Biodegradation of Phenanthrene by Mycobacterium sp. TJFP1: Genetic Basis and Environmental Validation
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Culture Media
2.2. Strain Resource
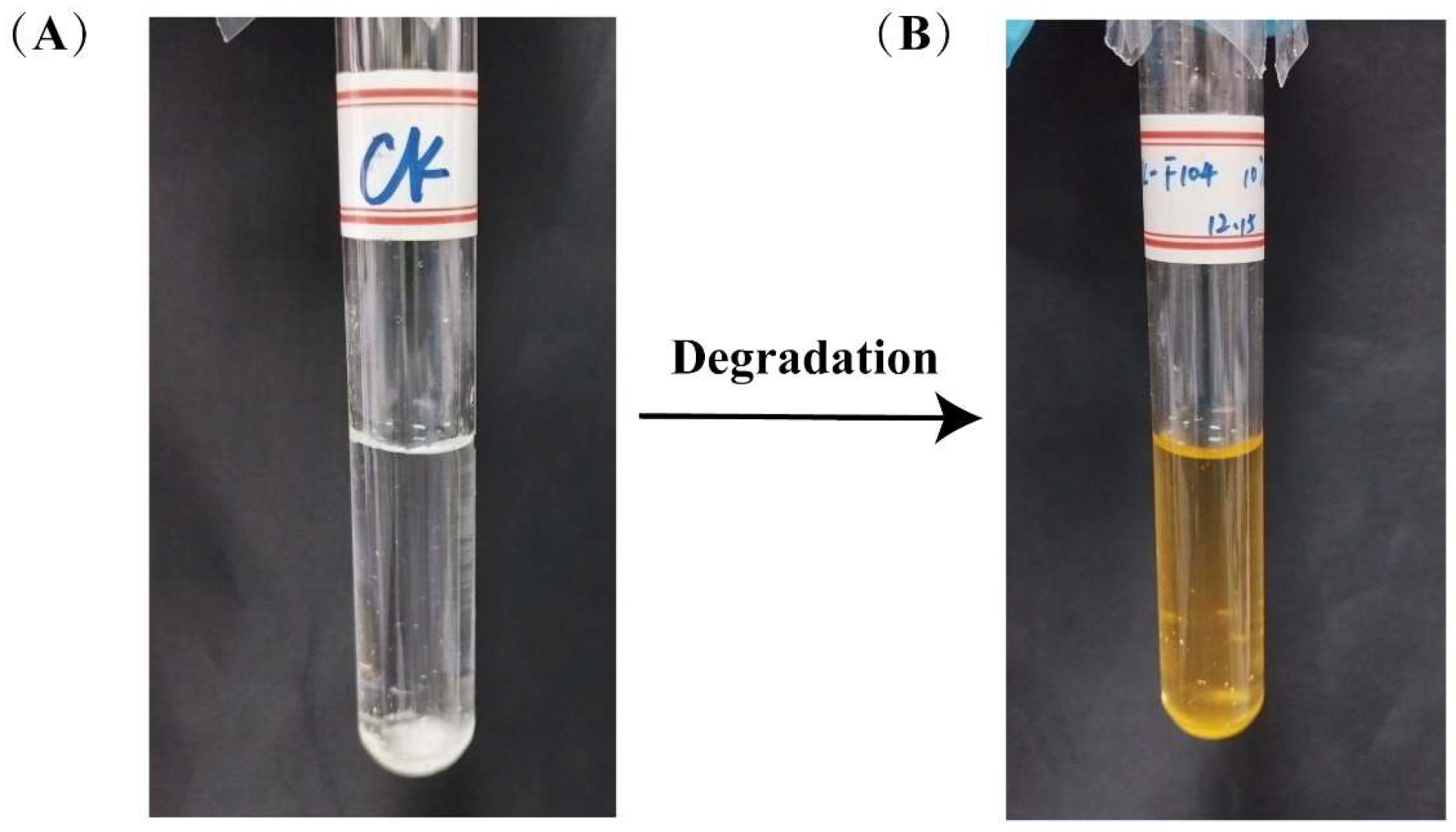
2.3. Validation of the Degradation Function of Phenanthrene and Other PAHs by Stain TJFP1
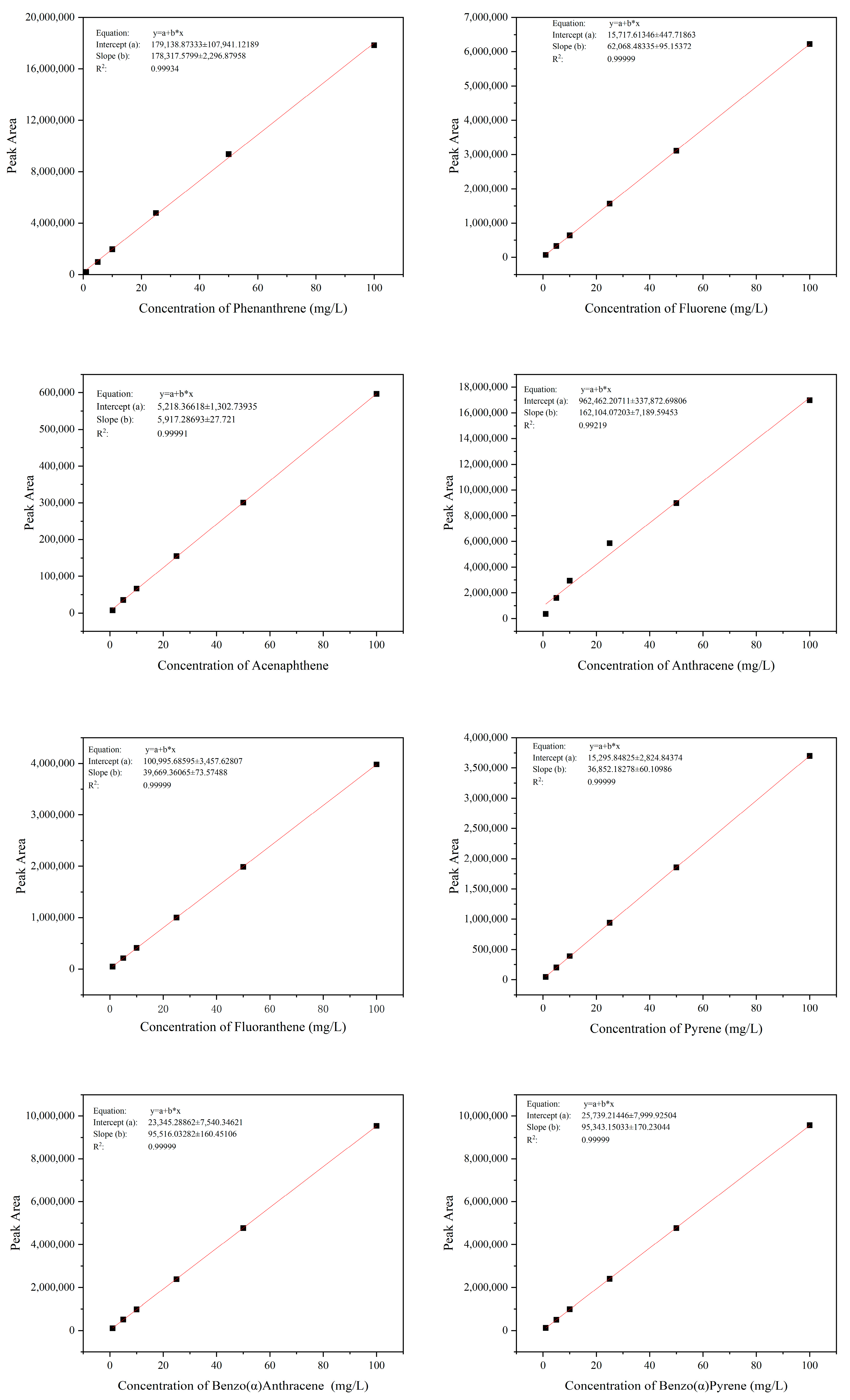
2.4. Detection and Quantification of Phenanthrene and Other PAHs
2.5. Effects of Environmental Conditions on the Degradation Performance of Stain TJFP1
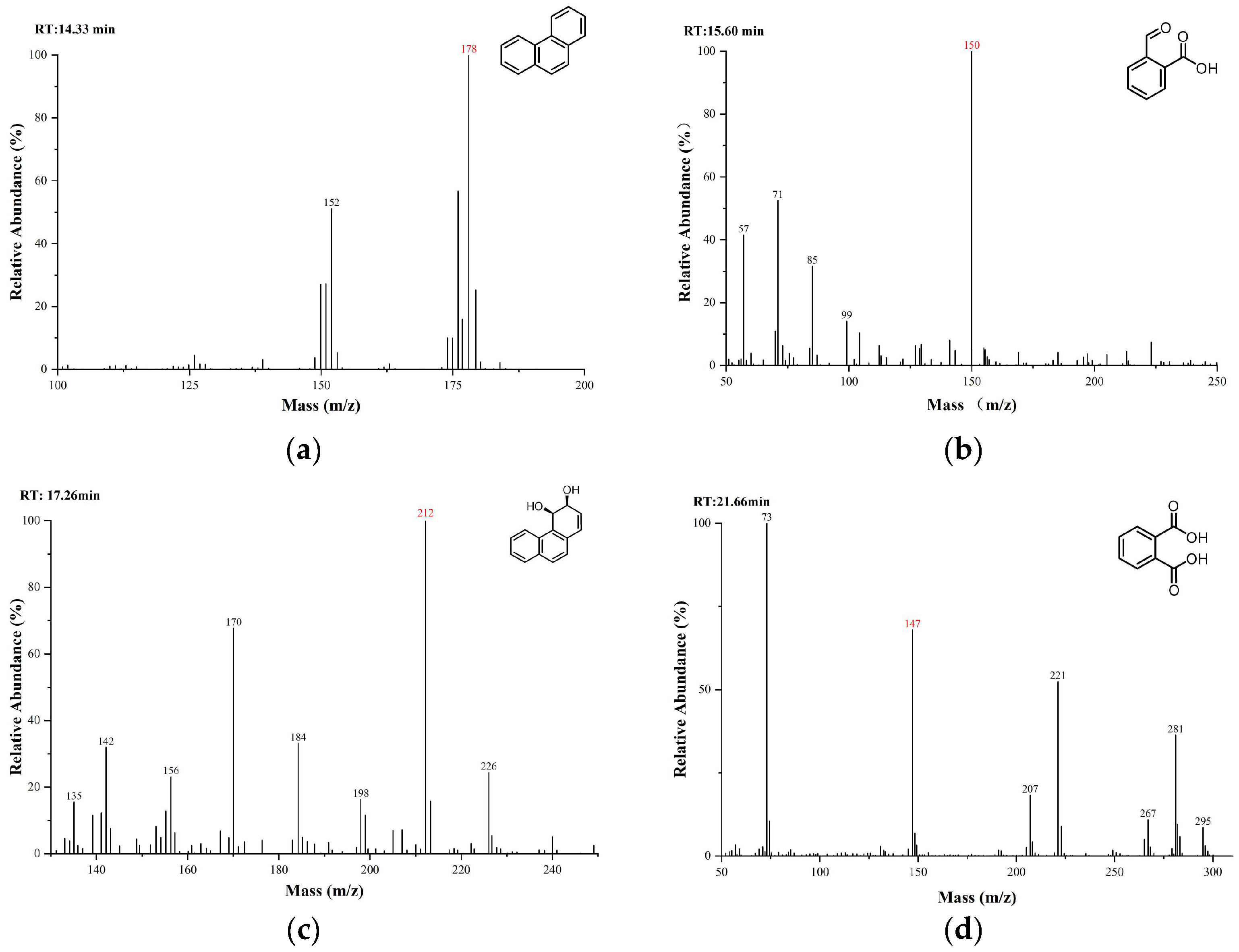
2.6. Detection of Phenanthrene Biodegradation Intermediates
2.7. Whole Genome Sequencing and Annotation Analysis
2.8. Simulation of Soil Remediation Experiments
3. Results
3.1. Determination of the Degradation Function of Strain TJFP1 on Phenanthrene and Other PAHs
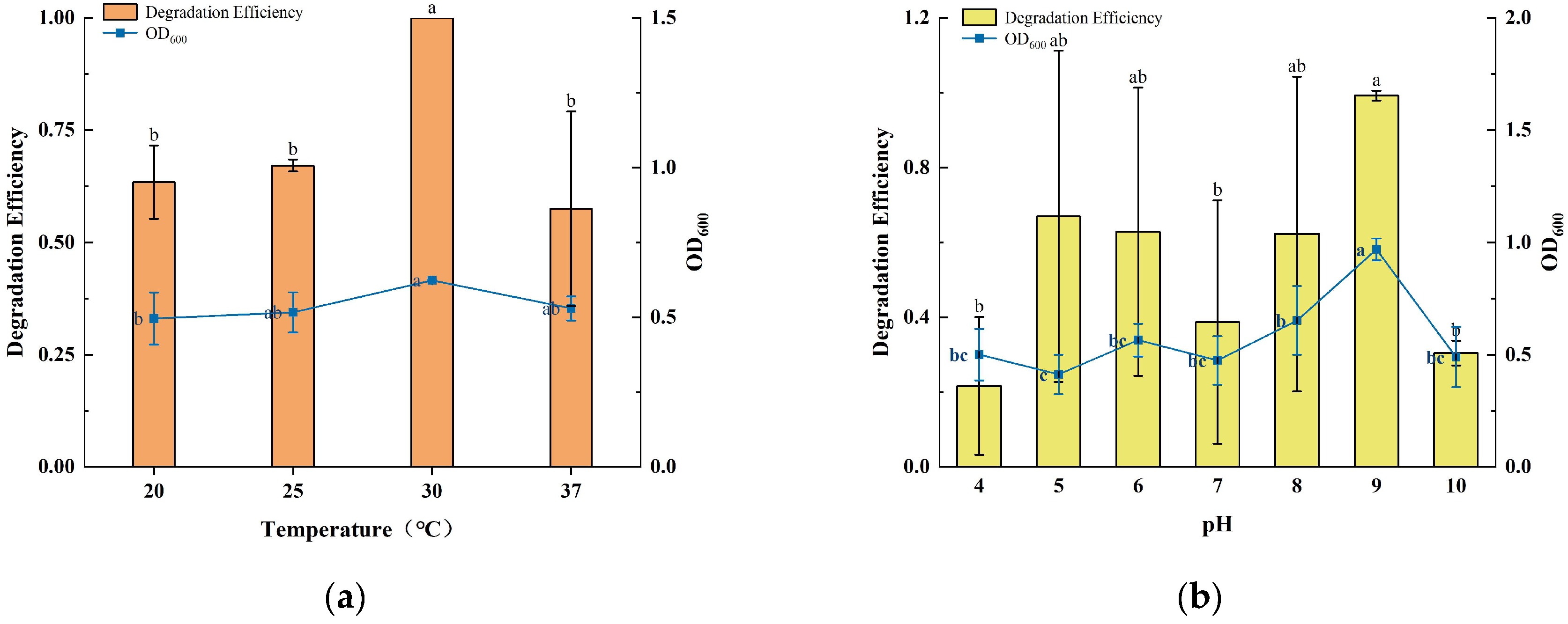
3.2. Effect of Temperature, pH, and Initial Concentration on Degradation
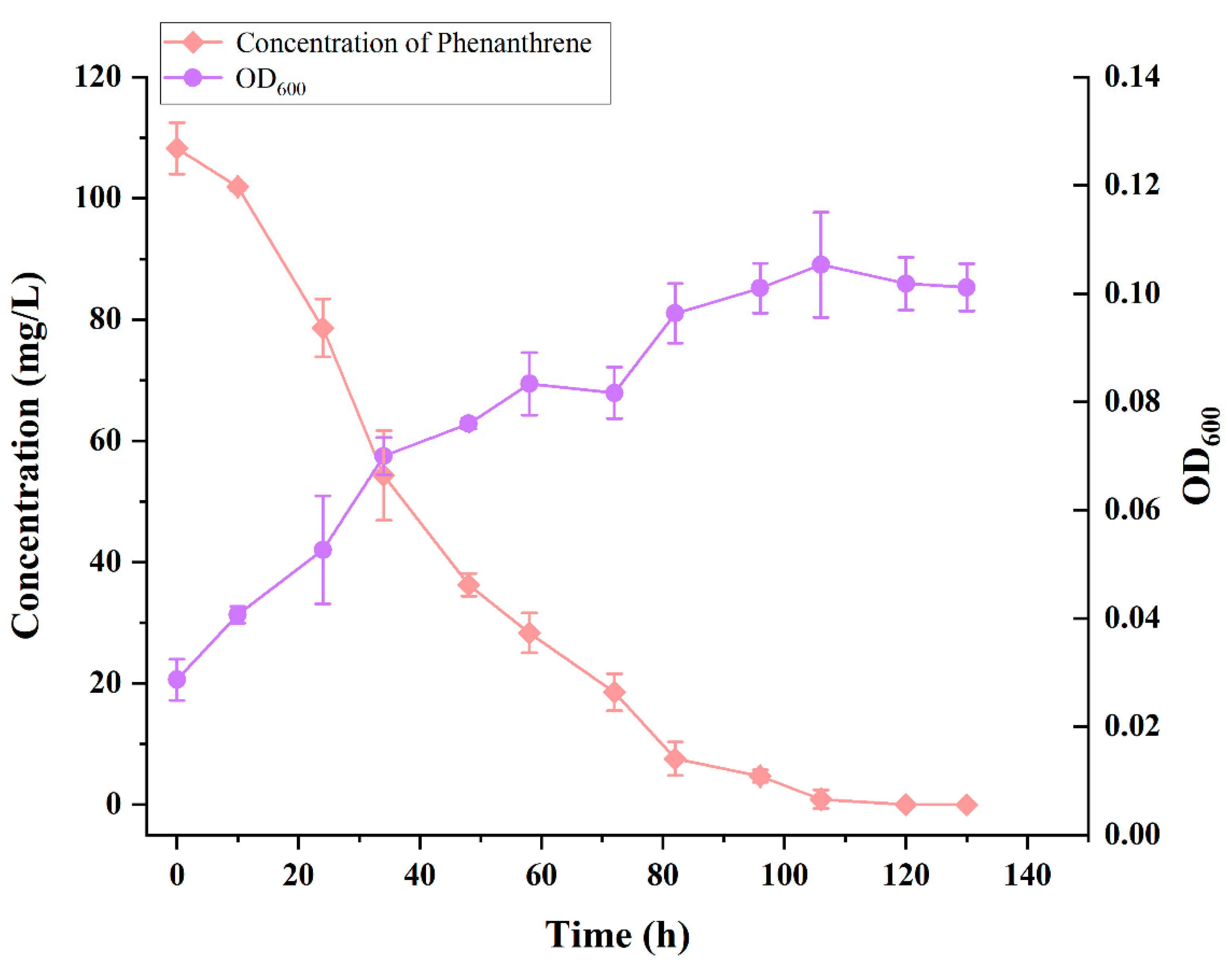
3.3. Growth and Degradation Curves of Strain TJFP1 for the Degradation of 100 mg/L Phenanthrene at Optimal Temperature and pH
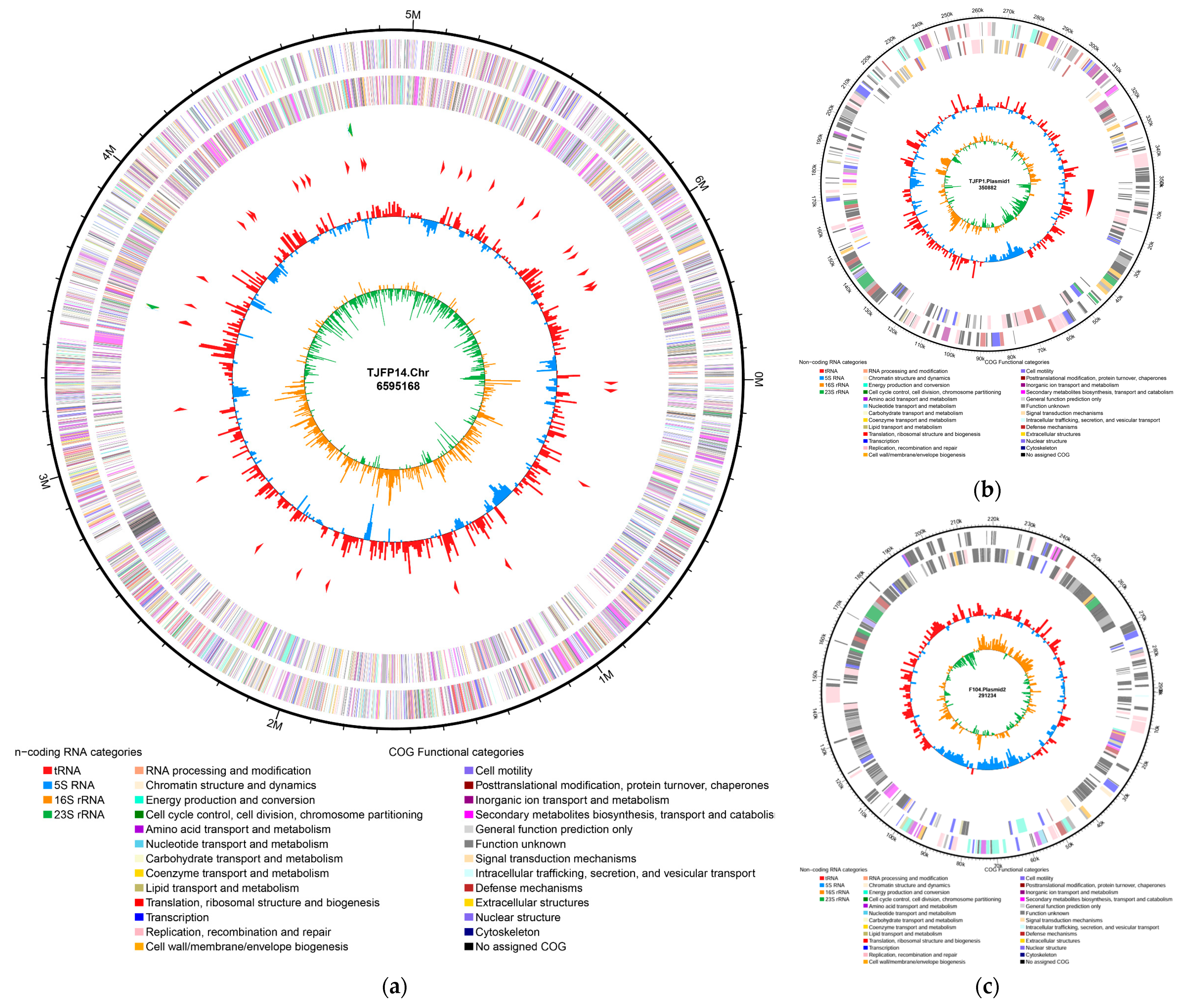
3.4. Whole Genome Sequencing Analysis
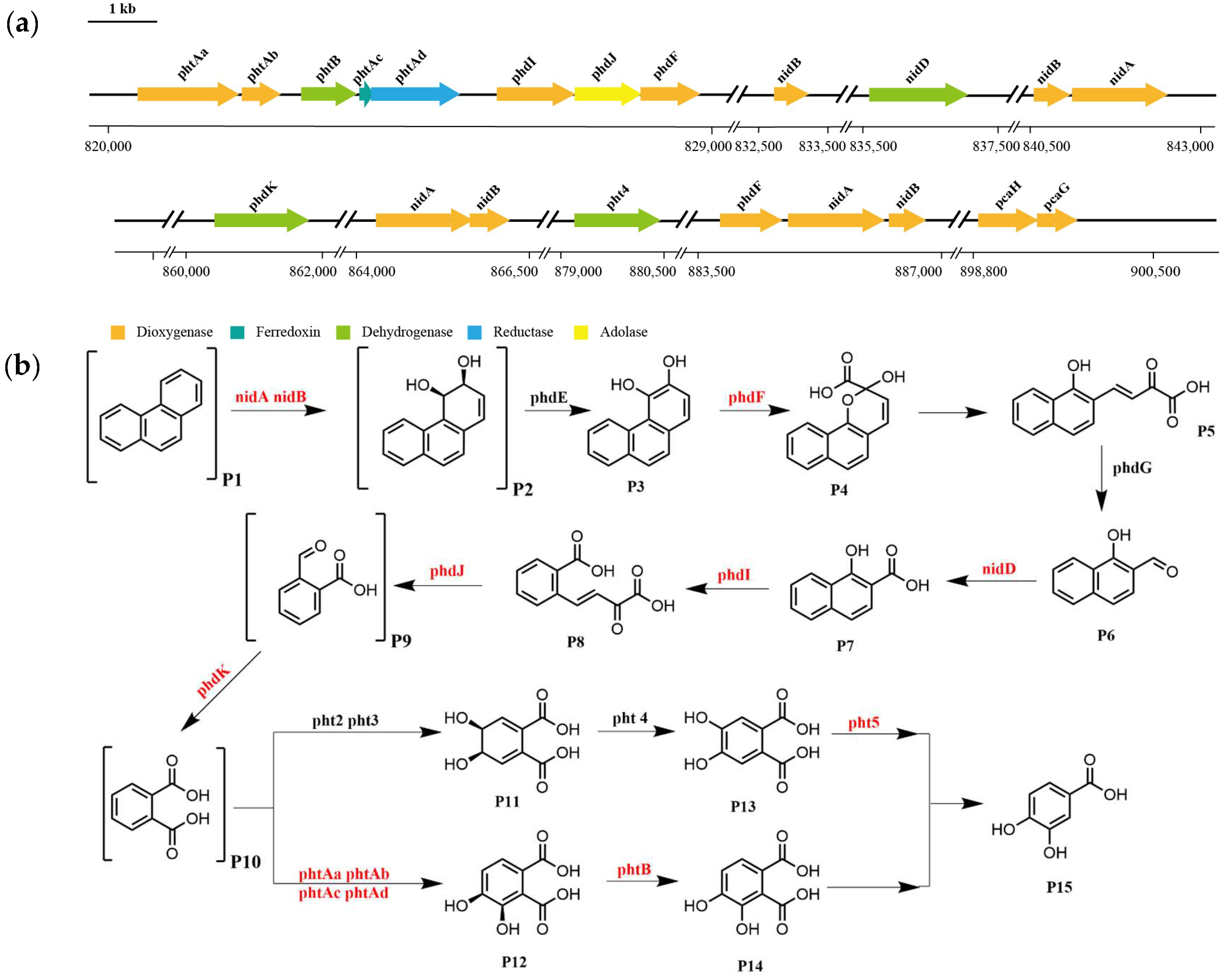
3.5. Pathway of Phenanthrene Degradation by Strain TJFP1
3.6. Assessment of Remediation Efficacy and Microbial Community Response to Phenanthrene-Contaminated Soil Driven by Strain TJFP1
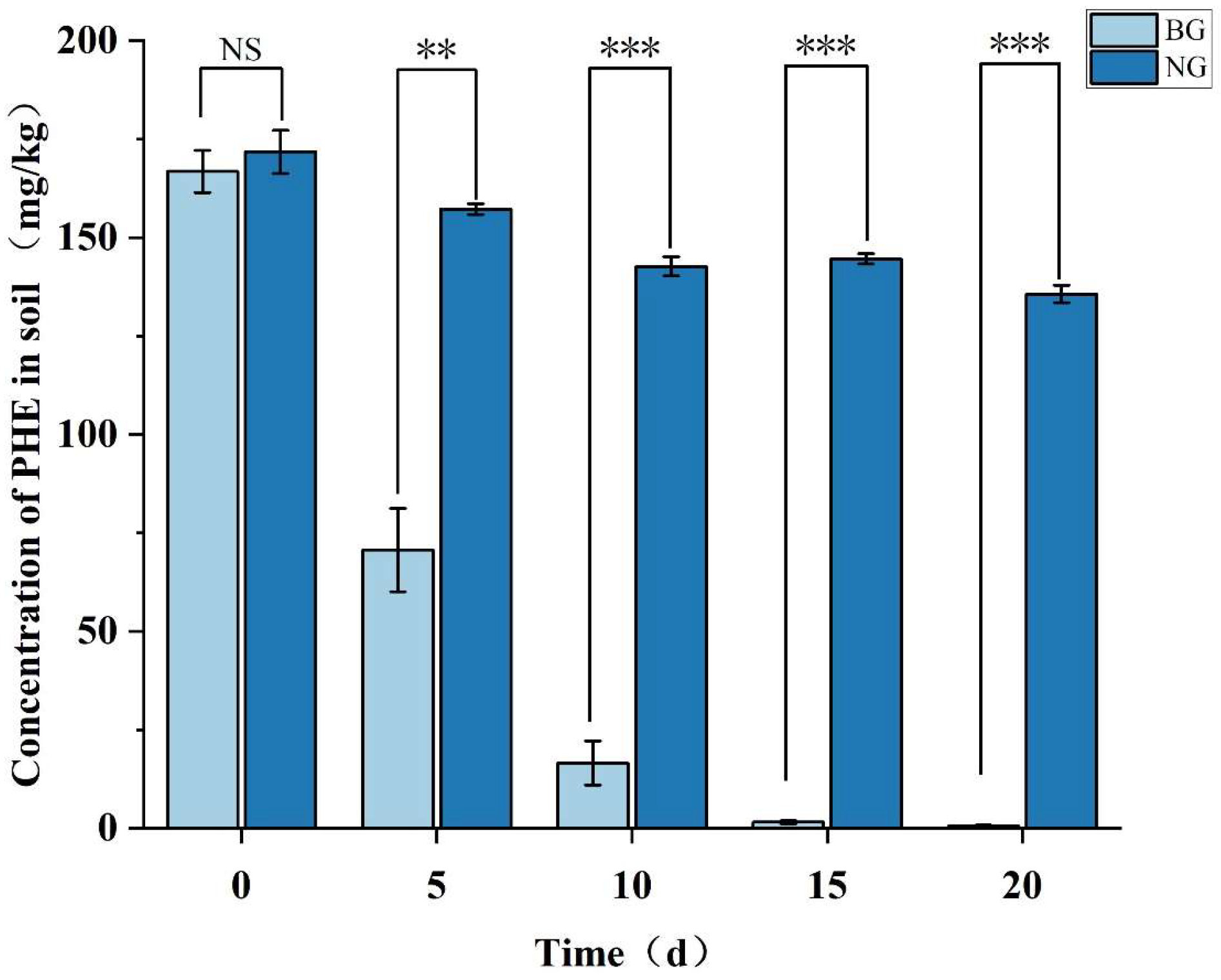
3.6.1. Strain TJFP1 Is Able to Produce Remediation Effects on Phenanthrene-Contaminated Soil
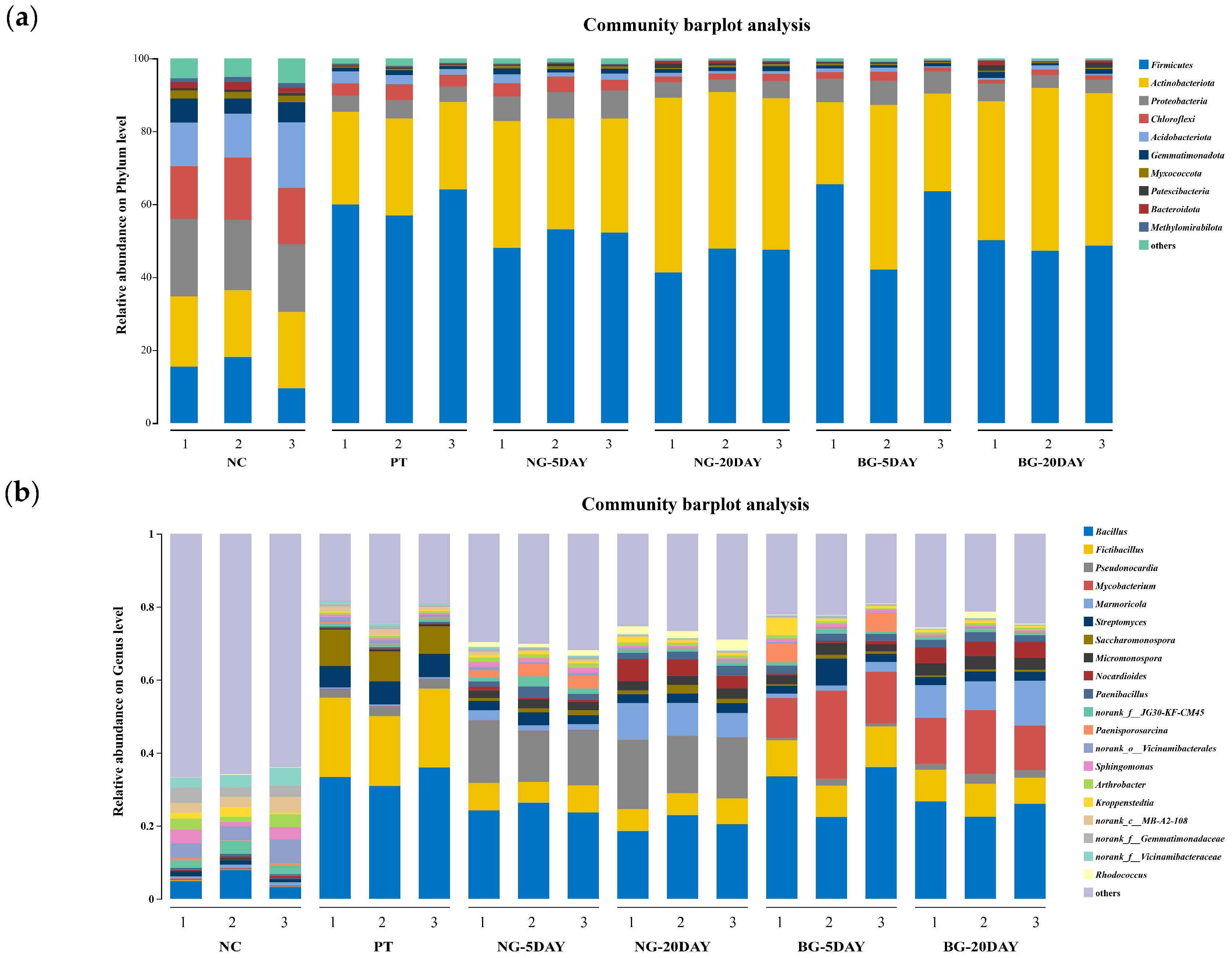
3.6.2. Changes in Microbial Community Structure and Diversity
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Gene ID | Gene | Position | Length | Predicted Function | Identity |
|---|---|---|---|---|---|
| TJFP1000810 | phtAa | 820437–821900 | 1463 | phthalate 3,4-dioxygenase, alpha subunit | 99% |
| TJFP1000811 | phtAb | 821945–822496 | 551 | phthalate dioxygenase small subunit | 98.9% |
| TJFP1000813 | phtB | 822795–823598 | 803 | phthalate 3,4-cis-dihydrodiol dehydrogenase | 100% |
| TJFP1000814 | phtAc | 823627–823827 | 200 | phthalate 3,4-dioxygenase ferredoxin component | 98.5% |
| TJFP1000815 | phtAd | 823824–825065 | 1241 | phthalate 3,4-dioxygenase ferredoxin reductase subunit | 97.8% |
| TJFP1000816 | phdI | 825643–826728 | 1085 | 1-hydroxy-2-naphthoate dioxygenase | 98.6% |
| TJFP1000817 | phdJ | 826747–827751 | 1004 | 4-(2-carboxyphenyl)-2-oxobut-3-enoate aldolase | 99.7% |
| TJFP1000818 | phdF | 827770–828660 | 890 | putative 2,3-dihydroxybiphenyl 1,2-dioxygenase | 99.7% |
| TJFP1000825 | nidB | 832716–833225 | 509 | aromatic-ring-hydroxylating dioxygenase subunit beta | 100% |
| TJFP1000828 | nidD | 835603–837063 | 1460 | aldehyde dehydrogenase | 100% |
| TJFP1000833 | nidB | 840539–841084 | 545 | aromatic-ring-hydroxylating dioxygenase subunit beta | 100% |
| TJFP1000834 | nidA | 841145–842512 | 1367 | aromatic-ring-hydroxylating dioxygenase subunit alpha | 100% |
| TJFP1000853 | phdK | 860418–861791 | 1373 | 2-formylbenzoate dehydrogenase | 100% |
| TJFP1000856 | nidA | 864270–865661 | 1391 | aromatic-ring-hydroxylating dioxygenase subunit alpha | 100% |
| TJFP1000857 | nidB | 865658–866200 | 542 | aromatic-ring-hydroxylating dioxygenase subunit beta | 100% |
| TJFP1000872 | pht4 | 879197–880426 | 1229 | phthalate 4,5-cis-dihydrodiol dehydrogenase | 99.3% |
| TJFP1000877 | phdF | 883813–884703 | 890 | putative 2,3-dihydroxybiphenyl 1,2-dioxygenase | 99.3% |
| TJFP1000878 | nidA | 884776–886176 | 1400 | aromatic-ring-hydroxylating dioxygenase subunit alpha | 99.6% |
| TJFP1000879 | nidB | 886241–886759 | 518 | aromatic-ring-hydroxylating dioxygenase subunit beta | 100% |
| TJFP1000892 | pcaH | 898863–899726 | 863 | protocatechuate 3,4-dioxygenase subunit beta | 99% |
| TJFP1000893 | pcaG | 899726–900295 | 569 | protocatechuate 3,4-dioxygenase subunit alpha | 97.9% |
| Sample Name | NC | PT | NG-5DAY | NG-20DAY | BG-5DAY | BG-20DAY |
|---|---|---|---|---|---|---|
| Firmicutes | 14.42 ± 2.21% abc | 60.32 ± 2.96% a | 51.16 ± 2.21% abc | 45.58 ± 3% c | 57.06 ± 10.58% ab | 48.72 ± 1.2% bc |
| Actinobacteria | 19.63 ± 1.89% bc | 25.36 ± 1.11% cd | 32.16 ± 1.89% bc | 44.11 ± 2.72% a | 31.49 ± 9.79% c | 41.45 ± 2.74% ab |
| Proteobacteria | 19.71 ± 0.38% b | 4.54 ± 0.34% c | 7.21 ± 0.38% b | 4.18 ± 0.49% c | 6.40 ± 0.22% b | 4.06 ± 0.6% c |
| Chloroflexi | 15.71 ± 0.47% b | 3.62 ± 0.49% b | 3.56 ± 0.47% b | 1.67 ± 0.16% c | 1.75 ± 0.57% c | 1.25 ± 0.21% c |
| Acidobacteriota | 13.95 ± 0.54% b | 2.48 ± 0.72% b | 1.77 ± 0.54% b | 0.82 ± 0.13% b | 0.81 ± 0.37% b | 0.68 ± 0.32% b |
| Gemmatimonadota | 5.42 ± 0.29% b | 1.07 ± 0.23% b | 1.29 ± 0.29% b | 1.13 ± 0.17% b | 0.81 ± 0.06% b | 1.17 ± 0.46% b |
| Myxococcata | 1.90 ± 0.06% b | 0.30 ± 0.01% c | 0.70 ± 0.06% b | 0.44 ± 0.09% c | 0.43 ± 0.07% c | 0.38 ± 0.04% c |
| Patescibacteria | 0.69 ± 0.21% a | 0.20 ± 0.07% a | 0.44 ± 0.21% a | 0.82 ± 0.29% a | 0.25 ± 0.06% a | 1.03 ± 0.63% a |
| Bacteroidota | 1.74 ± 0.02% b | 0.28 ± 0.05% b | 0.16 ± 0.02% b | 0.51 ± 0.25% b | 0.09 ± 0.01% b | 0.57 ± 0.44% b |
| Methylxorhablous | 1.25 ± 0.02% b | 0.27 ± 0.04% b | 0.24 ± 0.02% b | 0.15 ± 0.04% c | 0.14 ± 0.02% c | 0.12 ± 0.02% c |
| Others | 5.76 ± 0.25% bc | 1.55 ± 0.37% b | 1.34 ± 0.25% bc | 0.63 ± 0.04% cd | 0.78 ± 0.14% cd | 0.58 ± 0.09% d |
| Genus | NC | PT | NG-5DAY | NG-20DAY | BG-5DAY | BG-20DAY |
|---|---|---|---|---|---|---|
| Bacillus | 5.34 ± 1.92% d | 33.42 ± 2.07% a | 24.73 ± 1.13% bc | 20.66 ± 1.79% c | 30.71 ± 5.95% ab | 24.99 ± 1.95% bc |
| Fictibacillus | 0.11 ± 0.04% e | 20.83 ± 1.15% a | 6.9 ± 0.82% cd | 6.37 ± 0.49% d | 9.85 ± 1.01% b | 8.3 ± 0.82% bc |
| Pseudonocardia | 0.12 ± 0.04% d | 2.45 ± 0.14% c | 15.3 ± 1.3% b | 17.1 ± 1.37% a | 1.11 ± 0.48% cd | 2.13 ± 0.46% c |
| Mycobacterium | 0.33 ± 0.11% b | 0.15 ± 0.01% b | 0.11 ± 0.04% b | 0.06 ± 0.02% b | 16.42 ± 5.69% a | 14.07 ± 2.4% a |
| Marmoricola | 0.83 ± 0.17% b | 0.44 ± 0.03% b | 1.97 ± 0.6% b | 8.53 ± 1.43% a | 1.77 ± 0.64% b | 9.74 ± 1.89% a |
| Streptomyces | 1.1 ± 0.15% c | 6.16 ± 0.23% a | 2.86 ± 0.5% bc | 2.55 ± 0.14% bc | 3.89 ± 2.44% ab | 2.46 ± 0.2% bc |
| Saccharomonospora | 0.04 ± 0.01% d | 8.62 ± 1.02% a | 1.08 ± 0.25% bc | 1.56 ± 0.61% b | 0.73 ± 0.25% bcd | 0.52 ± 0.1% cd |
| Micromonospora | 0.33 ± 0.08% a | 0.46 ± 0.1% a | 2.21 ± 0.14% b | 2.59 ± 0.13% b | 2.49 ± 0.53% b | 3.38 ± 0.24% a |
| Nocardioides | 0.41 ± 0.1% b | 0.19 ± 0.02% b | 0.75 ± 0.25% b | 4.71 ± 1.11% a | 0.65 ± 0.26% b | 4.24 ± 0.24% a |
| Paenibacillus | 0.57 ± 0.1% b | 0.4 ± 0.04% b | 2.05 ± 0.77% a | 2.17 ± 0.42% a | 2.11 ± 0.19% a | 2.17 ± 0.33% a |
| norank_f__JG30-KF-CM45 | 2.66 ± 0.61% a | 0.88 ± 0.05% c | 1.77 ± 0.7% b | 0.81 ± 0.13% c | 0.88 ± 0.21% c | 0.68 ± 0.05% c |
| Paenisporosarcina | 0.46 ± 0.2% b | 0.35 ± 0.06% b | 2.96 ± 0.71% a | 0.18 ± 0.03% b | 3.48 ± 2.31% a | 0.18 ± 0.03% b |
| norank_o__Vicinamibacterales | 4.76 ± 1.23% a | 0.93 ± 0.34% b | 0.66 ± 0.17% b | 0.33 ± 0.06% b | 0.32 ± 0.16% b | 0.27 ± 0.18% b |
| Sphingomonas | 2.84 ± 1.09% a | 0.60 ± 0.04% b | 1.36 ± 0.15% b | 0.57 ± 0.06% b | 0.92 ± 0.04% b | 0.47 ± 0.06% b |
| Arthrobacter | 2.67 ± 0.95% a | 0.55 ± 0.02% b | 1.16 ± 0.1% b | 0.79 ± 0.06% b | 0.66 ± 0.22% b | 0.65 ± 0.02% b |
| Kroppenstedtia | 1.49 ± 0.92% a | 0.28 ± 0.03% a | 0.7 ± 0.09% a | 0.89 ± 0.42% a | 1.86 ± 1.92% a | 0.53 ± 0.07% a |
| norank_c__MB-A2-108 | 3.34 ± 0.7% a | 1.23 ± 0.35% b | 0.49 ± 0.15% c | 0.23 ± 0.02% c | 0.22 ± 0.11% c | 0.15 ± 0.07% c |
| norank_f__Gemmatimonadaceae | 3.22 ± 0.56% a | 0.66 ± 0.11% b | 0.66 ± 0.09% b | 0.31 ± 0.06% b | 0.43 ± 0.03% b | 0.24 ± 0.02% b |
| norank_f___Vicinamibacteraceae | 3.81 ± 0.86% a | 0.64 ± 0.19% b | 0.43 ± 0.13% b | 0.19 ± 0.03% b | 0.18 ± 0.09% b | 0.16 ± 0.08% b |
| Rhodococcus | 0.47 ± 0.52% bc | 0.03 ± 0.00% c | 1.26 ± 0.22% b | 2.41 ± 0.46% a | 0.14 ± 0.03% c | 0.75 ± 0.69% bc |
References
- Edwards, N.T. Polycyclic aromatic hydrocarbons (PAHs) in the terrestrial environment-A review. J. Environ. Qual. 1983, 12, 427–441. [Google Scholar] [CrossRef]
- Kim, K.-H.; Jahan, S.A.; Kabir, E.; Brown, R.J.C. A review of airborne polycyclic aromatic hydrocarbons (PAHs) and their human health effects. Environ. Int. 2013, 60, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Mojiri, A.; Zhou, J.L.; Ohashi, A.; Ozaki, N.; Kindaichi, T. Comprehensive review of polycyclic aromatic hydrocarbons in water sources, their effects and treatments. Sci. Total Environ. 2019, 696, 133971. [Google Scholar] [CrossRef] [PubMed]
- Zafra, G.; Absalon, A.E.; Angel Anducho-Reyes, M.; Fernandez, F.J.; Cortes-Espinosa, D.V. Construction of PAH-degrading mixed microbial consortia by induced selection in soil. Chemosphere 2017, 172, 120–126. [Google Scholar] [CrossRef]
- Gamboa, R.T.; Gamboa, A.R.; Bravo, A.H.; Ostrosky, W.P. Genotoxicity in child populations exposed to polycyclic aromatic hydrocarbons (PAHs) in the air from Tabasco, Mexico. Int. J. Environ. Res. Public Health 2008, 5, 349–355. [Google Scholar] [CrossRef]
- Patel, A.B.; Shaikh, S.; Jain, K.R.; Desai, C.; Madamwar, D. Polycyclic Aromatic Hydrocarbons: Sources, Toxicity, and Remediation Approaches. Front. Microbiol. 2020, 11, 562813. [Google Scholar] [CrossRef]
- Brozman, O.; Novak, J.; Bauer, A.K.; Babica, P. Airborne PAHs inhibit gap junctional intercellular communication and activate MAPKs in human bronchial epithelial cell line. Environ. Toxicol. Pharmacol. 2020, 79, 103422. [Google Scholar] [CrossRef]
- Yin, S.; Tang, M.; Chen, F.; Li, T.; Liu, W. Environmental exposure to polycyclic aromatic hydrocarbons (PAHs): The correlation with and impact on reproductive hormones in umbilical cord serum. Environ. Pollut. 2017, 220, 1429–1437. [Google Scholar] [CrossRef]
- Torres-Moreno, C.; Puente-DelaCruz, L.; Codling, G.; Villa, A.L.; Cobo, M.; Klanova, J.; Johnson-Restrepo, B. Polycyclic aromatic hydrocarbons (PAHs) in human breast milk from Colombia: Spatial occurrence, sources and probabilistic risk assessment. Environ. Res. 2022, 204, 111981. [Google Scholar] [CrossRef]
- Keith, L.H.; Telliard, W.A. Priority pollutantants-A perspective view. Environ. Sci. Technol. 1979, 13, 416–423. [Google Scholar] [CrossRef]
- Zhang, Y.; Tao, S. Global atmospheric emission inventory of polycyclic aromatic hydrocarbons (PAHs) for 2004. Atmos. Environ. 2009, 43, 812–819. [Google Scholar] [CrossRef]
- Kuppusamy, S.; Thavamani, P.; Venkateswarlu, K.; Lee, Y.B.; Naidu, R.; Megharaj, M. Remediation approaches for polycyclic aromatic hydrocarbons (PAHs) contaminated soils: Technological constraints, emerging trends and future directions. Chemosphere 2017, 168, 944–968. [Google Scholar] [CrossRef]
- Zhou, Z.; Liu, X.; Sun, K.; Lin, C.; Ma, J.; He, M.; Ouyang, W. Persulfate-based advanced oxidation processes (AOPs) for organic-contaminated soil remediation: A review. Chem. Eng. J. 2019, 372, 836–851. [Google Scholar] [CrossRef]
- Trellu, C.; Mousset, E.; Pechaud, Y.; Huguenot, D.; van Hullebusch, E.D.; Esposito, G.; Oturan, M.A. Removal of hydrophobic organic pollutants from soil washing/flushing solutions: A critical review. J. Hazard. Mater. 2016, 306, 149–174. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, Z.; Zuo, Y.; Zhang, J.; Zhang, P. Comparison of exogenous degrader-enhanced bioremediation with low-dose persulfate oxidation for polycyclic aromatic hydrocarbon removal in alkaline soil: Efficiency and influence on ecological health. Front. Environ. Sci. Eng. 2023, 17, 1–12. [Google Scholar] [CrossRef]
- Ali, M.; Song, X.; Ding, D.; Wang, Q.; Zhang, Z.; Tang, Z. Bioremediation of PAHs and heavy metals co-contaminated soils: Challenges and enhancement strategies. Environ. Pollut. 2022, 295, 118686. [Google Scholar] [CrossRef]
- Li, X.; Cao, X.; Zhang, Z.; Li, Y.; Zhang, Y.; Wang, C.; Fan, W. Mechanism of phenanthrene degradation by the halophilic Pelagerythrobacter sp. N7. Chemosphere 2024, 350, 141175. [Google Scholar] [CrossRef]
- Davletgildeeva, A.T.; Kuznetsov, N.A. Bioremediation of Polycyclic Aromatic Hydrocarbons by Means of Bacteria and Bacterial Enzymes. Microorganisms 2024, 12, 1814. [Google Scholar] [CrossRef]
- Lu, H.; Wei, J.-L.; Tang, G.-X.; Chen, Y.-S.; Huang, Y.-H.; Hu, R.; Mo, C.-H.; Zhao, H.-M.; Xiang, L.; Li, Y.-W.; et al. Microbial consortium degrading of organic pollutants: Source, degradation efficiency, pathway, mechanism and application. J. Clean. Prod. 2024, 451, 141913. [Google Scholar] [CrossRef]
- Seo, J.-S.; Keum, Y.-S.; Li, Q.X. Bacterial Degradation of Aromatic Compounds. Int. J. Environ. Res. Public Health 2009, 6, 278–309. [Google Scholar] [CrossRef]
- Ismail, N.A.; Kasmuri, N.; Hamzah, N. Microbial Bioremediation Techniques for Polycyclic Aromatic Hydrocarbon (PAHs)—A Review. Water Air Soil Pollut. 2022, 233, 1–27. [Google Scholar] [CrossRef]
- Manian, S.K.R. Bioremediation of polycyclic aromatic hydrocarbons contaminated soils: Recent progress, perspectives and challenges. Environ. Monit. Assess. 2023, 195, 1–17. [Google Scholar] [CrossRef]
- Salari, M.; Rahmanian, V.; Hashemi, S.A.; Chiang, W.-H.; Lai, C.W.; Mousavi, S.M.; Gholami, A. Bioremediation Treatment of Polyaromatic Hydrocarbons for Environmental Sustainability. Water 2022, 14, 3980. [Google Scholar] [CrossRef]
- Pandolfo, E.; Barra Caracciolo, A.; Rolando, L. Recent Advances in Bacterial Degradation of Hydrocarbons. Water 2023, 15, 375. [Google Scholar] [CrossRef]
- Huang, Y.; Li, L.; Yin, X.; Zhang, T. Polycyclic aromatic hydrocarbon (PAH) biodegradation capacity revealed by a genome-function relationship approach. Environ. Environ. Microbiome 2023, 18, 1–13. [Google Scholar] [CrossRef]
- Pagnout, C.; Frache, G.; Poupin, P.; Maunit, B.; Muller, J.-F.; Ferard, J.-F. Isolation and characterization of a gene cluster involved in PAH degradation in Mycobacterium sp. strain SNP11:Expression in Mycobacterium smegmatis mc2 155. Res. Microbiol. 2007, 158, 175–186. [Google Scholar] [CrossRef]
- Krivobok, S.; Kuony, S.; Meyer, C.; Louwagie, M.; Willison, J.C.; Jouanneau, Y. Identification of pyrene-induced proteins in Mycobacterium sp. strain 6PY1: Evidence for two ring-hydroxylating dioxygenases. J. Bacteriol. 2003, 185, 3828–3841. [Google Scholar] [CrossRef]
- Sawulski, P.; Clipson, N.; Doyle, E. Effects of polycyclic aromatic hydrocarbons on microbial community structure and PAH ring hydroxylating dioxygenase gene abundance in soil. Biodegradation 2014, 25, 835–847. [Google Scholar] [CrossRef]
- Uyttebroek, M.; Breugelmans, P.; Janssen, M.; Wattiau, P.; Joffe, B.; Karlson, U.; Ortega-Calvo, J.J.; Bastiaens, L.; Ryngaert, A.; Hausner, M.; et al. Distribution of the Mycobacterium community and polycyclic aromatic hydrocarbons (PAHs) among different size fractions of a long-term PAH-contaminated soil. Environ. Microbiol. 2006, 8, 836–847. [Google Scholar] [CrossRef]
- Jia, X.; He, Y.; Jiang, D.; Liu, C.; Lu, W. Construction and analysis of an engineered Escherichia coli-Pseudomonas aeruginosa co-culture consortium for phenanthrene bioremoval. Biochem. Eng. J. 2019, 148, 214–223. [Google Scholar] [CrossRef]
- Memory, J.D. Electrophilic super-delocalizability and carcinogenesis by polycyclic aromatic hydrocarbons: Pullman theory. Int. J. Quantum Chem. 1979, 15, 363–368. [Google Scholar] [CrossRef]
- Pahlman, R.; Pelkonen, O. Mutagenicity studies of different polycyclic aromatic hydrocarbons: The significance of enzymatic factors and molecular structure. Carcinogenesis 1987, 8, 773–778. [Google Scholar] [CrossRef]
- Weis, L.M. Determining a Correlation Between Structural Elements of Polycyclic Aromatic Hydrocarbons and Inhibition of Gap Junctional Intercellular Communication. Master’s Thesis, Michigan State University, East Lansing, MI, USA, 1996. [Google Scholar]
- Cavalieri, E.L.; Rogan, E.G. The Approach to Understanding Aromatic Hydrocarbon Carcinogenesis—The Central Role of Radical Cations in Metabolic-Activation. Pharmacol. Ther. 1992, 55, 183–199. [Google Scholar] [CrossRef]
- Yin, C.; Xiong, W.; Qiu, H.; Peng, W.; Deng, Z.; Lin, S.; Liang, R. Characterization of the Phenanthrene-Degrading Sphingobium yanoikuyae SJTF8 in Heavy Metal Co-Existing Liquid Medium and Analysis of Its Metabolic Pathway. Microorganisms 2020, 8, 946. [Google Scholar] [CrossRef]
- Ji, D.; Mao, Z.; He, J.; Peng, S.; Wen, H. Characterization and genomic function analysis of phenanthrene-degrading bacterium Pseudomonas sp. Lphe-2. J. Environ. Sci. Health Part A-Toxic/Hazard. Subst. Environ. Eng. 2020, 55, 549–562. [Google Scholar] [CrossRef]
- Lara-Moreno, A.; Morillo, E.; Merchan, F.; Gonzalez-Pimentel, J.L.; Villaverde, J. Genome sequence of Stenotrophomonas indicatrix CPHE1, a powerful phenanthrene-degrading bacterium. 3 Biotech 2023, 13, 1–15. [Google Scholar] [CrossRef]
- Qu, Y.; Ma, Q.; Liu, Z.; Wang, W.; Tang, H.; Zhou, J.; Xu, P. Unveiling the biotransformation mechanism of indole in a Cupriavidus sp. strain. Mol. Microbiol. 2017, 106, 905–918. [Google Scholar] [CrossRef]
- Wen, L.; Huang, Y.; Wang, W.; Zhang, L.; Xu, J.; Li, Z.; Xu, P.; Tang, H. A novel Diaphorobacter sp. strain isolated from saponification wastewater shows highly efficient phenanthrene degradation. Environ. Res. 2022, 214, 114047. [Google Scholar] [CrossRef]
- Zhang, Y.; Manzoor, A.; Dong, J.; Yang, Q.; Zhou, W.; Ling, J. Isolation, identification and degradation characteristics of a phenanthrene degrading bacteria derived from seagrass sediment. Microbiol. 2021, 48, 1841–1853. [Google Scholar]
- Li, J.; Peng, W.; Yin, X.; Wang, X.; Liu, Z.; Liu, Q.; Deng, Z.; Lin, S.; Liang, R. Identification of an efficient phenanthrene-degrading Pseudarthrobacter sp. L1SW and characterization of its metabolites and catabolic pathway. J. Hazard. Mater. 2024, 465, 133138. [Google Scholar] [CrossRef]
- Imam, A.; Suman, S.K.; Kanaujia, P.K.; Ray, A. Biological machinery for polycyclic aromatic hydrocarbons degradation: A review. Bioresour. Technol. 2022, 343, 126121. [Google Scholar] [CrossRef] [PubMed]
- Premnath, N.; Mohanrasu, K.; Rao, R.G.R.; Dinesh, G.H.; Prakash, G.S.; Ananthi, V.; Ponnuchamy, K.; Muthusamy, G.; Arun, A. A crucial review on polycyclic aromatic Hydrocarbons—Environmental occurrence and strategies for microbial degradation. Chemosphere 2021, 280, 130608. [Google Scholar] [CrossRef]
- Elyamine, A.M.; Kan, J.; Meng, S.; Tao, P.; Wang, H.; Hu, Z. Aerobic and Anaerobic Bacterial and Fungal Degradation of Pyrene: Mechanism Pathway Including Biochemical Reaction and Catabolic Genes. Int. J. Mol. Sci. 2021, 22, 8202. [Google Scholar] [CrossRef]
- Li, J.; Hong, M.; Tang, R.; Cui, T.; Yang, Y.; Lv, J.; Liu, N.; Lei, Y. Isolation of Diaphorobacter sp. LW2 capable of degrading Phenanthrene and its migration mediated by Pythium ultimum. Environ. Technol. 2024, 45, 1497–1507. [Google Scholar] [CrossRef]
- Zhao, B.; Jing, J.; Wang, T.; Yang, Y.; Qu, Y. Screening of low-molecular-weight polycyclic aromatic hydrocarbons degrading strains, construction of bacterial consortium, and degradation mechanism. Chin. J. Appl. Environ. Biol. 2024, 30, 317–323. [Google Scholar]
- Zhou, G.; Qiao, H.; Liu, Y.; Yu, X.; Niu, X. High phenanthrene degrading efficiency by different microbial compositions construction. Front. Microbiol. 2024, 15, 1439216. [Google Scholar] [CrossRef]
- Hou, N.; Zhang, N.; Jia, T.; Sun, Y.; Dai, Y.; Wang, Q.; Li, D.; Luo, Z.; Li, C. Biodegradation of phenanthrene by biodemulsifier-producing strain Achromobacter sp. LH-1 and the study on its metabolisms and fermentation kinetics. Ecotoxicol. Environ. Saf. 2018, 163, 205–214. [Google Scholar] [CrossRef]
- Woo, S.H.; Park, J.M. Microbial degradation and enhanced bioremediation of polycyclic aromatic hydrocarbons. J. Ind. Eng. Chem. 2004, 10, 16–23. [Google Scholar]
- Zhang, X.-X.; Cheng, S.-P.; Zhu, C.-J.; Sun, S.-L. Microbial PAH-degradation in soil: Degradation pathways and contributing factors. Pedosphere 2006, 16, 555–565. [Google Scholar] [CrossRef]
- Naloka, K.; Kuntaveesuk, A.; Muangchinda, C.; Chavanich, S.; Viyakarn, V.; Chen, B.; Pinyakong, O. Pseudomonas and Pseudarthrobacter are the key players in synergistic phenanthrene biodegradation at low temperatures. Sci. Rep. 2024, 14, 1–14. [Google Scholar] [CrossRef]
- Jindal, S.; Aggarwal, K.K. Assessment of Phenanthrene-Degrading Potential of Klebsiella pneumoniae SJK1 Isolated from an Oil-Contaminated Site. Microbiology 2023, 92, 572–586. [Google Scholar] [CrossRef]
- Sharma, G.; Sinha, P.G.; Verma, K.; Walia, D.; Lahiri, M.; Mathur, V. Isolation and characterization of phenanthrene-degrading bacteria from urban soil. Bioremediation J. 2024, 28, 354–367. [Google Scholar] [CrossRef]
- Kaplieva-Dudek, I.; Samak, N.A.; Bormann, J.; Kaschani, F.; Kaiser, M.; Meckenstock, R.U. Characterization of 2-phenanthroate: CoA ligase from the sulfate-reducing, phenanthrene-degrading enrichment culture TRIP. Appl. Environ. Microbiol. 2024, 90, e0129624. [Google Scholar] [CrossRef]
- Mawad, A.M.M.; Abdel-Mageed, W.S.; Hesham, A.E.L. Quantification of Naphthalene Dioxygenase (NahAC) and Catechol Dioxygenase (C23O) Catabolic Genes Produced by Phenanthrene-Degrading Pseudomonas fluorescens AH-40. Curr. Genom. 2020, 21, 111–118. [Google Scholar] [CrossRef]
- Zhao, H.; Gu, Y.; Liu, X.; Liu, J.; Waigi, M.G. Reducing Phenanthrene Contamination in Trifolium repens L. With Root-Associated Phenanthrene-Degrading Bacterium Diaphorobacter sp. Phe15. Front. Microbiol. 2021, 12, 792698. [Google Scholar] [CrossRef]
- Festa, S.; Coppotelli, B.M.; Morelli, I.S. Bacterial diversity and functional interactions between bacterial strains from a phenanthrene-degrading consortium obtained from a chronically contaminated-soil. Int. Biodeterior. Biodegrad. 2013, 85, 42–51. [Google Scholar] [CrossRef]
- Sun, K.; Liu, J.; Gao, Y.; Jin, L.; Gu, Y.; Wang, W. Isolation, plant colonization potential, and phenanthrene degradation performance of the endophytic bacterium Pseudomonas sp. Ph6-gfp. Sci. Rep. 2014, 4, 5462. [Google Scholar] [CrossRef]
- Liu, J.; Liu, S.; Sun, K.; Sheng, Y.; Gu, Y.; Gao, Y. Colonization on Root Surface by a Phenanthrene-Degrading Endophytic Bacterium and Its Application for Reducing Plant Phenanthrene Contamination. PLoS ONE 2014, 9, e108249. [Google Scholar] [CrossRef]
- Yuan, K.; Xie, X.; Wang, X.; Lin, L.; Yang, L.; Luan, T.; Chen, B. Transcriptional response of Mycobacterium sp. strain A1-PYR to multiple polycyclic aromatic hydrocarbon contaminations. Environ. Pollut. 2018, 243, 824–832. [Google Scholar] [CrossRef]
- Masakorala, K.; Yao, J.; Cai, M.; Chandankere, R.; Yuan, H.; Chen, H. Isolation and characterization of a novel phenanthrene (PHE) degrading strain Psuedomonas sp. USTB-RU from petroleum contaminated soil. J. Hazard. Mater. 2013, 263, 493–500. [Google Scholar] [CrossRef]
- Prakash, O.; Lal, R. Role of Unstable Phenanthrene-Degrading Pseudomonas species in Natural Attenuation of Phenanthrene- Contaminated Site. Microbiol. Biotechnol. Lett. 2013, 41, 79–87. [Google Scholar] [CrossRef]
- Moghadam, M.S.; Ebrahimipour, G.; Abtahi, B.; Ghassempour, A. Isolation, Identification and Optimization of Phenanthrene Degrading Bacteria from the Coastal Sediments of Nayband Bay. Jundishapur J. Microbiol. 2013, 6, e13816. [Google Scholar] [CrossRef]
- Li, H.; La, S.; Zhang, X.; Gao, L.; Tian, Y. Salt-induced recruitment of specific root-associated bacterial consortium capable of enhancing plant adaptability to salt stress. ISME J. 2021, 15, 2865–2882. [Google Scholar] [CrossRef]
- Li, X.; Qu, C.; Bian, Y.; Gu, C.; Jiang, X.; Song, Y. New insights into the responses of soil microorganisms to polycyclic aromatic hydrocarbon stress by combining enzyme activity and sequencing analysis with metabolomics. Environ. Pollut. 2019, 255, 113312. [Google Scholar] [CrossRef]
- Deveryshetty, J.; Phale, P.S. Biodegradation of phenanthrene by Alcaligenes sp. strain PPH: Partial purification and characterization of 1-hydroxy-2-naphthoic acid hydroxylase. FEMS Microbiol. Lett. 2010, 311, 93–101. [Google Scholar] [CrossRef]
- Li, Y.; You, X.; Tang, Z.; Zhu, T.; Liu, B.; Chen, M.-X.; Xu, Y.; Liu, T.-Y. Isolation and identification of plant growth-promoting rhizobacteria from tall fescue rhizosphere and their functions under salt stress. Physiol. Plant. 2022, 174, e13817. [Google Scholar] [CrossRef]
- Liang, C.; Huang, Y.; Wang, H. pahE, a Functional Marker Gene for Polycyclic Aromatic Hydrocarbon-Degrading Bacteria. Appl. Environ. Microbiol. 2019, 85, e02399-18. [Google Scholar] [CrossRef]
- Sun, S.; Wang, H.; Chen, Y.; Lou, J.; Wu, L.; Xu, J. Salicylate and phthalate pathways contributed differently on phenanthrene and pyrene degradations in Mycobacterium sp. WY10. J. Hazard. Mater. 2019, 364, 509–518. [Google Scholar] [CrossRef]
- Haritash, A.K.; Kaushik, C.P. Biodegradation aspects of Polycyclic Aromatic Hydrocarbons (PAHs): A review. J. Hazard. Mater. 2009, 169, 1–15. [Google Scholar] [CrossRef]
- Mutanda, I.; Sun, J.; Jiang, J.; Zhu, D. Bacterial membrane transporter systems for aromatic compounds: Regulation, engineering, and biotechnological applications. Biotechnol. Adv. 2022, 59, 107952. [Google Scholar] [CrossRef]
- Ghosal, D.; Ghosh, S.; Dutta, T.K.; Ahn, Y. Current State of Knowledge in Microbial Degradation of Polycyclic Aromatic Hydrocarbons (PAHs): A Review. Front. Microbiol. 2016, 7, 1369. [Google Scholar] [CrossRef] [PubMed]
- Diaz, E.; Jimenez, J.I.; Nogales, J. Aerobic degradation of aromatic compounds. Curr. Opin. Biotechnol. 2013, 24, 431–442. [Google Scholar] [CrossRef]
- Bacosa, H.P.; Inoue, C. Polycyclic aromatic hydrocarbons (PAHs) biodegradation potential and diversity of microbial consortia enriched from tsunami sediments in Miyagi, Japan. J. Hazard. Mater. 2015, 283, 689–697. [Google Scholar] [CrossRef]
- Lee, D.W.; Lee, H.; Lee, A.H.; Kwon, B.-O.; Khim, J.S.; Yim, U.H.; Kim, B.S.; Kim, J.-J. Microbial community composition and PAHs removal potential of indigenous bacteria in oil contaminated sediment of Taean coast, Korea. Environ. Pollut. 2018, 234, 503–512. [Google Scholar] [CrossRef]
- Tang, T.; Li, J.; Yang, Z.; Xiang, F. Research Progress on Biodegradation and Transformation Pathways of Polycyclic Aromatic Hydrocarbons. Acta Petrolei Sinica. Pet. Process. Sect. 2019, 35, 403–413. [Google Scholar]
- Gran-Scheuch, A.; Fuentes, E.; Bravo, D.M.; Cristobal Jimenez, J.; Perez-Donoso, J.M. Isolation and Characterization of Phenanthrene Degrading Bacteria from Diesel Fuel-Contaminated Antarctic Soils. Front. Microbiol. 2017, 8, 1634. [Google Scholar] [CrossRef]
- Feng, T.C.; Cui, C.Z.; Dong, F.; Feng, Y.Y.; Liu, Y.D.; Yang, X.M. Phenanthrene biodegradation by halophilic Martelella sp. AD-3. J. Appl. Microbiol. 2012, 113, 779–789. [Google Scholar] [CrossRef]
- Seo, J.-S.; Keum, Y.-S.; Hu, Y.; Lee, S.-E.; Li, Q.X. Phenanthrene degradation in Arthrobacter sp. P1-1: Initial 1,2-, 3,4- and 9,10-dioxygenation, and meta- and ortho-cleavages of naphthalene-1,2-diol after its formation from naphthalene-1,2-dicarboxylic acid and hydroxyl naphthoic acids. Chemosphere 2006, 65, 2388–2394. [Google Scholar] [CrossRef]
- Zeinali, M.; Vossoughi, M.; Ardestani, S.K. Degradation of phenanthrene and anthracene by Nocardia otitidiscaviarum strain TSH1, a moderately thermophilic bacterium. J. Appl. Microbiol. 2008, 105, 398–406. [Google Scholar] [CrossRef]
- Mallick, S.; Chakraborty, J.; Dutta, T.K. Role of oxygenases in guiding diverse metabolic pathways in the bacterial degradation of low-molecular-weight polycyclic aromatic hydrocarbons: A review. Crit. Rev. Microbiol. 2011, 37, 64–90. [Google Scholar] [CrossRef]
- Adachi, K.; Iwabuchi, T.; Sano, H.; Harayama, S. Structure of the ring cleavage product of 1-hydroxy-2-naphthoate, an intermediate of the phenanthrene-degradative pathway of Nocardioides sp. strain KP7. J. Bacteriol. 1999, 181, 757–763. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.-S.; Keum, Y.-S.; Hu, Y.; Lee, S.-E.; Li, Q.X. Degradation of phenanthrene by Burkholderia sp. C3:: Initial 1,2- and 3,4-dioxygenation and meta- and ortho-cleavage of naphthalene-1,2-diol. Biodegradation 2007, 18, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Habe, H.; Omori, T. Genetics of polycyclic aromatic hydrocarbon metabolism in diverse aerobic bacteria. Biosci. Biotechnol. Biochem. 2003, 67, 225–243. [Google Scholar] [CrossRef]
- Samanta, S.K.; Chakraborti, A.K.; Jain, R.K. Degradation of phenanthrene by different bacteria: Evidence for novel transformation sequences involving the formation of 1-naphthol. Appl. Microbiol. Biotechnol. 1999, 53, 98–107. [Google Scholar] [CrossRef]
- Zhou, N.; Guo, H.; Liu, Q.; Zhang, Z.; Sun, J.; Wang, H. Bioaugmentation of polycyclic aromatic hydrocarbon (PAH)-contaminated soil with the nitrate-reducing bacterium PheN7 under anaerobic condition. J. Hazard. Mater. 2022, 439, 129643. [Google Scholar] [CrossRef]
- Ma, X.; Li, X.; Liu, J.; Cheng, Y.; Zhai, F.; Sun, Z.; Han, L. Enhancing Salix viminalis L.-mediated phytoremediation of polycyclic aromatic hydrocarbon-contaminated soil by inoculation with Crucibulum leave (white-rot fungus). Environ. Sci. Pollut. Res. 2020, 27, 41326–41341. [Google Scholar] [CrossRef]
- Zhou, X.; Sun, Y.; Wang, T.; Tang, L.; Ling, W.; Mosa, A.; Wang, J.; Gao, Y. Remediation potential of an immobilized microbial consortium with corn straw as a carrier in polycyclic aromatic hydrocarbons contaminated soil. J. Hazard. Mater. 2024, 469, 134091. [Google Scholar] [CrossRef]
- Jia, W.; Cheng, L.; Tan, Q.; Liu, Y.; Dou, J.; Yang, K.; Yang, Q.; Wang, S.; Li, J.; Niu, G.; et al. Response of the soil microbial community to petroleum hydrocarbon stress shows a threshold effect: Research on aged realistic contaminated fields. Front. Microbiol. 2023, 14, 1188229. [Google Scholar] [CrossRef]
- Chauhan, A.; Fazlurrahman; Oakeshott, J.G.; Jain, R.K. Bacterial metabolism of polycyclic aromatic hydrocarbons: Strategies for bioremediation. Indian J. Microbiol. 2008, 48, 95–113. [Google Scholar] [CrossRef]
- Li, J.; Huang, Y.; Liu, L.; Li, M. Isolation, identification, and mechanism of the polycyclic aromatic hydrocarbons degrader Aquabacter sediminis P-9~T in mangrove sediment. Acta Microbiol. Sin. 2024, 64, 2115–2132. [Google Scholar]
- Yang, J.; Gu, Y.; Chen, Z.; Song, Y.; Sun, F.; Liu, J.; Waigi, M.G. Colonization and performance of a pyrene-degrading bacterium Mycolicibacterium sp. Pyr9 on root surfaces of white clover. Chemosphere 2021, 263, 127918. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Yan, K.; You, Q.; Chen, Y.; Pan, Y.; Wang, H.; Wu, L.; Xu, J. Soil indigenous microorganisms weaken the synergy of Massilia sp. WF1 and Phanerochaete chrysosporium in phenanthrene biodegradation. Sci. Total Environ. 2021, 781, 146655. [Google Scholar] [CrossRef]
- Masy, T.; Demanèche, S.; Tromme, O.; Thonart, P.; Jacques, P.; Hiligsmann, S.; Vogel, T.M. Hydrocarbon biostimulation and bioaugmentation in organic carbon and clay-rich soils. Soil Biol. Biochem. 2016, 99, 66–74. [Google Scholar] [CrossRef]
- Sun, Y.; Chen, W.; Wang, Y.; Guo, J.; Zhang, H.; Hu, X. Nutrient depletion is the main limiting factor in the crude oil bioaugmentation process. J. Environ. Sci. 2021, 100, 317–327. [Google Scholar] [CrossRef]
- Xun, W.; Li, W.; Xiong, W.; Ren, Y.; Liu, Y.; Miao, Y.; Xu, Z.; Zhang, N.; Shen, Q.; Zhang, R. Diversity-triggered deterministic bacterial assembly constrains community functions. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Wu, M.; Wu, J.; Zhang, X.; Ye, X. Effect of bioaugmentation and biostimulation on hydrocarbon degradation and microbial community composition in petroleum-contaminated loessal soil. Chemosphere 2019, 237, 124456. [Google Scholar] [CrossRef]
- Ahmad, M.; Liang, T.; Zhang, Y.; Wang, Y.; Gu, J.; Cheng, H.; Masmoudi, K.; Zhou, W.; Yang, Q.; Huang, X.; et al. Unraveling the genomic and metabolic mechanisms of pyrene and phenanthrene degradation by Mycolicibacterium sp. SCSIO 43805: A high-Efficiency bacterium isolated from seagrass sediment. Int. Biodeterior. Biodegrad. 2025, 202, 106101. [Google Scholar] [CrossRef]
- Zeng, J.; Lin, X.; Zhang, J.; Li, X. Isolation of polycyclic aromatic hydrocarbons (PAHs)-degrading Mycobacterium spp. and the degradation in soil. J. Hazard. Mater. 2010, 183, 718–723. [Google Scholar] [CrossRef]


| PAHs | Degradation Ability * | Degradation Efficiency ** | Medium Color Change *** |
|---|---|---|---|
| PHE | ++ | 89.82 ± 15.17% | + |
| ANT | + | 20.78 ± 18.36% | + |
| ACE | ++ | 81.3 ± 2.73% | + |
| FLN | ++ | 100% | + |
| FLU | + | 20.48 ± 4.36% | + |
| PYR | + | 23.72 ± 15.03% | + |
| BaA | - | 3.8 ± 2.87% | - |
| BaP | - | 2.7 ± 1.31% | - |
| Strain | Phenanthrene (mg/L) | Degradation Efficiency | Degradation Rate (mg/L/day) | Reference |
|---|---|---|---|---|
| TJFP1 | 100 | 100% (4.5 day) | 24.48 | This study |
| Pseudomonas sp. ANT13_1 | 50 | 22.4% (15 days) | 0.75 | [51] |
| Pseudarthrobacter sp. L1SW | 500 | 96.3% (72 h) | 160.50 | [41] |
| Klebsiella pneumoniae SJK1 | 1250 | 40.5(12 days) | 42.19 | [52] |
| Providencia rettgeri VMP5 | 50 | 98.6% (4 days) | 12.33 | [53] |
| Bacillus tropicus VMP4 | 50 | 89.9% (4 days) | 11.24 | [53] |
| Pseudarthrobacter phenanthrenivorans A-5 | 50 | 79.54% (7 days) | 5.68 | [54] |
| Stenotrophomonas in dicatrix CPHE1 | 10 | 100% (21 days) | 0.48 | [37] |
| Pseudomonas sp. Lphe-2 | 100 | 92.76% (7 days) | 13.25 | [36] |
| Pseudomonas fluorescens AH-40 | 150 | 97% (15 days) | 9.70 | [55] |
| Diaphorobacter sp. Phe15 | 50 | 22% (15 days) | 0.73 | [56] |
| Sphingobium sp. AM | 100 | 87.4% (7 days) | 12.49 | [57] |
| Pseudomonas sp. Ph6-gfp | 50 | 81.1% (15 days) | 2.70 | [58] |
| Massilia sp. Pn2 | 150 | 95% (48 h) | 71.25 | [59] |
| Mycolicibacterium sp. A1-PYR | 10 | 99% (2 days) | 4.95 | [60] |
| Pseudomonas sp. USTB-RU | 100 | 86.6% (8 days) | 10.83 | [61] |
| Pseudomonas sp. Ph-3 | 100 | 90% (7 days) | 12.86 | [62] |
| Roseovarius sp. SBU1 | 100 | 28.4% (10 days) | 2.84 | [63] |
| Pseudarthrobacter sp. Sphe3 | 400 | 90% (4 days) | 90.00 | [64] |
| Pseudarthrobacter sp. J015 | 100 | 90% (4 days) | 22.50 | [65] |
| Arthrobacter sp. P1–1 | 40 | 99% (7 days) | 5.66 | [66] |
| Arthrobacter sp. YC-RL1 | 50 | 82.3% (5 days) | 8.23 | [67] |
| Arthrobacter sp. K3 | 250 | 99% (5 days) | 49.50 | [68] |
| Mycolicibacterium. WY10 | 100 | 100% (60 h) | 40 | [69] |
| Databases | Number of Protein-Coding Genes | % |
|---|---|---|
| NR | 6777 | 96.70 |
| GO | 1413 | 20.85 |
| eggnog | 5320 | 376.50 |
| KEGG | 2832 | 53.23 |
| Swiss | 3671 | 129.63 |
| CAZy | 217 | 5.91 |
| CARD | 106 | 48.85 |
| Sample | Shannon | Simpson | ACE | Chao1 | Pielou | Coverage | |
|---|---|---|---|---|---|---|---|
| Treatment | Replicate | ||||||
| NC | 1 | 4.916584 | 0.015235 | 702.4928 | 704.6786 | 0.755206 | 0.999071 |
| 2 | 4.885554 | 0.017757 | 728.6357 | 721.00 | 0.747903 | 0.998949 | |
| 3 | 4.836199 | 0.018395 | 753.4264 | 752.3582 | 0.737588 | 0.998766 | |
| PT | 1 | 2.825297 | 0.173504 | 575.9003 | 577.1613 | 0.452611 | 0.998644 |
| 2 | 3.095102 | 0.145655 | 589.6721 | 590.4355 | 0.491655 | 0.998812 | |
| 3 | 2.77461 | 0.18746 | 589.3641 | 591.381 | 0.443802 | 0.998537 | |
| NG-5DAY | 1 | 3.467205 | 0.099935 | 553.1702 | 549.2542 | 0.555444 | 0.99901 |
| 2 | 3.483609 | 0.100303 | 608.2021 | 616.1724 | 0.553046 | 0.998598 | |
| 3 | 3.549557 | 0.091821 | 571.5692 | 565.9242 | 0.564177 | 0.999101 | |
| NG-20DAY | 1 | 3.19783 | 0.096232 | 528.1885 | 518.0845 | 0.518144 | 0.998857 |
| 2 | 3.182169 | 0.10002 | 508.6596 | 502.3871 | 0.51901 | 0.998888 | |
| 3 | 3.24315 | 0.094227 | 496.7473 | 497.06 | 0.52821 | 0.999116 | |
| BG-5DAY | 1 | 3.088348 | 0.142454 | 520.5665 | 514.25 | 0.501257 | 0.998933 |
| 2 | 3.09602 | 0.125491 | 565.0113 | 561.0909 | 0.497072 | 0.998705 | |
| 3 | 2.809184 | 0.169538 | 515.8918 | 525.1429 | 0.45916 | 0.99872 | |
| BG-20DAY | 1 | 3.123404 | 0.111245 | 515.0163 | 509.35 | 0.509788 | 0.998796 |
| 2 | 3.045859 | 0.106694 | 497.9351 | 489.5 | 0.497487 | 0.998979 | |
| 3 | 3.093912 | 0.111542 | 491.1816 | 497.1633 | 0.507359 | 0.998903 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Liu, J.; Fang, P. Biodegradation of Phenanthrene by Mycobacterium sp. TJFP1: Genetic Basis and Environmental Validation. Microorganisms 2025, 13, 1171. https://doi.org/10.3390/microorganisms13051171
Li S, Liu J, Fang P. Biodegradation of Phenanthrene by Mycobacterium sp. TJFP1: Genetic Basis and Environmental Validation. Microorganisms. 2025; 13(5):1171. https://doi.org/10.3390/microorganisms13051171
Chicago/Turabian StyleLi, Shuyun, Jiazhen Liu, and Ping Fang. 2025. "Biodegradation of Phenanthrene by Mycobacterium sp. TJFP1: Genetic Basis and Environmental Validation" Microorganisms 13, no. 5: 1171. https://doi.org/10.3390/microorganisms13051171
APA StyleLi, S., Liu, J., & Fang, P. (2025). Biodegradation of Phenanthrene by Mycobacterium sp. TJFP1: Genetic Basis and Environmental Validation. Microorganisms, 13(5), 1171. https://doi.org/10.3390/microorganisms13051171






