valine arylamidase | W | + | W | + | + | − | − | + | + |
| Cystine arylamidase | − | W | − | − | + | − | − | W | W |
| Trypsin | + | + | + | + | + | − | + | − | + |
| α-chymotrypsin | + | + | + | + | + | + | − | + | + |
| α-glucosidase | + | − | − | − | − | − | − | + | W |
| N-acetyl-β-glucosaminidase | − | − | − | − | + | − | − | + | + |
| Hydrolysis from: | |||||||||
| Urease | − | − | − | − | + | − | − | + | + |
| Aesculin | + | + | + | + | + | + | − | + | + |
| Gelatin | + | + | + | + | + | − | − | + | + |
| G+C content (mol %) | 73.0 | 69.1 | 69.7 | 68.3 | 67.0 | 69.2 | 68.3 | 64.4 | 64.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, T.; Liu, H.; Chen, Y.; Liu, J.; Piao, C.; Xue, H.; Xu, R.; Li, Y. Familial Reclassification Within Order Lysobacterales and Proposal of Four Novel Species. Microorganisms 2025, 13, 1212. https://doi.org/10.3390/microorganisms13061212
Ma T, Liu H, Chen Y, Liu J, Piao C, Xue H, Xu R, Li Y. Familial Reclassification Within Order Lysobacterales and Proposal of Four Novel Species. Microorganisms. 2025; 13(6):1212. https://doi.org/10.3390/microorganisms13061212
Chicago/Turabian StyleMa, Tengfei, Haijiao Liu, Yafei Chen, Juan Liu, Chungen Piao, Han Xue, Risheng Xu, and Yong Li. 2025. "Familial Reclassification Within Order Lysobacterales and Proposal of Four Novel Species" Microorganisms 13, no. 6: 1212. https://doi.org/10.3390/microorganisms13061212
APA StyleMa, T., Liu, H., Chen, Y., Liu, J., Piao, C., Xue, H., Xu, R., & Li, Y. (2025). Familial Reclassification Within Order Lysobacterales and Proposal of Four Novel Species. Microorganisms, 13(6), 1212. https://doi.org/10.3390/microorganisms13061212






