In-Feed vs. In-Water Chlortetracycline Administration on the Fecal Prevalence of Virulence Genes and Pathotypes of Escherichia coli Involved in Enteric Colibacillosis in Piglets
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Study Design
2.2. Escherichia coli Enrichment
2.3. Development and Validation of an Eleven-Plex PCR to Detect Major Virulence Genes of Enteric E. coli Pathotypes
2.3.1. Gene Targets
2.3.2. Assay Conditions
2.3.3. Sensitivity of the Assay
2.4. Isolation of E. coli by Direct Plating of Enriched Fecal Samples
2.5. Subtyping of stx2e Gene in STEC Isolates
2.6. Detection of Fimbrial Genes by PCR
2.7. Phenotypic and Genotypic Antimicrobial Susceptibility Determinations
2.8. Statistical Analysis
3. Results
3.1. Prevalence of Virulence Genes in Fecal Samples
3.2. Isolation of E. coli Positive for One or More of the Nine Virulence Genes
3.3. Prevalence of E. coli Pathotypes
3.4. Prevalence of Fimbrial Genes
3.5. Antimicrobial Susceptibility Testing
3.6. Prevalence of Tetracycline Resistance Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| E. coli | Escherichia coli |
| ETEC | Enterotoxigenic Escherichia coli |
| EPEC | Enteropathogenic Escherichia coli |
| STEC | Shigatoxigenic Escherichia coli |
| aEPEC | Atypical enteropathogenic Escherichia coli |
| CTC | Chlortetracycline |
| PCR | Polymerase chain reaction |
| tet | Tetracycline |
| AMR | Antimicrobial resistance |
| MAC | MacConkey |
| mPCR | Multiplex polymerase chain reaction |
| MIC | Minimum inhibitory concentration |
| CLSI | Clinical Laboratory and Standards Institute |
| PWD | Post-weaning diarrhea |
| elt | Heat-labile enterotoxin |
| estA | Heat-stable enterotoxin A |
| estB | Heat-stable enterotoxin B |
| astA | Enteroaggregative heat-stable enterotoxin (EAST) |
| stx 1 | Shiga toxin 1 |
| stx 2 | Shiga toxin 2 |
| eae | Intimin |
| hlyA | Hemolysin |
| ehxA | Enterohemolysin |
| aggA | Subunit of enteroaggregative adherence fimbria AAF/1 |
| bfpA | Bundle-forming pilus protein |
References
- Moxley, R.A.; Duhamel, G.E. Comparative pathology of bacterial enteric diseases of swine. Adv. Exp. Med. Biol. 1999, 473, 83–101. [Google Scholar] [PubMed]
- Holland, R.E. Some infectious causes of diarrhea in young farm animals. Clin. Microbiol. Rev. 1990, 3, 345–375. [Google Scholar] [CrossRef]
- Garcia-Menino, I.; Garcia, V.; Mora, A.; Diaz-Jimenez, D.; Flament-Simon, S.C.; Alonso, M.P.; Blanco, J.E.; Blanco, M.; Blanco, J. Swine enteric colibacillosis in Spain: Pathogenic potential of mcr-1 ST10 and ST131 E. coli isolates. Front. Microbiol. 2018, 9, 2659. [Google Scholar] [CrossRef]
- Nordeste, R.; Tessema, A.; Sharma, S.; Kovac, Z.; Wang, C.; Morales, R.; Griffiths, M.W. Molecules produced by probiotics prevent enteric colibacillosis in pigs. BMC Vet. Res. 2017, 13, 335. [Google Scholar] [CrossRef]
- Wills, R.W. Diarrhea in growing-finishing swine. Vet. Clin. N. Am. Food. Anim. Pract. 2000, 16, 135–161. [Google Scholar] [CrossRef]
- Sacristan, R.D.P.; Rodriguez, A.L.; Sierens, A.; Vranckx, K.; Boyen, F.; Dereu, A.; Haesebrouck, F.; Maes, D.G.D. Efficacy of in-feed medication with chlortetracycline in a farrow-to-finish herd against a clinical outbreak of respiratory disease in fattening pigs. Vet. Rec. 2012, 171, 645. [Google Scholar] [CrossRef]
- Mayer, C.; Borges, A.; Flament-Simon, S.-C.; Simões, M. Quorum sensing architecture network in Escherichia coli virulence and pathogenesis. FEMS Microbiol. Rev. 2023, 47, fuad031. [Google Scholar] [CrossRef]
- Holman, D.B.; Bearson, B.L.; Allen, H.K.; Shippy, D.C.; Loving, C.L.; Kerr, B.J.; Bearson, S.M.D.; Brunelle, B.W. Chlortetracycline enhances tonsil colonization and fecal shedding of multidrug-resistant Salmonella enterica serovar Typhimurium DT104 without major alterations to the porcine tonsillar and intestinal microbiota. Appl. Environ. Microbiol. 2019, 85, e02354-18. [Google Scholar] [CrossRef]
- Remfry, S.E.; Amachawadi, R.G.; Shi, X.; Bai, J.; Woodworth, J.C.; Tokach, M.D.; Dritz, S.S.; Goodband, R.D.; Derouchey, J.M.; Nagaraja, T.G. Polymerase chain reaction-based prevalence of serogroups of Escherichia coli known to carry Shiga toxin genes in feces of finisher pigs. Foodborne Pathog. Dis. 2020, 17, 782–791. [Google Scholar] [CrossRef]
- Williams, H.E.; Tokach, M.D.; Dritz, S.S.; Woodworth, J.C.; DeRouchey, J.M.; Nagaraja, T.G.; Goodband, R.D.; Pluske, J.R.; Chitakasempornkul, K.; Bello, N.M.; et al. Effects of chlortetracycline alone or in combination with direct-fed microbials on nursery pig growth performance and antimicrobial resistance of fecal Escherichia coli. J. Anim. Sci. 2018, 96, 5166–5178. [Google Scholar] [CrossRef]
- Agga, G.E.; Scott, H.M.; Amachawadi, R.G.; Nagaraja, T.G.; Vinasco, J.; Bai, J.; Norby, B.; Renter, D.G.; Dritz, S.S.; Nelssen, J.L.; et al. Effects of chlortetracycline and copper supplementation on antimicrobial resistance of fecal Escherichia coli from weaned pigs. Prev. Vet. Med. 2014, 114, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Shi, X.; Nagaraja, T.G. A multiplex PCR procedure for the detection of six major virulence genes in Escherichia coli O157:H7. J. Microbiol. Methods. 2010, 82, 85–89. [Google Scholar] [CrossRef]
- Paddock, Z.; Bai, J.; Shi, X.; Renter, D.G.; Nagaraja, T.G. Detection of Escherichia coli O104 in the feces of feedlot cattle by a multiplex PCR assay designed to target major genetic traits of the virulent hybrid strain responsible for the 2011 German outbreak. Appl. Environ. Microbiol. 2013, 79, 3522–3525. [Google Scholar] [CrossRef]
- Noll, L.W.; Shridhar, P.B.; Dewsbury, D.M.; Shi, X.; Cernicchiaro, N.; Renter, D.G.; Nagaraja, T.G. A comparison of culture and PCR based methods to detect six major non-O157 serogroups of Shiga toxin-producing Escherichia coli in cattle feces. PLoS ONE 2015, 10, e0135446. [Google Scholar] [CrossRef]
- Shridhar, P.B.; Noll, L.W.; Shi, X.; Cernicchiaro, N.; Renter, D.G.; Bai, J.; Nagaraja, T.G. Escherichia coli O104 in feedlot cattle feces: Prevalence, isolation and characterization. PLoS ONE 2016, 11, e0152101. [Google Scholar] [CrossRef]
- Ng, L.K.; Martin, I.; Alfa, M.; Mulvey, M. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 2001, 15, 209–215. [Google Scholar] [CrossRef]
- Aminov, R.I.; Chee-Sanford, J.C.; Garrigues, N.; Teferedegne, B.; Krapac, I.J.; White, B.A. Development, validation, and application of PCR Primers for detection of tetracycline efflux genes of gram-negative bacteria. Appl. Environ. Microbiol. 2002, 68, 1786–1793. [Google Scholar] [CrossRef]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Nataro, J.P.; Kaper, J.B. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 1998, 11, 142. [Google Scholar] [CrossRef]
- Blank, T.E.; Donnenberg, M.S. Novel topology of BfpE, a cytoplasmic membrane protein required for type IV fimbrial biogenesis in enteropathogenic Escherichia coli. J. Bacteriol. 2001, 183, 4435–4450. [Google Scholar] [CrossRef]
- Elias, W.P.; Czeczulin, J.R.; Henderson, I.R.; Trabulsi, L.R.; Nataro, J.P. Organization of biogenesis genes for aggregative adherence fimbria II defines a virulence gene cluster in enteroaggregative Escherichia coli. J. Bacteriol. 1999, 181, 1779–1785. [Google Scholar] [CrossRef] [PubMed]
- Frydendahl, K. Prevalence of serogroups and virulence genes in Escherichia coli associated with postweaning diarrhoea and edema disease in pigs and a comparison of diagnostic approaches. Vet. Microbiol. 2002, 85, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Fairbrother, J.M.; Nadeau, E. Colibacillosis. In Diseases of Swine, 11th ed.; Zimmerman, J.J., Karriker, L.A., Ramirez, A., Schwartz, K.J., Stevenson, G.W., Zhang, J., Eds.; Wiley Blackwell: Hoboken, NJ, USA, 2019; pp. 807–834. [Google Scholar]
- Fairbrother, J.M.; Nadeau, E.; Gyles, C.L. Escherichia. coli in postweaning diarrhea in pigs: An update on bacterial types, pathogenesis, and prevention strategies. Anim. Health Res. Rev. 2005, 6, 17–39. [Google Scholar] [CrossRef]
- Smith, H.W.; Linggood, M.A. Observations on the pathogenic properties of the K88, Hly and Ent plasmids of Escherichia coli with particular reference to porcine diarrhoea. J. Med. Microbiol. 1971, 4, 467–485. [Google Scholar] [CrossRef]
- Lorenz, S.C.; Son, I.; Maounounen-Laasri, A.; Lin, A.; Fischer, M.; Kase, J.A. Prevalence of hemolysin genes and comparison of ehxA subtype patterns in Shiga Toxin-Producing Escherichia coli (STEC) and Non-STEC Strains from clinical, food, and animal sources. Appl. Environ. Microbiol. 2013, 79, 6301–6311. [Google Scholar] [CrossRef]
- Beutin, L.; Montenegro, M.A.; Orskov, I.; Orskov, F.; Prada, J.; Zimmermann, S.; Stephan, R. Close association of verotoxin (Shiga-like toxin) production with enterohemolysin production in strains of Escherichia coli. J. Clin. Microbiol. 1989, 27, 2559–2564. [Google Scholar] [CrossRef]
- Frydendahl, K.; Imberechts, H.; Lehmann, S. Automated 5’ nuclease assay for detection of virulence factors in porcine Escherichia coli. Mol. Cell. Probes 2001, 15, 151–160. [Google Scholar] [CrossRef]
- Do, T.; Stephens, C.; Townsend, K.; Wu, X.; Chapman, T.; Chin, J.; McCormick, B.; Bara, M.; Trott, D. Rapid identification of virulence genes in enterotoxigenic Escherichia coli isolates associated with diarrhea in Queensland piggeries. Aust. Vet. J. 2005, 83, 293–299. [Google Scholar] [CrossRef]
- Taras, D.; Vahjen, W.; Macha, M.; Simon, O. Performance, diarrhea incidence, and occurrence of Escherichia coli virulence genes during long-term administration of a probiotic Enterococcus faecium strain to sows and piglets. J. Anim. Sci. 2006, 84, 608–617. [Google Scholar] [CrossRef]
- Wu, X.Y.; Chapman, T.; Trott, D.J.; Bettelheim, K.; Do, T.N.; Driesen, S.; Walker, M.J.; Chin, J. Comparative analysis of virulence genes, genetic diversity, and phylogeny of commensal and enterotoxigenic Escherichia coli isolates from weaned pigs. Appl. Environ. Microbiol. 2007, 73, 83–91. [Google Scholar] [CrossRef]
- Lee, S.I.; Kang, S.G.; Kang, M.L.; Yoo, H.S. Development of multiplex polymerase chain reaction assays for detecting enterotoxigenic Escherichia coli and their application to field isolates from piglets with diarrhoea. J. Vet. Diagn. Investig. 2008, 20, 492–496. [Google Scholar] [CrossRef] [PubMed]
- Casey, T.A.; Bosworth, B.T. Design and evaluation of a multiplex polymerase chain reaction assay for the simultaneous identification of genes for nine different virulence factors associated with Escherichia coli that cause diarrhea and edema disease in swine. J. Vet. Diagn. Investig. 2009, 21, 25–30. [Google Scholar] [CrossRef]
- Mohlatlole, R.P.; Madoroba, E.; Muchadeyi, F.C.; Chimonyo, M.; Kanengoni, A.T.; Dzomba, E.F. Virulence profiles of enterotoxigenic, Shiga toxin and enteroaggregative Escherichia coli in South African pigs. Trop. Anim. Health Prod. 2013, 45, 1399–1405. [Google Scholar] [CrossRef]
- Luppi, A.; Gibellini, M.; Gin, T.; Vangroenweghe, F.; Vandenbroucke, V.; Bauerfeind, R.; Bonilauri, P.; Labarque, G.; Hidalgo, A. Prevalence of virulence factors in enterotoxigenic Escherichia coli isolated from pigs with post-weaning diarrhoea in Europe. Porc. Health Manag. 2016, 2, 20. [Google Scholar] [CrossRef]
- Chen, H.D.; Frankel, G. Enteropathogenic Escherichia coli: Unravelling pathogenesis. FEMS Microbiol. Rev. 2005, 29, 83–98. [Google Scholar] [CrossRef]
- Suzart, S.; Guth, B.E.; Pedroso, M.Z.; Okafor, U.M.; Gomes, T.A. Diversity of surface structures and virulence genetic markers among enteroaggregative Escherichia coli (EAEC) strains with and without the EAEC DNA probe sequence. FEMS Microbiol. Lett. 2001, 201, 163–168. [Google Scholar] [CrossRef]
- Jonsson, R.; Struve, C.; Boisen, N.; Mateiu, R.V.; Santiago, A.E.; Jenssen, H.; Nataro, J.P.; Krogfelt, K.A. Novel aggregative adherence fimbria variant of enteroaggregative Escherichia coli. Infect. Immun. 2015, 83, 1396–1405. [Google Scholar] [CrossRef]
- Okeke, I.N.; Nataro, J.P. Enteroaggregative Escherichia coli. Lancet Infect. Dis. 2001, 1, 304–313. [Google Scholar] [CrossRef]
- Croxen, M.A.; Law, R.J.; Scholz, R.; Keeney, K.M.; Wlodarska, M.; Finlay, B.B. Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 2013, 26, 822–880. [Google Scholar] [CrossRef]
- Zhang, R.; Gu, D.; Huang, Y.; Chan, E.W.; Chen, G.; Chen, S. Comparative genetic characterization of enteroaggregative Escherichia coli strains recovered from clinical and non-clinical settings. Sci. Rep. 2016, 6, 24321. [Google Scholar] [CrossRef]
- Tang, F.; Wang, J.; Li, D.; Gao, S.; Ren, J.; Ma, L.; Liu, F.; Zhuge, X.; Yan, G.; Lu, Y.; et al. Comparative genomic analysis of 127 Escherichia coli strains isolated from domestic animals with diarrhea in China. BMC Genom. 2019, 20, 212. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Nakazawa, M. Detection and sequences of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene in enterotoxigenic E. coli strains isolated from piglets and calves with diarrhea. J. Clin. Microbiol. 1997, 35, 223–227. [Google Scholar] [CrossRef]
- Choi, C.; Cho, W.S.; Chung, H.K.; Jung, T.; Kim, J.; Chae, C. Prevalence of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 (EAST1) gene in isolates in weaned pigs with diarrhea and/or edema disease. Vet. Microbiol. 2001, 81, 65–71. [Google Scholar] [CrossRef]
- Menard, L.P.; Dubreuil, J.D. Enteroaggregative Escherichia coli heat-stable enterotoxin 1 (EAST1): A new toxin with an old twist. Crit. Rev. Microbiol. 2002, 28, 43–60. [Google Scholar] [CrossRef]
- Osek, J. Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 (EAST1) gene and its relationship with fimbrial and enterotoxin markers in E. coli isolates from pigs with diarrhoea. Vet. Micrbiol. 2003, 91, 65–72. [Google Scholar] [CrossRef]
- Turner, S.M.; Scott-Tucker, A.; Cooper, L.M.; Henderson, I.R. Weapons of mass destruction: Virulence factors of the global killer enterotoxigenic Escherichia coli. FEMS Microbiol. Lett. 2006, 263, 10–20. [Google Scholar] [CrossRef]
- Berberov, E.M.; Zhou, Y.; Francis, D.H.; Scott, M.A.; Kachman, S.D.; Moxley, R.A. Relative importance of heat-labile enterotoxin in the causation of severe diarrheal disease in the gnotobiotic piglet model by a strain of enterotoxigenic Escherichia coli that produces multiple enterotoxins. Infect. Immun. 2004, 72, 3914–3924. [Google Scholar] [CrossRef]
- Silva, L.E.P.; Souza, T.B.; Silva, N.P.; Scaletsky, I.C.A. Detection and genetic analysis of the enteroaggregative Escherichia coli heat-stable enterotoxin (EAST1) gene in clinical isolates of enteropathogenic Escherichia coli (EPEC) strains. BMC Microbiol. 2014, 14, 135. [Google Scholar] [CrossRef]
- Blanco, M.; Lazo, L.; Blanco, J.E.; Dahbi, G.; Mora, A.; Lopez, C.; Gonzalez, E.A.; Blanco, J. Serotypes, virulence genes, and PFGE patterns of enteropathogenic Escherichia coli isolated from Cuban pigs with diarrhea. Int. Microbiol. 2006, 9, 53–60. [Google Scholar]
- Zhang, W.; Zhao, M.; Ruesch, L.; Omot, A.; Francis, D. Prevalence of virulence genes in Escherichia coli strains recently isolated from young pigs with diarrhea in the US. Vet. Microbiol. 2007, 123, 145–152. [Google Scholar] [CrossRef]
- Wang, X.M.; Liao, X.P.; Liu, S.G.; Zhang, W.J.; Jiang, H.X.; Zhang, M.J.; Zhu, H.Q.; Sun, Y.; Sun, J.; Li, A.X.; et al. Serotypes, virulence genes, and antimicrobial susceptibility of Escherichia coli isolates from pigs. Foodborne Pathog. Dis. 2011, 8, 687–692. [Google Scholar] [CrossRef]
- Byun, J.W.; Moon, B.Y.; Do, K.H.; Lee, K.H.; Lee, Y.; Kim, W.I.; So, B.J.; Lee, W.K. O-serogroups and pathovirotypes of Escherichia coli Isolated from post-weaning piglets showing diarrhoea and/or oedema in South Korea. Vet. Sci. 2022, 9, 1. [Google Scholar] [CrossRef]
- Tesh, V.L.; O’Brien, A.D. Adherence and colonization mechanisms of enteropathogenic and enterohemorrhagic Escherichia coli. Microb. Pathog. 1992, 12, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.K.; Choi, C.; Jung, K.; Kim, J.; Han, D.U.; Ha, Y.; Lee, S.D.; Kim, S.H.; Chae, C. Genotypic prevalence of the adhesin involved in diffuse adherence in Escherichia coli isolates in pre-weaned pigs with diarrhoea in Korea. J. Vet. Med. B 2004, 51, 166–168. [Google Scholar] [CrossRef] [PubMed]
- An, H.; Fairbrother, J.M.; Desautels, C.; Harel, J. Distribution of a novel locus called Paa (porcine attaching and effacing associated) among enteric Escherichia coli. Adv. Exp. Med. Biol. 1999, 473, 179–184. [Google Scholar]
- Macleod, D.L.; Gyles, C.L. Immunization of pigs with a purified Shiga-like toxin II variant toxoid. Vet. Microbiol. 1991, 29, 309–318. [Google Scholar] [CrossRef]
- Shridhar, P.B.; Siepker, C.; Noll, L.W.; Shi, X.; Nagaraja, T.G.; Bai, J. Shiga toxin subtypes of non-O157 Escherichia coli serogroups isolated from cattle feces. Front. Cell. Infect. Microbiol. 2017, 7, 121. [Google Scholar] [CrossRef]
- Tseng, M.; Fratamico, P.M.; Bagi, L.; Manzinger, D.; Funk, J.A. Shiga toxin producing E. coli (STEC) in swine: Prevalence over the finishing period and characteristics of the STEC isolates. Epidemiol. Infect. 2014, 143, 505–514. [Google Scholar] [CrossRef]
- Moxley, R.A. Edema disease. Vet. Clin. N. Am. Food Anim. Pract. 2000, 16, 175–185. [Google Scholar] [CrossRef]
- Langlois, B.; Dawson, K.; Leak, I.; Aaron, D. Effect of age and housing location on antibiotic resistance of fecal coliforms from pigs in a non-antibiotic-exposed herd. Appl. Environ. Microbiol. 1988, 54, 1341–1344. [Google Scholar] [CrossRef]
- Tadesse, D.A.; Zhao, S.; Tong, E.; Ayers, S.; Singh, A.; Bartholomew, M.J.; McDermott, P.F. Antimicrobial drug resistance in Escherichia coli from humans and food animals, United States, 1950–2002. Emerg. Infect. Dis. 2012, 18, 741–749. [Google Scholar] [CrossRef]
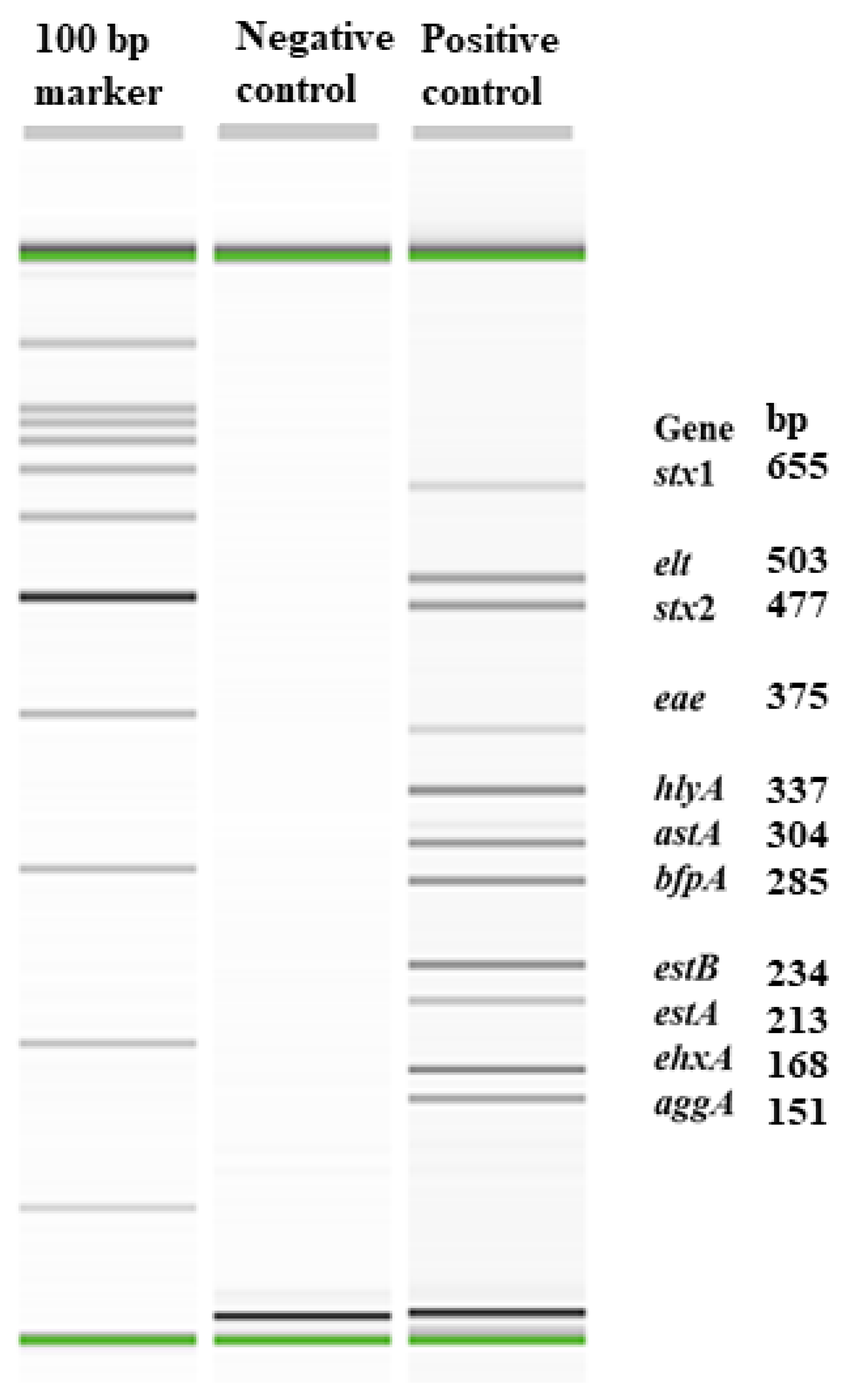
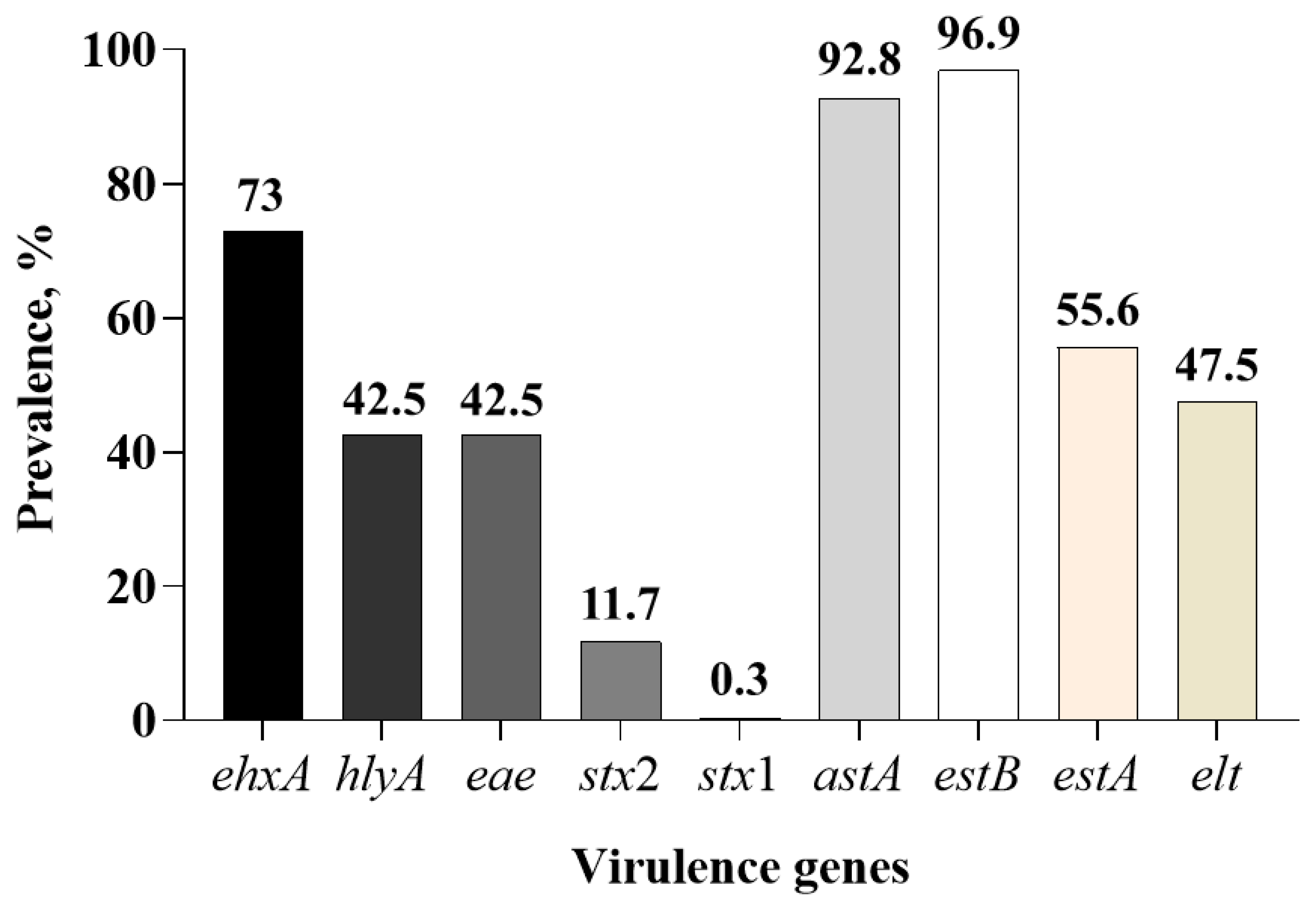
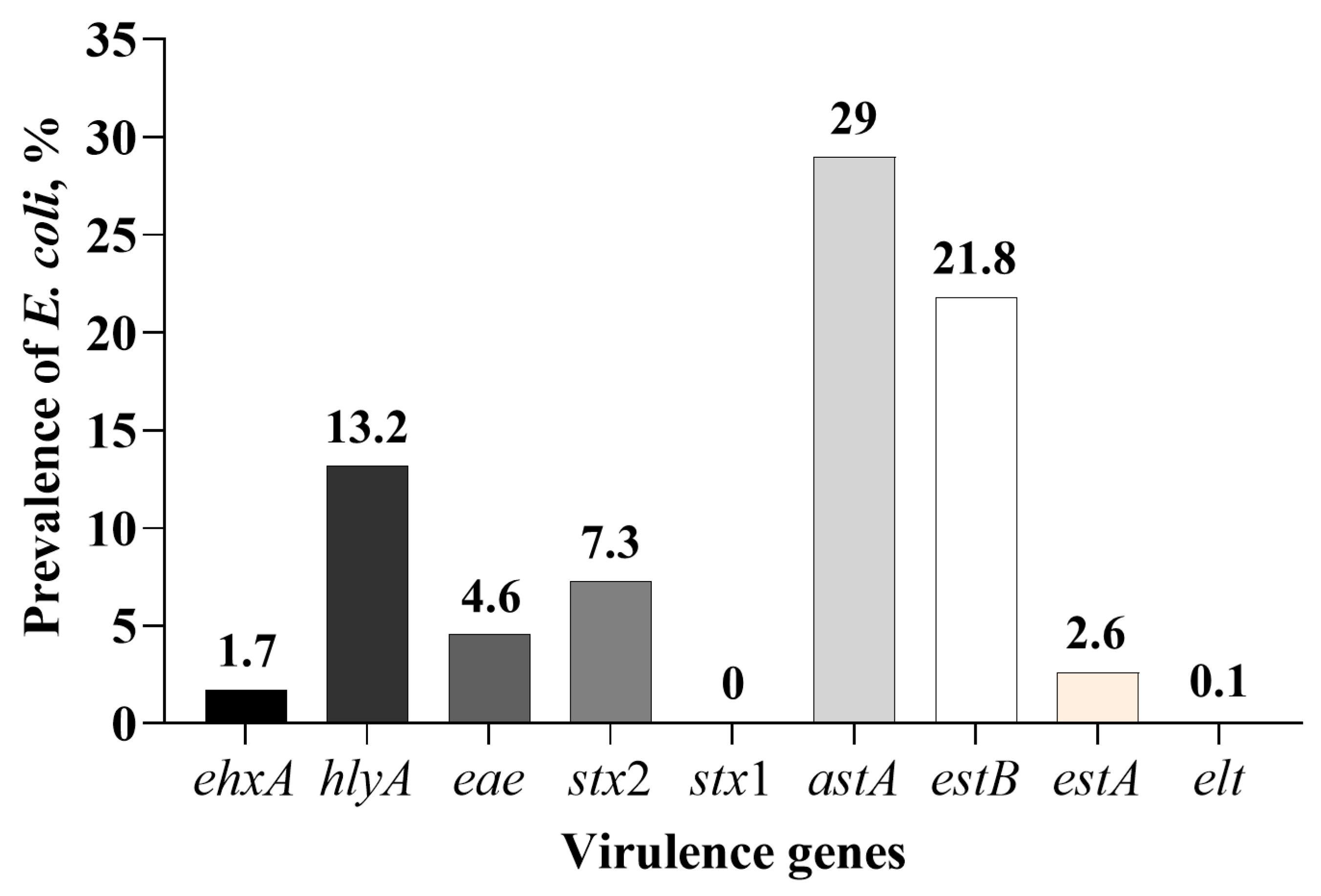
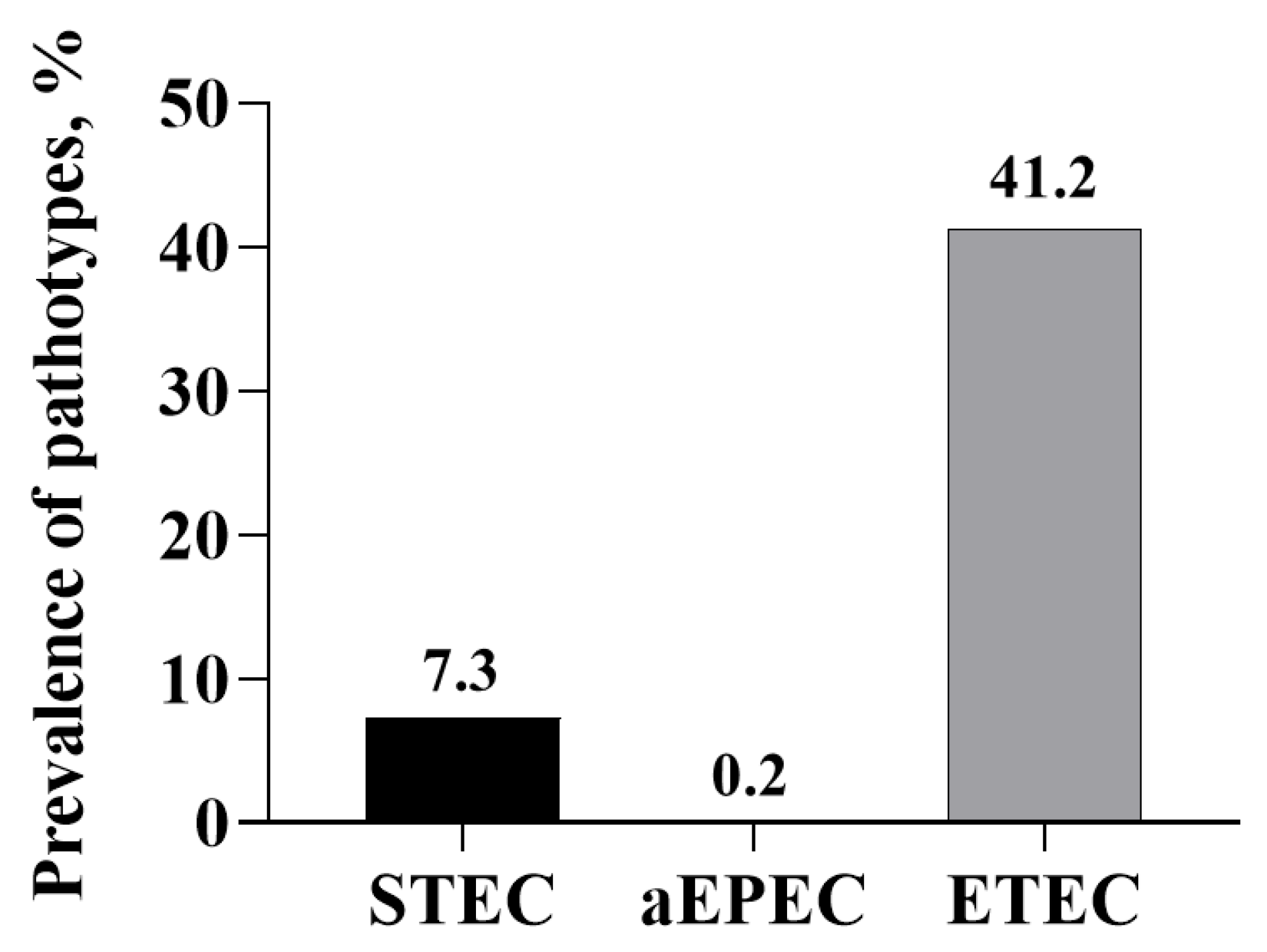

| Virulence Genes | Virulence Factors | Primers | Primer Sequences (5′ to 3′) | Amplicon Size, bp | Source |
|---|---|---|---|---|---|
| aggA | Subunit of enteroaggregative adherence fimbria AAF/1 | aggA-F1 | CGTTACAAATGATTGTCCTGTTACTAT | 151 | Paddock et al., 2013 [13] |
| aggA-R1 | ACCTGTTCCCCATAACCAGAC | ||||
| ehxA | Enterohemolysin | ehxA-F | GCGAGCTAAGCAGCTTGAAT | 168 | Bai et al., 2010 [12] |
| ehxA-R | CTGGAGGCTGCACTAACTCC | ||||
| estA | Heat-stable enterotoxin A | estA-F2 | CATGACGGGAGGTAACATGA | 213 | This study |
| estA-R2 | GGATTACAACAAAGTTCACAGCA | ||||
| estB | Heat-stable enterotoxin B | estB-F2 | CTTGACTCATATAAAAGCCCACTG | 234 | This study |
| estB-R2 | GCAGTACCATCTCTAACCCCTAAA | ||||
| bfpA | Bundle-forming pilus protein | bfpA-F2 | CAGAAGTAATGAGCGCAACG | 285 | Shridhar et al., 2016 [15] |
| bfpA-R2 | CGTAGCCTTTCGCTGAAGTA | ||||
| astA | Enteroaggregative heat-stable enterotoxin (EAST) | astA-F | GGCTCAATGTGCTGACTGAA | 304 | This study |
| astA-R | TGCCAGCTTCGGCTTATC | ||||
| hlyA | Hemolysin | hlyA-F4 | ACGAAAGTACTGGGTAATGTTGG | 337 | This study |
| hlyA-R4 | ATGTCGTTGCAGCAGCACT | ||||
| eae | Intimin | eae-F2 | TACGCGAAAGATACCGCTCT | 375 | Noll et al., 2015 [14] |
| eae-R2 | CATGCGGAAATAGCCGTTA | ||||
| stx2 | Shiga toxin 2 | stx1-F | CCATGACAACGGACAGCAGTT | 477 | Bai et al., 2010 [12] |
| stx1-R | TGTCGCCAGTTATCTGACATTC | ||||
| elt | Heat-labile enterotoxin | elt-F2 | TTATGATCACGCGAGAGGAA | 503 | This study |
| elt-R2 | TTGTGCTCAGATTCTGGGTCT | ||||
| stx1 | Shiga toxin 1 | stx1-F | TGTCGCATAGTGGAACCTCA | 655 | Bai et al., 2010 [12] |
| stx1-R | TGCGCACTGAGAAGAAGAGA |
| Genes Targeted | Encoded Protein | Primers | Primer Sequences | Amplicon Size, bp | Source |
|---|---|---|---|---|---|
| clpB | Caseinolytic protease B (A heat-shock protein) | clpB-F# | CATACGAATGCTGGATGCTG | 449 | This study |
| clpB-R | TTTGAAGAACGTTTAAAAGGCG | ||||
| uidA | Beta-glucuronidase | uidA-F | ACCACGGTGATATCGTCCAC | 449 | This study |
| uidA-R | TACAAGAAAGCCGGGCAAT | ||||
| ybbW | Putative allantoin permease | ybbW-F | AATCTGGCCGGGATTTTT | 449 | This study |
| ybbW-R | TGGCTCCGGCAATAATACAT | ||||
| faeG | F4 fimbrial adhesin | F4-F | ATTTCAATGGTTCGGTCGAT | 416 | This study |
| F4-R | CGCAGAAGTAACCCCACCT | ||||
| fanC | F5 fimbrial adhesin | F5-F | CAGGAAATACTGCTGCTAAAG | 150 | This study |
| F5-R | GCTGGGCTGAATAGTTAAATGAC | ||||
| fasA | F6 fimbrial adhesin | F6-F | ACCAGCCAGGCAAATTTAGA | 492 | This study |
| F6-R | TGTACCTGCTGAACGAATAGTCA | ||||
| fedA | F18 fimbrial adhesin | F18-F | CAGCAAGGGGATGTTAAATTC | 218 | This study |
| F18-R | AACTGCCCGCTCCAAGTTA | ||||
| f41 | F41 fimbrial adhesin | F41-F | TGATTGGACGGAAGGTCAAC | 561 | This study |
| F41-R | CCTGGCATTAACTTTTCTACATAACC | ||||
| _ | Putative glycosyltransferases | O8/O9-F | GTCTTCATCCGGGACATAGC | 735 | This study |
| O8/O9-R | CGTGAAATCGAAGAGCTGAA |
| Virulence Factor | Day 0 | Day 14 | Day 28 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control (n = 40) | In-Feed CTC (n = 40) | In-Water CTC (n = 40) | Total, n = 120 (%) | Control (n = 40) | In-Feed CTC (n = 40) | In-Water CTC (n = 40) | Total, n = 120 (%) | Control (n = 40) | In-Feed CTC (n = 40) | In-Water CTC (n = 40) | Total, n = 120 (%) | |
| aggA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| bfpA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| eae | 5 | 3 | 2 | 10 (8.3%) | 31 | 29 | 35 | 95 (79.2%) | 15 | 15 | 18 | 48 (40%) |
| elt | 0 | 0 | 0 | 15 | 23 | 19 | 57 (47.5%) | 0 | 0 | 0 | 0 | |
| estA | 2 | 1 | 0 | 3 (2.5%) | 36 | 28 | 37 | 101 (84.2%) | 33 | 32 | 31 | 96 (80%) |
| estB | 38 | 37 | 36 | 111 (92.5%) | 40 | 40 | 40 | 120 (100%) | 39 | 40 | 39 | 118 (98.3%) |
| astA | 39 | 40 | 39 | 118 (98.3%) | 33 | 38 | 33 | 104 (86.7%) | 37 | 38 | 37 | 112 (93.3%) |
| stx1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 (0.83%) |
| stx2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13 | 16 | 13 | 42 (35%) |
| hlyA | 10 | 9 | 9 | 28 (23.3%) | 31 | 29 | 35 | 95 (79.2%) | 15 | 15 | 18 | 48 (40%) |
| ehxA | 14 | 15 | 9 | 38 (31.7%) | 40 | 40 | 40 | 120 (100%) | 33 | 34 | 37 | 104 (86.7%) |
| Virulence Factor | Day 0 | Day 14 | Day 28 | Trt | Day | Trt × Day | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control (n = 8) | In-Feed CTC (n = 8) | In-Water CTC (n = 8) | Control (n = 8) | In-Feed CTC (n = 8) | In-Water CTC (n = 8) | Control (n = 8) | In-Feed CTC (n = 8) | In-Water CTC (n = 8) | ||||
| Colonization factor | ||||||||||||
| eae | 4 (50%) | 2 (25%) | 2 (25%) | 8 (100%) | 8 (100%) | 8 (100%) | 6 (75%) | 6 (75%) | 6 (75%) | 0.658 | <0.001 | 0.769 |
| Enterotoxins | ||||||||||||
| elt | 0 | 0 | 0 | 5 (62.5%) | 7 (87.5%) | 7 (87.5%) | 0 | 0 | 0 | 0.522 | <0.001 | 0.623 |
| estA | 1 (12.5%) | 1 (12.5%) | 0 | 8 (100%) | 7 (87.5%) | 8 (100%) | 8 (100%) | 8 (100%) | 8 (100%) | 0.301 | <0.001 | 0.311 |
| estB | 8 (100%) | 8 (100%) | 8 (100%) | 8 (100%) | 8 (100%) | 8 (100%) | 8 (100%) | 8 (100%) | 8 (100%) | - | - | - |
| astA | 8 (100%) | 8 (100%) | 8 (100%) | 8 (100%) | 8 (100%) | 8 (100%) | 8 (100%) | 8 (100%) | 8 (100%) | - | - | - |
| Cytotoxins | ||||||||||||
| stx1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (12.5) | 0 | 0.376 | 0.377 | 0.418 |
| stx2 | 0 | 0 | 0 | 0 | 0 | 0 | 4 (50%) | 5 (62.5%) | 3 (37.5%) | 0.926 | <0.001 | 0.989 |
| hlyA | 6 (75%) | 5 (62.5%) | 8 (100%) | 8 (100%) | 8 (100%) | 8 (100%) | 8 (100%) | 7 (87.5%) | 8 (100%) | 0.707 | <0.001 | 0.982 |
| ehxA | 5 (62.5%) | 8 (100%) | 6 (75%) | 8 (100%) | 8 (100%) | 8 (100%) | 8 (100%) | 8 (100%) | 8 (100%) | 0.920 | <0.001 | 0.337 |
| Virulence Factor | Day 0 | Day 14 | Day 28 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control (n = 40) | In-Feed CTC (n = 40) | In-Water CTC (n = 40) | Total, (n = 120) | Control (n = 40) | In-Feed CTC (n = 40) | In-Water CTC (n = 40) | Total (n = 120) | Control (n = 40) | In-Feed CTC (n = 40) | In-Water CTC (n = 40) | Total (n = 120) | |
| No. of isolates | 310 | 280 | 279 | 869 | 394 | 400 | 400 | 1194 | 400 | 400 | 400 | 1200 |
| eae | 8 (2.6) a | 21 (7.5) | 41 (14.7) | 70 (8.1) | 21 (5.3) | 23 (5.8) | 22 (5.5) | 66 (5.5) | 8 (2) | 2 (0.5) | 5 (1.3) | 15 (1.3) |
| elt | 0 | 0 | 0 | 0 | 2 (0.5) | 0 | 0 | 2 (0.2) | 0 | 0 | 0 | 0 |
| estA | 7 (2.3) | 0 | 0 | 7 (0.8) | 21 (5.3) | 12 (3) | 16 (4) | 49 (4.1) | 3 (0.8) | 12 (3) | 13 (3.3) | 28 (2.3) |
| estB | 99 (31.9) | 112 (40) | 83 (29.7) | 294 (33.8) | 78 (19.8) | 154 (38.5) | 145 (36.3) | 377 (31.6) | 2 (0.5) | 16 (4) | 25 (6.3) | 43 (6) |
| astA | 98 (32) | 102 (36.4) | 92 (33) | 292 (33.6) | 31 (7.9) | 40 (10) | 34 (8.5) | 105 (8.8) | 163 (40.8) | 209 (52.3) | 178 (44.5) | 550 (45.8) |
| stx1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| stx2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 90 (22.5) | 75 (18.7) | 73 (18.2) | 238 (19.8) |
| hlyA | 15 (4.8) | 9 (3.2) | 9 (3.2) | 33 (3.8) | 73 (18.5) | 29 (7.3) | 35 (8.8) | 137 (11.5) | 97 (24.3) | 87 (21.8) | 76 (19) | 260 (21.6) |
| ehxA | 12 (3.9) | 0 | 0 | 12 (1.4) | 9 (2.3) | 15 (3.8) | 10 (2.5) | 34 (2.8) | 5 (1.3) | 1 (0.3) | 4 (1) | 10 (0.8) |
| Virulence Genes | Day 0 | Day 14 | Day 28 | p-value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | In-Feed CTC | In- Water CTC | Control | In-Feed CTC | In- Water CTC | Control | In-Feed CTC | In- Water CTC | Trt | Day | Trt × Day | |
| Colonization factor | ||||||||||||
| eae | 2 (25%) | 3 (37.5%) | 4 (50%) | 6 (75%) | 6 (75%) | 6 (75%) | 3 (37.5%) | 2 (25%) | 2 (25%) | 0.257 | 0.030 | 0.102 |
| Enterotoxins | ||||||||||||
| elt | 0 | 0 | 0 | 1 (12.5%) | 0 | 0 | 0 | 0 | 0 | 0.377 | 0.377 | 0.418 |
| estA | 1 (12.5%) | 0 | 0 | 5 (62.5%) | 4 (50%) | 6 (75%) | 3 (37.5%) | 3 (37.5%) | 2 (25%) | 0.881 | 0.054 | 0.525 |
| estB | 6 (75%) | 7 (87.5%) | 6 (75%) | 6 (75%) | 7 (87.5%) | 7 (87.5%) | 2 (25%) | 3 (37.5%) | 4 (50%) | 0.268 | < 0.001 | 0.829 |
| astA | 6 (75%) | 7 (87.5%) | 6 (75%) | 6 (75%) | 6 (75%) | 5(62.5%) | 7 (87.5%) | 7 (87.5%) | 7 (87.5%) | 0.737 | < 0.001 | 0.976 |
| Cytotoxins | ||||||||||||
| stx1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| stx2 | 0 | 0 | 0 | 0 | 0 | 0 | 3 (37.5%) | 3 (37.5%) | 3 (37.5%) | 0.964 | 0.001 | 0.997 |
| hlyA | 2 (25%) | 3 (37.5%) | 2 (25%) | 8 (100%) | 4 (50%) | 6 (75%) | 6 (75%) | 5 (62.5%) | 6 (75%) | 0.576 | 0.016 | 0.963 |
| ehxA | 2 (25%) | 0 | 0 | 4 (50%) | 6 (75%) | 4 (50%) | 2 (25%) | 1 (12.5%) | 2 (25%) | 0.387 | 0.069 | 0.256 |
| Pathotypes | Day 0 | Day 14 | Day 28 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control (n = 40) | In-Feed CTC (n = 40) | In-Water CTC (n = 40) | Total (n = 120) | Control (n = 40) | In-Feed CTC (n = 40) | In-Water CTC (n = 40) | Total (n = 120) | Control (n = 40) | In-Feed CTC (n = 40) | In-Water CTC (n = 40) | Total (n = 120) | |
| No. of isolates | 310 | 280 | 279 | 869 | 394 | 400 | 400 | 1194 | 400 | 400 | 400 | 1200 |
| ETEC a | 107 | 111 | 93 | 311 | 104 | 190 | 152 | 446 | 168 | 220 | 199 | 587 |
| Heat-labile (elt) | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| Heat-stable A (estA) | 7 | 0 | 0 | 7 | 21 | 12 | 16 | 49 | 3 | 12 | 13 | 28 |
| Heat-stable B (estB) | 99 | 111 | 83 | 294 | 78 | 154 | 145 | 377 | 2 | 16 | 25 | 43 |
| Enteroaggregative heat-stable (astA) | 98 | 102 | 92 | 292 | 31 | 40 | 34 | 105 | 163 | 209 | 178 | 550 |
| elt + estB | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| estA + estB | 7 | 0 | 0 | 7 | 21 | 11 | 16 | 48 | 0 | 8 | 10 | 18 |
| estA + astA | 0 | 0 | 0 | 0 | 2 | 1 | 1 | 4 | 0 | 8 | 7 | 15 |
| estB + astA | 90 | 102 | 82 | 274 | 4 | 5 | 23 | 32 | 0 | 8 | 7 | 15 |
| estA + estB + astA | 0 | 0 | 0 | 0 | 2 | 1 | 1 | 4 | 0 | 8 | 7 | 15 |
| aEPEC b | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 4 | 0 | 1 | 0 | 1 |
| STEC c | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 90 | 75 | 73 | 238 |
| Pathotype | Day 0 | Day 14 | Day 28 | Trt | Day | Trt × Day | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control (n = 8) | In-Feed CTC (n = 8) | In-Water CTC (n = 8) | Control (n = 8) | In-Feed CTC (n = 8) | In-Water CTC (n = 8) | Control (n = 8) | In-Feed CTC (n = 8) | In-Water CTC (n = 8) | ||||
| Enterotoxigenic E. coli a | 6 (75%) | 7 (87.5%) | 6 (75%) | 7 (87.5%) | 7 (87.5%) | 7 (87.5%) | 7 (87.5%) | 7 (87.5%) | 7 (87.5%) | 0.929 | 0.640 | 0.977 |
| Atypical Enteropathogenic E. coli b | 0 | 0 | 0 | 1 (12.5%) | 1 (12.5%) | 2 (25%) | 0 | 1 (12.5%) | 0 | 0.815 | 0.004 | 0.895 |
| Shigatoxigenic E. coli c | 0 | 0 | 0 | 0 | 0 | 0 | 3 (37.5%) | 3 (37.5%) | 3 (37.5%) | 1.000 | <0.001 | 1.000 |
| Fimbriae Genes | Day 0 | Day 14 | Day 28 | Overall Total, n = 1344 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control (n = 107) | In-Feed CTC (n = 111) | In-Water CTC (n = 93) | Total, n = 311 | Control (n = 104) | In-Feed CTC (n = 190) | In-Water CTC (n = 152) | Total, n = 446 | Control (n = 168) | In-Feed CTC (n = 220) | In-Water CTC (n = 199) | Total, n = 587 | ||
| F4-faeG | 7 | 0 | 0 | 7 | 1 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 9 * |
| F5-fanC | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F6-fasA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F18-fedA | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 2 ** |
| F41-f41 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Treatment Groups | Pathotypes | No. of Isolates | Chlortetracycline, µg/mL | Tetracycline Resistance Genes (%, Proportion) | |||
|---|---|---|---|---|---|---|---|
| MIC | 95% Confidence Interval | tetA | tetB | tetD | |||
| Control | ETEC a | 35 | 54.2 | [44.9–63.6] | 31 (88.5%) | 5 (14.3%) | 4 (11.4%) |
| STEC b | 3 | 45.8 | [27.9–63.7] | 0 | 3 (100%) | 0 | |
| Others c | 14 | 62.2 | [43.3–81.2] | 14 (100%) | 1 (7.1%) | 4 (28.5%) | |
| Total | 52 | 50 | [37.5–62.5] | 45 (86.5%) | 9 (17.3%) | 8 (15.4%) | |
| In-feed CTC | ETEC a | 41 | 53.9 | [46.6–61.2] | 40 (97.5%) | 5 (12.2%) | 3 (7.3%) |
| STEC b | 3 | 43.7 | [28.2–59.3] | 0 | 3 (100%) | 0 | |
| Others c | 13 | 75.5 | [58.3–92.8] | 13 (100%) | 1 (7.7%) | 2 (15.4%) | |
| Total | 57 | 50 | [44.7–60.5] | 53 (93%) | 9 (15.8%) | 5 (8.7%) | |
| In-water CTC | ETEC a | 36 | 58.8 | [49.6–68.0] | 35 (97.2%) | 5 (13.8%) | 5 (13.8%) |
| STEC b | 3 | 45.8 | [36.8–54.7] | 2 (66.6%) | 3 (100%) | 0 | |
| Others c | 17 | 74.4 | [58.2–90.6] | 16 (94.1%) | 2 (11.7%) | 3 (17.6%) | |
| Total | 56 | 62.5 | [50–75] | 53 (94.6%) | 10 (17.8%) | 8 (14.3%) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalam, R.; Amachawadi, R.G.; Shi, X.; Bai, J.; Abbasi, M.; Tokach, M.D.; Nagaraja, T.G. In-Feed vs. In-Water Chlortetracycline Administration on the Fecal Prevalence of Virulence Genes and Pathotypes of Escherichia coli Involved in Enteric Colibacillosis in Piglets. Microorganisms 2025, 13, 1185. https://doi.org/10.3390/microorganisms13061185
Kalam R, Amachawadi RG, Shi X, Bai J, Abbasi M, Tokach MD, Nagaraja TG. In-Feed vs. In-Water Chlortetracycline Administration on the Fecal Prevalence of Virulence Genes and Pathotypes of Escherichia coli Involved in Enteric Colibacillosis in Piglets. Microorganisms. 2025; 13(6):1185. https://doi.org/10.3390/microorganisms13061185
Chicago/Turabian StyleKalam, Ramya, Raghavendra G. Amachawadi, Xiaorong Shi, Jianfa Bai, Mina Abbasi, Mike D. Tokach, and Tiruvoor G. Nagaraja. 2025. "In-Feed vs. In-Water Chlortetracycline Administration on the Fecal Prevalence of Virulence Genes and Pathotypes of Escherichia coli Involved in Enteric Colibacillosis in Piglets" Microorganisms 13, no. 6: 1185. https://doi.org/10.3390/microorganisms13061185
APA StyleKalam, R., Amachawadi, R. G., Shi, X., Bai, J., Abbasi, M., Tokach, M. D., & Nagaraja, T. G. (2025). In-Feed vs. In-Water Chlortetracycline Administration on the Fecal Prevalence of Virulence Genes and Pathotypes of Escherichia coli Involved in Enteric Colibacillosis in Piglets. Microorganisms, 13(6), 1185. https://doi.org/10.3390/microorganisms13061185






