Advances in Wastewater-Based Epidemiology for Pandemic Surveillance: Methodological Frameworks and Future Perspectives
Abstract
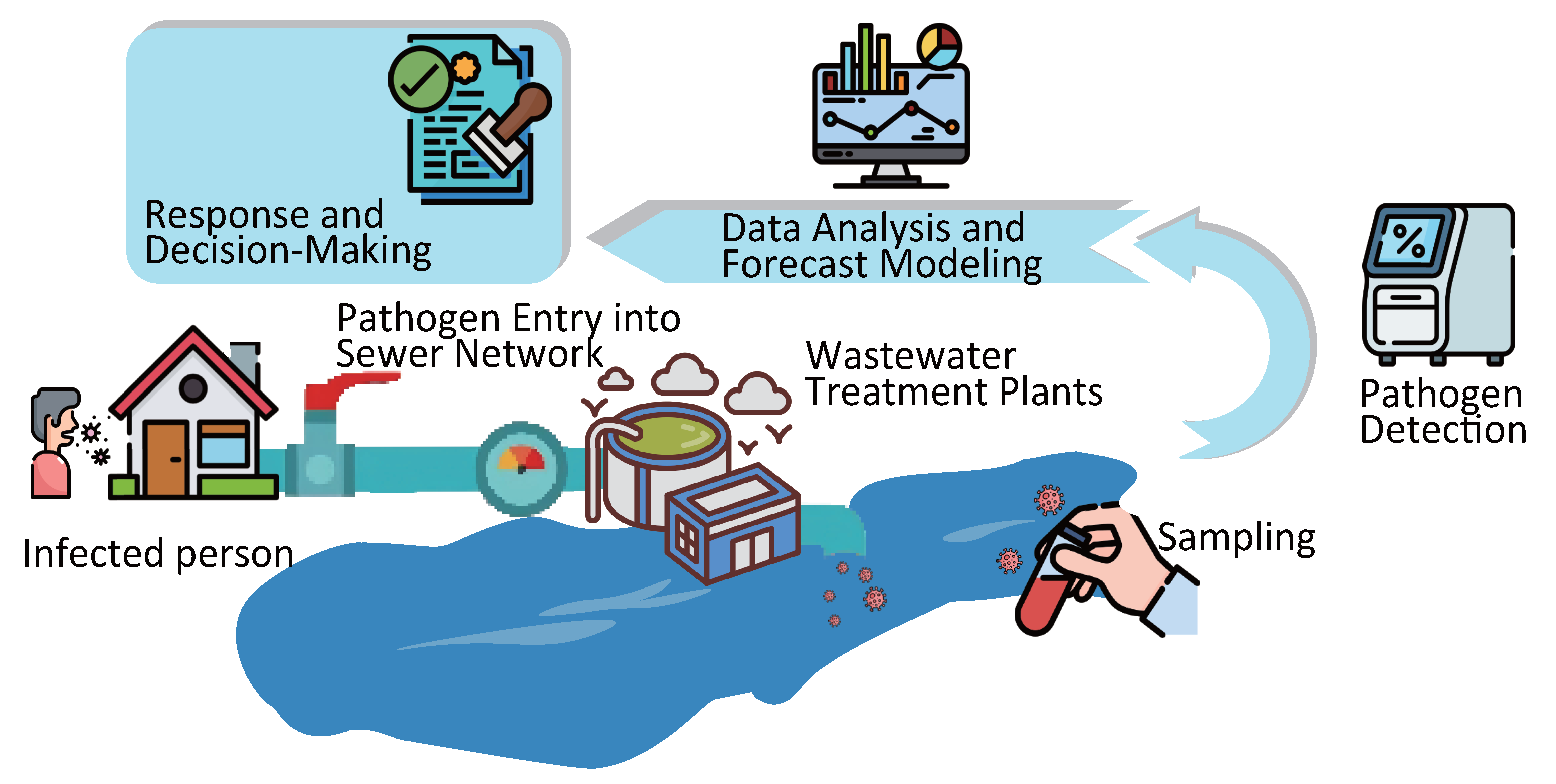
1. Introduction
2. Sampling in WBE
2.1. Sampling Locations
2.2. Sampling Timing
2.3. Sampling Frequency
2.4. Sampling Methods
3. Pathogen Detection Techniques in WBE
3.1. qPCR: The Gold Standard for Pathogen Detection
3.2. Sequencing: Beyond the Gold Standard
3.2.1. Amplicon Sequencing
3.2.2. Metagenomic Sequencing
3.2.3. Sequencing Platforms: Short-Read vs. Long-Read
3.2.4. Challenges of Sequencing in WBE
3.3. Biosensors: Emerging Tools for Flexible Detection
3.3.1. Types of Biosensors in WBE
3.3.2. Applications of Biosensors in WBE
3.3.3. Emerging Innovations of Biosensors in WBE
3.3.4. Challenges of Biosensors in WBE
4. Data Preprocessing and Epidemic Forecast Modeling
4.1. Data Preprocessing
4.1.1. Data Normalization
4.1.2. Data Evaluation and Adjustment
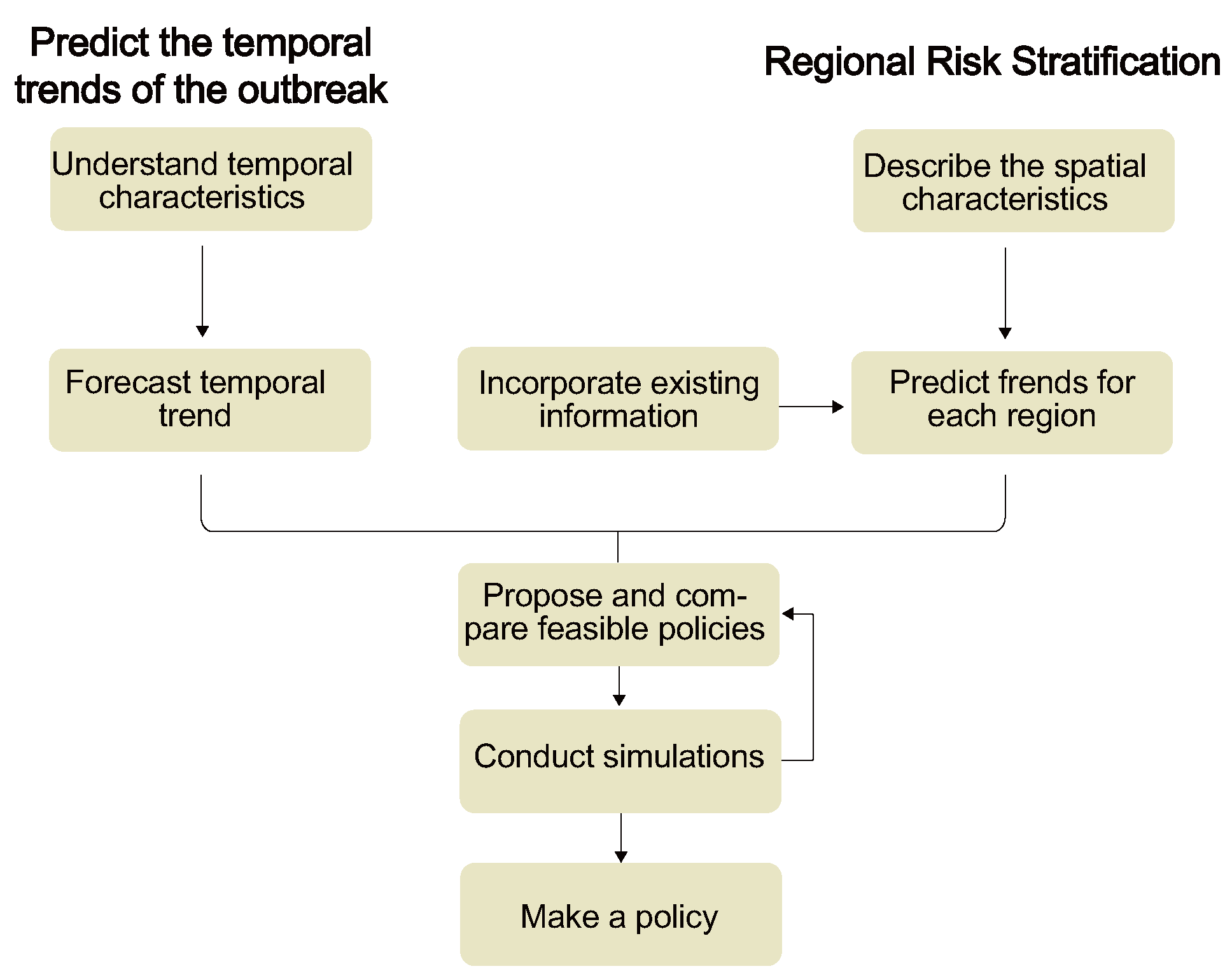
4.2. Epidemic Forecast Modeling
4.2.1. Modeling for Predicting the Temporal Trends of Epidemics
4.2.2. Modeling for Regional Risk Stratification
4.2.3. Machine Learning Models Used in WBE
4.3. Decision Making Based on Predictive Results
5. Future Perspectives for WBE
5.1. Improving WBE Accuracy
5.2. Increasing Speed and Efficiency in WBE
5.3. Reducing Costs for Global WBE Accessibility
5.4. Standardizing Global WBE Practices
5.5. Ethical and Privacy Considerations in WBE
5.6. Expanding WBE Targets
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Abbreviation | Full Form |
| WBE | Wastewater-based epidemiology |
| LC-MS | Liquid chromatography-mass spectrometry |
| qPCR | Quantitative polymerase chain reaction |
| GIS | Geographic information system |
| VOCs | Variants of concerns |
| NGS | Next-generation sequencing |
| ddPCR | Droplet digital polymerase chain reaction |
| MST | Microbial source tracking |
| HTS | High-throughput sequencing |
| RT-LAMP | Reverse transcription loop-mediated isothermal amplification |
| NASBA | Nucleic acid sequence-based amplification |
| FET | Field-effect transistors |
| mHealth | Mobile health |
| SVR | Support vector regression |
| ANN | Artificial neural network |
| RF | Random forest |
| SVM | Support vector machine |
| SMA | Simple moving average |
| LOESS | Locally weighted regression |
| PSAP | Porous superabsorbent polymer |
| SRT | Sewer residence time |
| VSV | Vesicular stomatitis virus |
| CDC | Center for Disease Control and Prevention |
| NWSS | National Wastewater Surveillance System |
| ARGs | Antibiotic resistance genes |
References
- World Health Organization. Public Health Surveillance for COVID-19: Interim Guidance, 14 February 2022; Report; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- World Health Organization. Support Tool to Strengthen Health Information Systems: Guidance for Health Information System Assessment and Strategy Development, 2nd ed.; Web Annex: Assessment Item Sheets; Report; Regional Office for Europe: Copenhagen, Denmark; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- Hewitt, J.; Leonard, M.; Greening, G.E.; Lewis, G.D. Influence of wastewater treatment process and the population size on human virus profiles in wastewater. Water Res. 2011, 45, 6267–6276. [Google Scholar] [CrossRef] [PubMed]
- Banta-Green, C.J.; Brewer, A.J.; Ort, C.; Helsel, D.R.; Williams, J.R.; Field, J.A. Using wastewater-based epidemiology to estimate drug consumption-Statistical analyses and data presentation. Sci. Total Environ. 2016, 568, 856–863. [Google Scholar] [CrossRef] [PubMed]
- Xagoraraki, I.; O’Brien, E. Wastewater-Based Epidemiology for Early Detection of Viral Outbreaks. In Women in Water Quality; Women in Engineering and Science; Springer: Cham, Switzerland, 2020; Chapter 5; pp. 75–97. [Google Scholar] [CrossRef]
- Hart, O.E.; Halden, R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: Feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020, 730, 138875. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, G.; Bonadonna, L.; Lucentini, L.; Kenmoe, S.; Suffredini, E. Coronavirus in water environments: Occurrence, persistence and concentration methods—A scoping review. Water Res. 2020, 179, 115899. [Google Scholar] [CrossRef]
- Xu, Y.; Li, X.; Zhu, B.; Liang, H.; Fang, C.; Gong, Y.; Guo, Q.; Sun, X.; Zhao, D.; Shen, J.; et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020, 26, 502–505. [Google Scholar] [CrossRef]
- Jafferali, M.H.; Khatami, K.; Atasoy, M.; Birgersson, M.; Williams, C.; Cetecioglu, Z. Benchmarking virus concentration methods for quantification of SARS-CoV-2 in raw wastewater. Sci. Total Environ. 2021, 755, 142939. [Google Scholar] [CrossRef]
- Klapsa, D.; Wilton, T.; Zealand, A.; Bujaki, E.; Saxentoff, E.; Troman, C.; Shaw, A.G.; Tedcastle, A.; Majumdar, M.; Mate, R.; et al. Sustained detection of type 2 poliovirus in London sewage between February and July, 2022, by enhanced environmental surveillance. Lancet 2022, 400, 1531–1538. [Google Scholar] [CrossRef]
- Pöyry, T.; Stenvik, M.; Hovi, T. Viruses in sewage waters during and after a poliomyelitis outbreak and subsequent nationwide oral poliovirus vaccination campaign in Finland. Appl. Environ. Microbiol. 1988, 54, 371–374. [Google Scholar] [CrossRef]
- van der Avoort, H.G.A.M.; Reimerink, J.H.J.; Ras, A.; Mulders, M.N.; van Loon, A.M. Isolation of epidemic poliovirus from sewage during the 1992–3 type 3 outbreak in the Netherlands. Epidemiol. Infect. 1995, 114, 481–491. [Google Scholar] [CrossRef]
- Daughton, C.G.; Ternes, T.A. Pharmaceuticals and personal care products in the environment: Agents of subtle change? Environ. Health Perspect 1999, 107 (Suppl. S6), 907–938. [Google Scholar] [CrossRef]
- Zuccato, E.; Chiabrando, C.; Castiglioni, S.; Calamari, D.; Bagnati, R.; Schiarea, S.; Fanelli, R. Cocaine in surface waters: A new evidence-based tool to monitor community drug abuse. Environ. Health 2005, 4, 14. [Google Scholar] [CrossRef] [PubMed]
- Corpuz, M.V.A.; Buonerba, A.; Vigliotta, G.; Zarra, T.; Ballesteros, F.J.; Campiglia, P.; Belgiorno, V.; Korshin, G.; Naddeo, V. Viruses in wastewater: Occurrence, abundance and detection methods. Sci. Total Environ. 2020, 745, 140910. [Google Scholar] [CrossRef]
- Fan, X.; Zhang, J.; Fu, X.; Zhou, B.; Xu, Z.; Huang, H.; Han, S.; Li, X. Analysis of synthetic cannabinoids in wastewater of major cities in China. Sci. Total Environ. 2022, 827, 154267. [Google Scholar] [CrossRef]
- Wei, Y.; Li, Y.; Wang, Y.; Luo, X.; Du, F.; Liu, W.; Xie, L.; Chen, J.; Ren, Z.; Hou, S.; et al. The microbial diversity in industrial effluents makes high-throughput sequencing-based source tracking of the effluents possible. Environ. Res. 2022, 212, 113640. [Google Scholar] [CrossRef]
- Prado, T.; Rey-Benito, G.; Miagostovich, M.P.; Sato, M.I.Z.; Rajal, V.B.; Filho, C.R.M.; Pereira, A.D.; Barbosa, M.R.F.; Mannarino, C.F.; da Silva, A.S. Wastewater-based epidemiology for preventing outbreaks and epidemics in Latin America—Lessons from the past and a look to the future. Sci. Total Environ. 2023, 865, 161210. [Google Scholar] [CrossRef]
- Zafeiriadou, A.; Kaltsis, L.; Kostakis, M.; Kapes, V.; Thomaidis, N.S.; Markou, A. Wastewater surveillance of the most common circulating respiratory viruses in Athens: The impact of COVID-19 on their seasonality. Sci. Total Environ. 2023, 900, 166136. [Google Scholar] [CrossRef]
- Ando, H.; Ahmed, W.; Iwamoto, R.; Ando, Y.; Okabe, S.; Kitajima, M. Impact of the COVID-19 pandemic on the prevalence of influenza A and respiratory syncytial viruses elucidated by wastewater-based epidemiology. Sci. Total Environ. 2023, 880, 162694. [Google Scholar] [CrossRef]
- Boehm, A.B.; Hughes, B.; Duong, D.; Chan-Herur, V.; Buchman, A.; Wolfe, M.K.; White, B.J. Wastewater concentrations of human influenza, metapneumovirus, parainfluenza, respiratory syncytial virus, rhinovirus, and seasonal coronavirus nucleic-acids during the COVID-19 pandemic: A surveillance study. Lancet Microbe 2023, 4, e340–e348. [Google Scholar] [CrossRef]
- Ort, C.; Eppler, J.M.; Scheidegger, A.; Rieckermann, J.; Kinzig, M.; Sörgel, F. Challenges of surveying wastewater drug loads of small populations and generalizable aspects on optimizing monitoring design. Addiction 2014, 109, 472–481. [Google Scholar] [CrossRef]
- Medema, G.; Heijnen, L.; Elsinga, G.; Italiaander, R.; Brouwer, A. Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020, 7, 511–516. [Google Scholar] [CrossRef]
- Randazzo, W.; Truchado, P.; Cuevas-Ferrando, E.; Simon, P.; Allende, A.; Sanchez, G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020, 181, 115942. [Google Scholar] [CrossRef] [PubMed]
- Fuschi, C.; Pu, H.; Negri, M.; Colwell, R.; Chen, J. Wastewater-Based Epidemiology for Managing the COVID-19 Pandemic. ACS ES&T Water 2021, 1, 1352–1362. [Google Scholar] [CrossRef]
- Singer, A.C.; Thompson, J.R.; Filho, C.R.M.; Street, R.; Li, X.; Castiglioni, S.; Thomas, K.V. A world of wastewater-based epidemiology. Nat. Water 2023, 1, 408–415. [Google Scholar] [CrossRef]
- Jones, D.L.; Baluja, M.Q.; Graham, D.W.; Corbishley, A.; McDonald, J.E.; Malham, S.K.; Hillary, L.S.; Connor, T.R.; Gaze, W.H.; Moura, I.B.; et al. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci. Total Environ. 2020, 749, 141364. [Google Scholar] [CrossRef]
- Tiwari, A.; Radu, E.; Kreuzinger, N.; Ahmed, W.; Pitkanen, T. Key considerations for pathogen surveillance in wastewater. Sci. Total Environ. 2024, 945, 173862. [Google Scholar] [CrossRef]
- Huizer, M.; ter Laak, T.L.; de Voogt, P.; van Wezel, A.P. Wastewater-based epidemiology for illicit drugs: A critical review on global data. Water Res. 2021, 207, 117789. [Google Scholar] [CrossRef]
- Rauch, W.; Schenk, H.; Insam, H.; Markt, R.; Kreuzinger, N. Data modelling recipes for SARS-CoV-2 wastewater-based epidemiology. Environ. Res. 2022, 214, 113809. [Google Scholar] [CrossRef]
- Keshaviah, A.; Hu Xindi, C.; Henry, M. Developing a Flexible National Wastewater Surveillance System for COVID-19 and Beyond. Environ. Health Perspect. 2021, 129, 045002. [Google Scholar] [CrossRef]
- Hewitt, J.; Trowsdale, S.; Armstrong, B.A.; Chapman, J.R.; Carter, K.M.; Croucher, D.M.; Trent, C.R.; Sim, R.E.; Gilpin, B.J. Sensitivity of wastewater-based epidemiology for detection of SARS-CoV-2 RNA in a low prevalence setting. Water Res. 2022, 211, 118032. [Google Scholar] [CrossRef]
- Wurtzer, S.; Waldman, P.; Ferrier-Rembert, A.; Frenois-Veyrat, G.; Mouchel, J.; Boni, M.; Maday, Y.; Marechal, V.; Moulin, L. Several forms of SARS-CoV-2 RNA can be detected in wastewaters: Implication for wastewater-based epidemiology and risk assessment. Water Res. 2021, 198, 117183. [Google Scholar] [CrossRef]
- Bivins, A.; Greaves, J.; Fischer, R.; Yinda, K.C.; Ahmed, W.; Kitajima, M.; Munster, V.J.; Bibby, K. Persistence of SARS-CoV-2 in Water and Wastewater. Environ. Sci. Technol. Lett. 2020, 7, 937–942. [Google Scholar] [CrossRef] [PubMed]
- Sala-Comorera, L.; Reynolds, L.J.; Martin, N.A.; O’Sullivan, J.J.; Meijer, W.G.; Fletcher, N.F. Decay of infectious SARS-CoV-2 and surrogates in aquatic environments. Water Res. 2021, 201, 117090. [Google Scholar] [CrossRef]
- Kasprzyk-Hordern, B.; Adams, B.; Adewale, I.D.; Agunbiade, F.O.; Akinyemi, M.I.; Archer, E.; Badru, F.A.; Barnett, J.; Bishop, I.J.; Di Lorenzo, M.; et al. Wastewater-based epidemiology in hazard forecasting and early-warning systems for global health risks. Environ. Int. 2022, 161, 107143. [Google Scholar] [CrossRef]
- Zehnder, C.; Been, F.; Vojinovic, Z.; Savic, D.; Torres, A.S.; Mark, O.; Zlatanovic, L.; Abebe, Y.A. Machine Learning for Detecting Virus Infection Hotspots Via Wastewater-Based Epidemiology: The Case of SARS-CoV-2 RNA. Geohealth 2023, 7, e2023GH000866. [Google Scholar] [CrossRef]
- Vaughan, L.; Zhang, M.; Gu, H.; Rose, J.B.; Naughton, C.C.; Medema, G.; Allan, V.; Roiko, A.; Blackall, L.; Zamyadi, A. An exploration of challenges associated with machine learning for time series forecasting of COVID-19 community spread using wastewater-based epidemiological data. Sci. Total Environ. 2023, 858, 159748. [Google Scholar] [CrossRef]
- Saingam, P.; Jain, T.; Woicik, A.; Li, B.; Candry, P.; Redcorn, R.; Wang, S.; Himmelfarb, J.; Bryan, A.; Winkler, M.K.H.; et al. Integrating socio-economic vulnerability factors improves neighborhood-scale wastewater-based epidemiology for public health applications. Water Res. 2024, 254, 121415. [Google Scholar] [CrossRef]
- Ye, Y.; Ellenberg, R.M.; Graham, K.E.; Wigginton, K.R. Survivability, Partitioning, and Recovery of Enveloped Viruses in Untreated Municipal Wastewater. Environ. Sci. Technol. 2016, 50, 5077–5085. [Google Scholar] [CrossRef]
- Fu, S.; He, F.; Wang, R.; Song, W.; Wang, Q.; Xia, W.; Qiu, Z. Development of quantitative wastewater surveillance models facilitated the precise epidemic management of COVID-19. Sci. Total Environ. 2023, 857, 159357. [Google Scholar] [CrossRef]
- Sherchan, S.P.; Shahin, S.; Ward, L.M.; Tandukar, S.; Aw, T.G.; Schmitz, B.; Ahmed, W.; Kitajima, M. First detection of SARS-CoV-2 RNA in wastewater in North America: A study in Louisiana, USA. Sci. Total Environ. 2020, 743, 140621. [Google Scholar] [CrossRef]
- Miura, F.; Kitajima, M.; Omori, R. Duration of SARS-CoV-2 viral shedding in faeces as a parameter for wastewater-based epidemiology: Re-analysis of patient data using a shedding dynamics model. Sci. Total Environ. 2021, 769, 144549. [Google Scholar] [CrossRef]
- Mahlangeni, N.; Street, R.; Horn, S.; Mathee, A.; Mangwana, N.; Dias, S.; Sharma, J.R.; Ramharack, P.; Louw, J.; Reddy, T.; et al. Using Wastewater Surveillance to Compare COVID-19 Outbreaks during the Easter Holidays over a 2-Year Period in Cape Town, South Africa. Viruses 2023, 15, 162. [Google Scholar] [CrossRef] [PubMed]
- Bibby, K.; Peccia, J. Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environ. Sci. Technol. 2013, 47, 1945–1951. [Google Scholar] [CrossRef]
- Maal-Bared, R.; Brisolara, K.; Knight, M.; Mansfeldt, C. To sample or not to sample: A governance-focused decision tree for wastewater service providers considering participation in wastewater-based epidemiology (WBE) in support of public health programs. Sci. Total Environ. 2023, 905, 167128. [Google Scholar] [CrossRef]
- Kitamura, K.; Sadamasu, K.; Muramatsu, M.; Yoshida, H. Efficient detection of SARS-CoV-2 RNA in the solid fraction of wastewater. Sci. Total Environ. 2021, 763, 144587. [Google Scholar] [CrossRef]
- Babler, K.M.; Sharkey, M.E.; Abelson, S.; Amirali, A.; Benitez, A.; Cosculluela, G.A.; Grills, G.S.; Kumar, N.; Laine, J.; Lamar, W.; et al. Degradation rates influence the ability of composite samples to represent 24-hourly means of SARS-CoV-2 and other microbiological target measures in wastewater. Sci. Total Environ. 2023, 867, 161423. [Google Scholar] [CrossRef]
- D’Aoust, P.M.; Mercier, E.; Montpetit, D.; Jia, J.J.; Alexandrov, I.; Neault, N.; Baig, A.T.; Mayne, J.; Zhang, X.; Alain, T.; et al. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021, 188, 116560. [Google Scholar] [CrossRef]
- Williams, R.C.; Perry, W.B.; Lambert-Slosarska, K.; Futcher, B.; Pellett, C.; Richardson-O’Neill, I.; Paterson, S.; Grimsley, J.M.S.; Wade, M.J.; Weightman, A.J.; et al. Examining the stability of viral RNA and DNA in wastewater: Effects of storage time, temperature, and freeze-thaw cycles. Water Res. 2024, 259, 121879. [Google Scholar] [CrossRef]
- Guo, Y.; Sivakumar, M.; Jiang, G. Decay of four enteric pathogens and implications to wastewater-based epidemiology: Effects of temperature and wastewater dilutions. Sci. Total Environ. 2022, 819, 152000. [Google Scholar] [CrossRef]
- Hassard, F.; Singh, S.; Coulon, F.; Yang, Z. Can wastewater monitoring protect public health in schools? Lancet Reg. Health—Am. 2023, 20, 100475. [Google Scholar] [CrossRef]
- Ahmed, W.; Bertsch, P.M.; Bivins, A.; Bibby, K.; Farkas, K.; Gathercole, A.; Haramoto, E.; Gyawali, P.; Korajkic, A.; McMinn, B.R.; et al. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020, 739, 139960. [Google Scholar] [CrossRef]
- Chaqroun, A.; Bertrand, I.; Wurtzer, S.; Moulin, L.; Boni, M.; Soubies, S.; Boudaud, N.; Gantzer, C. Assessing infectivity of emerging enveloped viruses in wastewater and sewage sludge: Relevance and procedures. Sci. Total Environ. 2024, 943, 173648. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Classification of Omicron (B.1.1.529) SARS-CoV-2 Variant of Concern; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Ousset, M.J.; Pianciola, L.A.; Mazzeo, M.; Oteiza, J.M.; Jaureguiberry, M.S.; Venturino, A.; Barril, P.A. Improved SARS-CoV-2 RNA recovery in wastewater matrices using a CTAB-based extraction method. J. Virol. Methods 2024, 327, 114918. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Angel, N.; Edson, J.; Bibby, K.; Bivins, A.; O’Brien, J.W.; Choi, P.M.; Kitajima, M.; Simpson, S.L.; Li, J.; et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020, 728, 138764. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zheng, X.; Li, S.; Lam, N.S.; Wang, Y.; Chu, D.K.W.; Poon, L.L.M.; Tun, H.M.; Peiris, M.; Deng, Y.; et al. The first case study of wastewater-based epidemiology of COVID-19 in Hong Kong. Sci. Total Environ. 2021, 790, 148000. [Google Scholar] [CrossRef]
- Ho, J.; Stange, C.; Suhrborg, R.; Wurzbacher, C.; Drewes, J.E.; Tiehm, A. SARS-CoV-2 wastewater surveillance in Germany: Long-term RT-digital droplet PCR monitoring, suitability of primer/probe combinations and biomarker stability. Water Res. 2022, 210, 117977. [Google Scholar] [CrossRef]
- Ahmed, W.; Simpson, S.L.; Bertsch, P.M.; Bibby, K.; Bivins, A.; Blackall, L.L.; Bofill-Mas, S.; Bosch, A.; Brandão, J.; Choi, P.M.; et al. Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Sci. Total Environ. 2022, 805, 149877. [Google Scholar] [CrossRef]
- Suo, T.; Liu, X.; Feng, J.; Guo, M.; Hu, W.; Guo, D.; Ullah, H.; Yang, Y.; Zhang, Q.; Wang, X.; et al. ddPCR: A more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes Infect. 2020, 9, 1259–1268. [Google Scholar] [CrossRef]
- Deng, Y.; Xu, X.; Zheng, X.; Ding, J.; Li, S.; Chui, H.K.; Wong, T.K.; Poon, L.L.M.; Zhang, T. Use of sewage surveillance for COVID-19 to guide public health response: A case study in Hong Kong. Sci. Total Environ. 2022, 821, 153250. [Google Scholar] [CrossRef]
- Balleste, E.; Pascual-Benito, M.; Martin-Diaz, J.; Blanch, A.R.; Lucena, F.; Muniesa, M.; Jofre, J.; Garcia-Aljaro, C. Dynamics of crAssphage as a human source tracking marker in potentially faecally polluted environments. Water Res. 2019, 155, 233–244. [Google Scholar] [CrossRef]
- Bisseux, M.; Debroas, D.; Mirand, A.; Archimbaud, C.; Peigue-Lafeuille, H.; Bailly, J.L.; Henquell, C. Monitoring of enterovirus diversity in wastewater by ultra-deep sequencing: An effective complementary tool for clinical enterovirus surveillance. Water Res. 2020, 169, 115246. [Google Scholar] [CrossRef]
- McCall, C.; Wu, H.; Miyani, B.; Xagoraraki, I. Identification of multiple potential viral diseases in a large urban center using wastewater surveillance. Water Res. 2020, 184, 116160. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ni, G.; Tian, W.; Yang, L.; Hosegood, I.; Newell, R.; Woodcroft, B.J.; Yang, B.; Hu, S.; Guo, J. Detection of SARS-CoV-2 Variants of Concern with Tiling Amplicon Sequencing from Wastewater. ACS ES&T Water 2022, 2, 2185–2193. [Google Scholar] [CrossRef]
- Xu, X.; Deng, Y.; Ding, J.; Zheng, X.; Wang, C.; Wang, D.; Liu, L.; Gu, H.; Peiris, M.; Poon, L.L.M.; et al. Wastewater genomic sequencing for SARS-CoV-2 variants surveillance in wastewater-based epidemiology applications. Water Res. 2023, 244, 120444. [Google Scholar] [CrossRef]
- Timme, R.E.; Woods, J.; Jones, J.L.; Calci, K.R.; Rodriguez, R.; Barnes, C.; Leard, E.; Craven, M.; Chen, H.; Boerner, C.; et al. SARS-CoV-2 wastewater variant surveillance: Pandemic response leveraging FDA’s GenomeTrakr network. mSystems 2024, 9, e01415-23. [Google Scholar] [CrossRef]
- Wang, Y.; Ni, G.; Tian, W.; Wang, H.; Li, J.; Thai, P.; Choi, P.M.; Jackson, G.; Hu, S.; Yang, B.; et al. Wastewater tiling amplicon sequencing in sentinel sites reveals longitudinal dynamics of SARS-CoV-2 variants prevalence. Water Res. X 2024, 23, 100224. [Google Scholar] [CrossRef]
- Bibby, K.; Crank, K.; Greaves, J.; Li, X.; Wu, Z.; Hamza, I.A.; Stachler, E. Metagenomics and the development of viral water quality tools. npj Clean Water 2019, 2, 9. [Google Scholar] [CrossRef]
- Child, H.T.; Airey, G.; Maloney, D.M.; Parker, A.; Wild, J.; McGinley, S.; Evens, N.; Porter, J.; Templeton, K.; Paterson, S.; et al. Comparison of metagenomic and targeted methods for sequencing human pathogenic viruses from wastewater. mBio 2023, 14, e01468-23. [Google Scholar] [CrossRef]
- Zhang, Z.; He, F.; Yi, L.; Deng, Z.; Wang, R.; Shen, L.; Fu, S. Wastewater surveillance together with metaviromic data revealed the unusual resurgence of infectious diseases after the first wave of the COVID-19 outbreak. J. Hazard. Mater. 2024, 473, 134635. [Google Scholar] [CrossRef]
- Tao, Z.; Chen, P.; Cui, N.; Lin, X.; Ji, F.; Liu, Y.; Xiong, P.; Zhang, L.; Xu, Q.; Song, Y.; et al. Detection of enteroviruses in urban sewage by next generation sequencing and its application in environmental surveillance. Sci. Total. Environ. 2020, 728, 138818. [Google Scholar] [CrossRef]
- Quail, M.A.; Kozarewa, I.; Smith, F.; Scally, A.; Stephens, P.J.; Durbin, R.; Swerdlow, H.; Turner, D.J. A large genome center’s improvements to the Illumina sequencing system. Nat. Methods 2008, 5, 1005–1010. [Google Scholar] [CrossRef]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, S.; Yan, Q.; Guo, R.; Zhang, Y.; Chen, F.; Tian, X.; Lv, Q.; Jin, H.; Ma, X.; et al. Optimization and evaluation of viral metagenomic amplification and sequencing procedures toward a genome-level resolution of the human fecal DNA virome. J. Adv. Res. 2023, 48, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Spurbeck, R.R.; Minard-Smith, A.; Catlin, L. Feasibility of neighborhood and building scale wastewater-based genomic epidemiology for pathogen surveillance. Sci. Total. Environ. 2021, 789, 147829. [Google Scholar] [CrossRef]
- Fernandez-Cassi, X.; Timoneda, N.; Martínez-Puchol, S.; Rusiñol, M.; Rodriguez-Manzano, J.; Figuerola, N.; Bofill-Mas, S.; Abril, J.F.; Girones, R. Metagenomics for the study of viruses in urban sewage as a tool for public health surveillance. Sci. Total Environ. 2018, 618, 870–880. [Google Scholar] [CrossRef]
- Ko, K.K.K.; Chng, K.R.; Nagarajan, N. Metagenomics-enabled microbial surveillance. Nat. Microbiol. 2022, 7, 486–496. [Google Scholar] [CrossRef]
- Kovaka, S.; Fan, Y.; Ni, B.; Timp, W.; Schatz, M.C. Targeted nanopore sequencing by real-time mapping of raw electrical signal with UNCALLED. Nat. Biotechnol. 2021, 39, 431–441. [Google Scholar] [CrossRef]
- Zhang, J.; Tang, A.; Jin, T.; Sun, D.; Guo, F.; Lei, H.; Lin, L.; Shu, W.; Yu, P.; Li, X.; et al. A panoramic view of the virosphere in three wastewater treatment plants by integrating viral-like particle-concentrated and traditional non-concentrated metagenomic approaches. iMeta 2024, 3, e188. [Google Scholar] [CrossRef]
- Xia, Y.; Li, X.; Wu, Z.; Nie, C.; Cheng, Z.; Sun, Y.; Liu, L.; Zhang, T. Strategies and tools in illumina and nanopore-integrated metagenomic analysis of microbiome data. iMeta 2023, 2, e72. [Google Scholar] [CrossRef]
- Rios, G.; Lacoux, C.; Leclercq, V.; Diamant, A.; Lebrigand, K.; Lazuka, A.; Soyeux, E.; Lacroix, S.; Fassy, J.; Couesnon, A.; et al. Monitoring SARS-CoV-2 variants alterations in Nice neighborhoods by wastewater nanopore sequencing. Lancet Reg. Health Eur. 2021, 10, 100202. [Google Scholar] [CrossRef]
- Barbe, L.; Schaeffer, J.; Besnard, A.; Jousse, S.; Wurtzer, S.; Moulin, L.; Consortium, O.; Le Guyader, F.S.; Desdouits, M. SARS-CoV-2 Whole-Genome Sequencing Using Oxford Nanopore Technology for Variant Monitoring in Wastewaters. Front. Microbiol. 2022, 13, 889811. [Google Scholar] [CrossRef]
- Kang, S.; Choi, P.; Maile-Moskowitz, A.; Brown, C.L.; Gonzalez, R.A.; Pruden, A.; Vikesland, P.J. Highly Multiplexed Reverse-Transcription Loop-Mediated Isothermal Amplification and Nanopore Sequencing (LAMPore) for Wastewater-Based Surveillance. ACS ES&T Water 2024, 4, 1629–1636. [Google Scholar] [CrossRef]
- van Dijk, E.L.; Jaszczyszyn, Y.; Naquin, D.; Thermes, C. The Third Revolution in Sequencing Technology. Trends Genet. 2018, 34, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Alafeef, M.; Dighe, K.; Moitra, P.; Pan, D. Rapid, Ultrasensitive, and Quantitative Detection of SARS-CoV-2 Using Antisense Oligonucleotides Directed Electrochemical Biosensor Chip. ACS Nano 2020, 14, 17028–17045. [Google Scholar] [CrossRef]
- Devianto, L.A.; Sano, D. Systematic review and meta-analysis of human health-related protein markers for realizing real-time wastewater-based epidemiology. Sci. Total Environ. 2023, 897, 165304. [Google Scholar] [CrossRef]
- Torres-Salvador, F.; Ojeda, J.; Castro, C.; Gerasimova, Y.; Chumbimuni-Torres, K. A Single Electrochemical Biosensor Designed to Detect Any Virus. Anal. Chem. 2024, 96, 5752–5756. [Google Scholar] [CrossRef]
- Boza, J.M.; Amirali, A.; Williams, S.L.; Currall, B.B.; Grills, G.S.; Mason, C.E.; Solo-Gabriele, H.M.; Erickson, D.C. Evaluation of a field deployable, high-throughput RT-LAMP device as an early warning system for COVID-19 through SARS-CoV-2 measurements in wastewater. Sci. Total Environ. 2024, 944, 173744. [Google Scholar] [CrossRef]
- Ahn, G.; Lee, S.; Lee, S.H.; Baek, Y.H.; Song, M.S.; Kim, Y.H.; Ahn, J.Y. Zika virus lateral flow assays using reverse transcription-loop-mediated isothermal amplification. RSC Adv. 2021, 11, 17800–17808. [Google Scholar] [CrossRef]
- Ramírez-Chavarría, R.G.; Castillo-Villanueva, E.; Alvarez-Serna, B.E.; Carrillo-Reyes, J.; Ramírez-Zamora, R.M.; Buitrón, G.; Alvarez-Icaza, L. Loop-mediated isothermal amplification-based electrochemical sensor for detecting SARS-CoV-2 in wastewater samples. J. Environ. Chem. Eng. 2022, 10, 107488. [Google Scholar] [CrossRef]
- Yang, Z. Low-cost and rapid sensors for wastewater surveillance at low-resource settings. Nat. Water 2023, 1, 405–407. [Google Scholar] [CrossRef]
- Pan, Y.; Wang, B.; Cooper, J.M.; Yang, Z. Paper microfluidic sentinel sensors enable rapid and on-site wastewater surveillance in community settings. Cell Rep. Phys. Sci. 2024, 5, 102154. [Google Scholar] [CrossRef]
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.H.; Choi, M.; Ku, K.B.; Lee, C.S.; Jun, S.; Park, D.; Kim, H.G.; et al. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, N.; Pan, Y.; Yang, Z.; Payam, A.F. Opportunities and Challenges for Biosensors and Nanoscale Analytical Tools for Pandemics: COVID-19. ACS Nano 2020, 14, 7783–7807. [Google Scholar] [CrossRef]
- Kang, H.; Wang, X.; Guo, M.; Dai, C.; Chen, R.; Yang, L.; Wu, Y.; Ying, T.; Zhu, Z.; Wei, D.; et al. Ultrasensitive Detection of SARS-CoV-2 Antibody by Graphene Field-Effect Transistors. Nano Lett. 2021, 21, 7897–7904. [Google Scholar] [CrossRef]
- Jiménez-Rodríguez, M.G.; Silva-Lance, F.; Parra-Arroyo, L.; Medina-Salazar, D.A.; Martínez-Ruiz, M.; Melchor-Martínez, E.M.; Martínez-Prado, M.A.; Iqbal, H.M.N.; Parra-Saldívar, R.; Barceló, D.; et al. Biosensors for the detection of disease outbreaks through wastewater-based epidemiology. TrAC Trends Anal. Chem. 2022, 155, 116585. [Google Scholar] [CrossRef]
- Goswami, N.; He, Y.R.; Deng, Y.H.; Oh, C.; Sobh, N.; Valera, E.; Bashir, R.; Ismail, N.; Kong, H.; Nguyen, T.H.; et al. Label-free SARS-CoV-2 detection and classification using phase imaging with computational specificity. Light Sci. Appl. 2021, 10, 176. [Google Scholar] [CrossRef]
- Kadadou, D.; Tizani, L.; Wadi, V.S.; Banat, F.; Alsafar, H.; Yousef, A.F.; Hasan, S.W. Detection of SARS-CoV-2 in clinical and environmental samples using highly sensitive reduced graphene oxide (rGO)-based biosensor. Chem. Eng. J. 2023, 453, 139750. [Google Scholar] [CrossRef]
- Mao, K.; Zhang, H.; Wang, Z.; Cao, H.; Zhang, K.; Li, X.; Yang, Z. Nanomaterial-based aptamer sensors for arsenic detection. Biosens Bioelectron 2020, 148, 111785. [Google Scholar] [CrossRef]
- Mao, K.; Zhang, H.; Yang, Z. An integrated biosensor system with mobile health and wastewater-based epidemiology (iBMW) for COVID-19 pandemic. Biosens Bioelectron 2020, 169, 112617. [Google Scholar] [CrossRef]
- Mao, K.; Zhang, H.; Pan, Y.; Yang, Z. Biosensors for wastewater-based epidemiology for monitoring public health. Water Res. 2021, 191, 116787. [Google Scholar] [CrossRef]
- Yang, Z.; Kasprzyk-Hordern, B.; Frost, C.G.; Estrela, P.; Thomas, K.V. Community sewage sensors for monitoring public health. Environ. Sci. Technol. 2015, 49, 5845–5846. [Google Scholar] [CrossRef]
- Hai, X.; Li, Y.; Zhu, C.; Song, W.; Cao, J.; Bi, S. DNA-based label-free electrochemical biosensors: From principles to applications. TrAC Trends Anal. Chem. 2020, 133, 116098. [Google Scholar] [CrossRef]
- Pilevar, M.; Kim, K.T.; Lee, W.H. Recent advances in biosensors for detecting viruses in water and wastewater. J. Hazard. Mater. 2021, 410, 124656. [Google Scholar] [CrossRef] [PubMed]
- Leisman, K.P.; Owen, C.; Warns, M.M.; Tiwari, A.; Bian, G.; Owens, S.M.; Catlett, C.; Shrestha, A.; Poretsky, R.; Packman, A.I.; et al. A modeling pipeline to relate municipal wastewater surveillance and regional public health data. Water Res. 2024, 252, 121178. [Google Scholar] [CrossRef]
- Schenk, H.; Heidinger, P.; Insam, H.; Kreuzinger, N.; Markt, R.; Nagele, F.; Oberacher, H.; Scheffknecht, C.; Steinlechner, M.; Vogl, G.; et al. Prediction of hospitalisations based on wastewater-based SARS-CoV-2 epidemiology. Sci. Total Environ. 2023, 873, 162149. [Google Scholar] [CrossRef]
- Rahm, E.; Do, H. Data Cleaning: Problems and Current Approaches. IEEE Data Eng. Bull. 2000, 23, 3–13. [Google Scholar]
- Niranjanamurthy, M.; Sheoran, K.; Dhand, G.; Kaur, P. Data Wrangling: Concepts, Applications and Tools; Wiley: Hoboken, NJ, USA, 2023. [Google Scholar]
- Zhuang, F.; Qi, Z.; Duan, K.; Xi, D.; Zhu, Y.; Zhu, H.; Xiong, H.; He, Q. A Comprehensive Survey on Transfer Learning. Proc. IEEE 2021, 109, 43–76. [Google Scholar] [CrossRef]
- Karthikeyan, S.; Levy, J.I.; De Hoff, P.; Humphrey, G.; Birmingham, A.; Jepsen, K.; Farmer, S.; Tubb, H.M.; Valles, T.; Tribelhorn, C.E.; et al. Wastewater sequencing reveals early cryptic SARS-CoV-2 variant transmission. Nature 2022, 609, 101–108. [Google Scholar] [CrossRef]
- Amirali, A.; Babler, K.M.; Sharkey, M.E.; Beaver, C.C.; Boone, M.M.; Comerford, S.; Cooper, D.; Currall, B.B.; Goodman, K.W.; Grills, G.S.; et al. Wastewater based surveillance can be used to reduce clinical testing intensity on a university campus. Sci. Total Environ. 2024, 918, 170452. [Google Scholar] [CrossRef]
- Jeng, H.A.; Singh, R.; Diawara, N.; Curtis, K.; Gonzalez, R.; Welch, N.; Jackson, C.; Jurgens, D.; Adikari, S. Application of wastewater-based surveillance and copula time-series model for COVID-19 forecasts. Sci. Total Environ. 2023, 885, 163655. [Google Scholar] [CrossRef]
- Krivonakova, N.; Soltysova, A.; Tamas, M.; Takac, Z.; Krahulec, J.; Ficek, A.; Gal, M.; Gall, M.; Feher, M.; Krivjanska, A.; et al. Mathematical modeling based on RT-qPCR analysis of SARS-CoV-2 in wastewater as a tool for epidemiology. Sci. Rep. 2021, 11, 19456. [Google Scholar] [CrossRef]
- Jiang, G.; Wu, J.; Weidhaas, J.; Li, X.; Chen, Y.; Mueller, J.; Li, J.; Kumar, M.; Zhou, X.; Arora, S.; et al. Artificial neural network-based estimation of COVID-19 case numbers and effective reproduction rate using wastewater-based epidemiology. Water Res. 2022, 218, 118451. [Google Scholar] [CrossRef]
- Lai, M.; Cao, Y.; Wulff, S.S.; Robinson, T.J.; McGuire, A.; Bisha, B. A time series based machine learning strategy for wastewater-based forecasting and nowcasting of COVID-19 dynamics. Sci. Total Environ. 2023, 897, 165105. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, H.; Gao, L.; Sherchan, S.P.; Zhou, T.; Khan, S.J.; van Loosdrecht, M.C.M.; Wang, Q. Wastewater-based epidemiology predicts COVID-19-induced weekly new hospital admissions in over 150 USA counties. Nat. Commun. 2023, 14, 4548. [Google Scholar] [CrossRef] [PubMed]
- Greff, K.; Srivastava, R.K.; Koutník, J.; Steunebrink, B.R.; Schmidhuber, J. LSTM: A Search Space Odyssey. IEEE Trans. Neural Netw. Learn. Syst. 2017, 28, 2222–2232. [Google Scholar] [CrossRef]
- Senjyu, T.; Mahalle, P.; Perumal, T.; Joshi, A. (Eds.) Smart Innovation, Systems and Technologies. In IOT with Smart Systems, Proceedings of ICTIS 2021, Ahmedabad, India, 23–24 April 2021; Springer: Singapore, 2022; Volume 2, XVI; 829p. [Google Scholar] [CrossRef]
- Hamerly, G.; Elkan, C. Learning the k in k-means. In Advances in Neural Information Processing Systems; Thrun, S., Saul, L., Schölkopf, B., Eds.; MIT Press: Cambridge, MA, USA, 2003; Volume 16. [Google Scholar]
- Krishna, K.; Narasimha Murty, M. Genetic K-means algorithm. IEEE Trans. Syst. Man, Cybern. Part B 1999, 29, 433–439. [Google Scholar] [CrossRef]
- Likas, A.; Vlassis, N.; Verbeek, J. The global k-means clustering algorithm. Pattern Recognit. 2003, 36, 451–461. [Google Scholar] [CrossRef]
- Snee, R.D. Validation of Regression Models: Methods and Examples. Technometrics 1977, 19, 415–428. [Google Scholar] [CrossRef]
- Cleveland, W.S.; Grosse, E.; Shyu, W.M. Local regression models. In Statistical models in S; Routledge: London, UK, 2017; pp. 309–376. [Google Scholar]
- Fahrmeir, L.; Kneib, T.; Lang, S.; Marx, B.D. Regression Models. In Regression: Models, Methods and Applications; Fahrmeir, L., Kneib, T., Lang, S., Marx, B.D., Eds.; Springer: Berlin/Heidelberg, Germany, 2021; pp. 23–84. [Google Scholar] [CrossRef]
- Harrell, F.E.; Harrell, F. Regression Modeling Strategies; Springer: Berlin/Heidelberg, Germany, 2001; Volume 54. [Google Scholar]
- Abdeldayem, O.M.; Dabbish, A.M.; Habashy, M.M.; Mostafa, M.K.; Elhefnawy, M.; Amin, L.; Al-Sakkari, E.G.; Ragab, A.; Rene, E.R. Viral outbreaks detection and surveillance using wastewater-based epidemiology, viral air sampling, and machine learning techniques: A comprehensive review and outlook. Sci. Total Environ. 2022, 803, 149834. [Google Scholar] [CrossRef]
- Thai, P.K.; Zheng, Q.; Phung, D.; Gartner, C.; Hall, W.; Ren, Y.; Mueller, J.F.; Thomas, K.V. The use of asthma and allergy medicines is associated with exposure to smoking. Nat. Water 2023, 1, 443–450. [Google Scholar] [CrossRef]
- Haak, L.; Delic, B.; Li, L.; Guarin, T.; Mazurowski, L.; Dastjerdi, N.G.; Dewan, A.; Pagilla, K. Spatial and temporal variability and data bias in wastewater surveillance of SARS-CoV-2 in a sewer system. Sci. Total Environ. 2022, 805, 150390. [Google Scholar] [CrossRef]
- Zoabi, Y.; Deri-Rozov, S.; Shomron, N. Machine learning-based prediction of COVID-19 diagnosis based on symptoms. npj Digit. Med. 2021, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Diamond, M.B.; Keshaviah, A.; Bento, A.I.; Conroy-Ben, O.; Driver, E.M.; Ensor, K.B.; Halden, R.U.; Hopkins, L.P.; Kuhn, K.G.; Moe, C.L.; et al. Wastewater surveillance of pathogens can inform public health responses. Nat. Med. 2022, 28, 1992–1995. [Google Scholar] [CrossRef] [PubMed]
- Devaux, Y.; Zhang, L.; Lumley, A.I.; Karaduzovic-Hadziabdic, K.; Mooser, V.; Rousseau, S.; Shoaib, M.; Satagopam, V.; Adilovic, M.; Srivastava, P.K.; et al. Development of a long noncoding RNA-based machine learning model to predict COVID-19 in-hospital mortality. Nat. Commun. 2024, 15, 4259. [Google Scholar] [CrossRef] [PubMed]
- Rallapalli, S.; Aggarwal, S.; Singh, A.P. Detecting SARS-CoV-2 RNA prone clusters in a municipal wastewater network using fuzzy-Bayesian optimization model to facilitate wastewater-based epidemiology. Sci. Total Environ. 2021, 778, 146294. [Google Scholar] [CrossRef]
- Wade, M.J.; Lo Jacomo, A.; Armenise, E.; Brown, M.R.; Bunce, J.T.; Cameron, G.J.; Fang, Z.; Farkas, K.; Gilpin, D.F.; Graham, D.W.; et al. Understanding and managing uncertainty and variability for wastewater monitoring beyond the pandemic: Lessons learned from the United Kingdom national COVID-19 surveillance programmes. J. Hazard. Mater. 2022, 424, 127456. [Google Scholar] [CrossRef]
- Dayan, I.; Roth, H.R.; Zhong, A.; Harouni, A.; Gentili, A.; Abidin, A.Z.; Liu, A.; Costa, A.B.; Wood, B.J.; Tsai, C.S.; et al. Federated learning for predicting clinical outcomes in patients with COVID-19. Nat. Med. 2021, 27, 1735–1743. [Google Scholar] [CrossRef]
- Lalmuanawma, S.; Hussain, J.; Chhakchhuak, L. Applications of machine learning and artificial intelligence for COVID-19 (SARS-CoV-2) pandemic: A review. Chaos Solitons Fractals 2020, 139, 110059. [Google Scholar] [CrossRef]
- Maseleno, A.; Hasan, M.M.; Tuah, N.; Tabbu, C.R. Fuzzy Logic and Mathematical Theory of Evidence to Detect the Risk of Disease Spreading of Highly Pathogenic Avian Influenza H5N1. Procedia Comput. Sci. 2015, 57, 348–357. [Google Scholar] [CrossRef]
- Kanchan, S.; Ogden, E.; Kesheri, M.; Skinner, A.; Miliken, E.; Lyman, D.; Armstrong, J.; Sciglitano, L.; Hampikian, G. COVID-19 hospitalizations and deaths predicted by SARS-CoV-2 levels in Boise, Idaho wastewater. Sci. Total Environ. 2024, 907, 167742. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Fu, S.; Zhang, Y.; Li, Y.; Zhang, Z.; Du, C.; Wang, R.; Peng, Y.; Yue, Z.; Xu, Z.; Hu, Q. Estimating epidemic trajectories of SARS-CoV-2 and influenza A virus based on wastewater monitoring and a novel machine learning algorithm. Sci. Total Environ. 2024, 951, 175830. [Google Scholar] [CrossRef] [PubMed]
- Bouchra, Z.; Mustapha, A.; Sbiti, N. Using Bayesian Networks for Risk Assessment in Healthcare System. In Bayesian Networks; Douglas, M., Ed.; IntechOpen: Rijeka, Croatia, 2019. [Google Scholar] [CrossRef]
- Zhang, L.M.; Xu, Y.; Jia, J.S.; Zhao, C. Diagnosis of embankment dam distresses using Bayesian networks. Part I. Global-level characteristics based on a dam distress database. Can. Geotech. J. 2011, 48, 1630–1644. [Google Scholar] [CrossRef]
- Ferguson, N.M.; Cummings, D.A.T.; Fraser, C.; Cajka, J.C.; Cooley, P.C.; Burke, D.S. Strategies for mitigating an influenza pandemic. Nature 2006, 442, 448–452. [Google Scholar] [CrossRef]
- Bivins, A.; North, D.; Ahmad, A.; Ahmed, W.; Alm, E.; Been, F.; Bhattacharya, P.; Bijlsma, L.; Boehm, A.B.; Brown, J.; et al. Wastewater-Based Epidemiology: Global Collaborative to Maximize Contributions in the Fight Against COVID-19. Environ. Sci. Technol. 2020, 54, 7754–7757. [Google Scholar] [CrossRef]
- Lin, T.; Karthikeyan, S.; Satterlund, A.; Schooley, R.; Knight, R.; De Gruttola, V.; Martin, N.; Zou, J. Optimizing campus-wide COVID-19 test notifications with interpretable wastewater time-series features using machine learning models. Sci. Rep. 2023, 13, 20670. [Google Scholar] [CrossRef]
- Schang, C.; Crosbie, N.D.; Nolan, M.; Poon, R.; Wang, M.; Jex, A.; John, N.; Baker, L.; Scales, P.; Schmidt, J.; et al. Passive Sampling of SARS-CoV-2 for Wastewater Surveillance. Environ. Sci. Technol. 2021, 55, 10432–10441. [Google Scholar] [CrossRef]
- Bogler, A.; Packman, A.; Furman, A.; Gross, A.; Kushmaro, A.; Ronen, A.; Dagot, C.; Hill, C.; Vaizel-Ohayon, D.; Morgenroth, E.; et al. Rethinking wastewater risks and monitoring in light of the COVID-19 pandemic. Nat. Sustain. 2020, 3, 981–990. [Google Scholar] [CrossRef]
- Shlyakhter, A.; Mirny, L.; Vlasov, A.; Wilson, R. Monte Carlo modeling of epidemiological studies. Hum. Ecol. Risk Assess. Int. J. 1996, 2, 920–938. [Google Scholar] [CrossRef]
- Ahmed, W.; Bertsch, P.M.; Bibby, K.; Haramoto, E.; Hewitt, J.; Huygens, F.; Gyawali, P.; Korajkic, A.; Riddell, S.; Sherchan, S.P.; et al. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020, 191, 110092. [Google Scholar] [CrossRef]
- Ahmed, W.; Bivins, A.; Bertsch, P.M.; Bibby, K.; Choi, P.M.; Gyawali, P.; Hamilton, K.A.; Haramoto, E.; Kitajima, M.; Simpson, S.L.; et al. Surveillance of SARS-CoV-2 RNA in wastewater: Methods optimization and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health 2020, 17, 82–93. [Google Scholar] [CrossRef]
- Kantor, R.S.; Nelson, K.L.; Greenwald, H.D.; Kennedy, L.C. Challenges in Measuring the Recovery of SARS-CoV-2 from Wastewater. Environ. Sci. Technol. 2021, 55, 3514–3519. [Google Scholar] [CrossRef] [PubMed]
- Cha, G.; Huang, Y.; Graham, K.E.; Luo, A.; Chen, W.; Hatt, J.K.; Konstantinidis, K.T.; Xie, X. Cold-chain free nucleic acid preservation using porous super-absorbent polymer (PSAP) beads to facilitate wastewater surveillance. Sci. Total Environ. 2024, 939, 173468. [Google Scholar] [CrossRef]
- Capone, D.; Berendes, D.; Cumming, O.; Knee, J.; Nala, R.; Risk, B.B.; Stauber, C.; Zhu, K.; Brown, J. Analysis of fecal sludges reveals common enteric pathogens in urban Maputo, Mozambique. Environ. Sci. Technol. Lett. 2020, 7, 889–895. [Google Scholar] [CrossRef]
- Schoen, M.E.; Bidwell, A.L.; Wolfe, M.K.; Boehm, A.B. United States Influenza 2022–2023 Season Characteristics as Inferred from Wastewater Solids, Influenza Hospitalization, and Syndromic Data. Environ. Sci. Technol. 2023, 57, 20542–20550. [Google Scholar] [CrossRef]
- Zhu, Y.; Oishi, W.; Maruo, C.; Saito, M.; Chen, R.; Kitajima, M.; Sano, D. Early warning of COVID-19 via wastewater-based epidemiology: Potential and bottlenecks. Sci. Total Environ. 2021, 767, 145124. [Google Scholar] [CrossRef]
- Lara-Jacobo, L.R.; Islam, G.; Desaulniers, J.P.; Kirkwood, A.E.; Simmons, D.B.D. Detection of SARS-CoV-2 Proteins in Wastewater Samples by Mass Spectrometry. Environ. Sci. Technol. 2022, 56, 5062–5070. [Google Scholar] [CrossRef]
- Yu, Q.; Olesen, S.W.; Duvallet, C.; Grad, Y.H. Assessment of sewer connectivity in the United States and its implications for equity in wastewater-based epidemiology. PLOS Glob. Public Health 2024, 4, e0003039. [Google Scholar] [CrossRef]
- Delgado Vela, J.; Philo, S.E.; Brown, J.; Taniuchi, M.; Cantrell, M.; Kossik, A.; Ramaswamy, M.; Ajjampur, S.S.R.; Guerfali, F.Z.; Holm, R.H.; et al. Moving beyond Wastewater: Perspectives on Environmental Surveillance of Infectious Diseases for Public Health Action in Low-Resource Settings. Environ. Health 2024, 2, 684–687. [Google Scholar] [CrossRef]
- Kapo, K.E.; Paschka, M.; Vamshi, R.; Sebasky, M.; McDonough, K. Estimation of U.S. sewer residence time distributions for national-scale risk assessment of down-the-drain chemicals. Sci. Total Environ. 2017, 603–604, 445–452. [Google Scholar] [CrossRef]
- Han, J.; He, S. Urban flooding events pose risks of virus spread during the novel coronavirus (COVID-19) pandemic. Sci. Total Environ. 2021, 755, 142491. [Google Scholar] [CrossRef]
- Boogaerts, T.; Van Wichelen, N.; Quireyns, M.; Burgard, D.; Bijlsma, L.; Delputte, P.; Gys, C.; Covaci, A.; van Nuijs, A.L.N. Current state and future perspectives on de facto population markers for normalization in wastewater-based epidemiology: A systematic literature review. Sci. Total Environ. 2024, 935, 173223. [Google Scholar] [CrossRef] [PubMed]
- McQuade, E.T.R.; Blake, I.M.; Brennhofer, S.A.; Islam, M.O.; Sony, S.S.S.; Rahman, T.; Bhuiyan, M.H.; Resha, S.K.; Wettstone, E.G.; Hughlett, L.; et al. Real-time sewage surveillance for SARS-CoV-2 in Dhaka, Bangladesh versus clinical COVID-19 surveillance: A longitudinal environmental surveillance study (December, 2019–December, 2021). Lancet Microbe 2023, 4, e442–e451. [Google Scholar] [CrossRef] [PubMed]
- Hoar, C.; McClary-Gutierrez, J.; Wolfe Marlene, K.; Bivins, A.; Bibby, K.; Silverman Andrea, I.; McLellan Sandra, L. Looking Forward: The Role of Academic Researchers in Building Sustainable Wastewater Surveillance Programs. Environ. Health Perspect. 2022, 130, 125002. [Google Scholar] [CrossRef]
- Daughton, C.G. Wastewater surveillance for population-wide COVID-19: The present and future. Sci. Total Environ. 2020, 736, 139631. [Google Scholar] [CrossRef]
- Pruden, A.; Vikesland, P.J.; Davis, B.C.; de Roda Husman, A.M. Seizing the moment: Now is the time for integrated global surveillance of antimicrobial resistance in wastewater environments. Curr. Opin. Microbiol. 2021, 64, 91–99. [Google Scholar] [CrossRef]
- Bayati, M.; Hsieh, H.Y.; Hsu, S.Y.; Qasim, S.; Li, C.; Belenchia, A.; Klutts, J.; Zemmer, S.A.; Sibley, K.; Reynolds, M.; et al. The different adsorption-degradation behaviors of SARS-CoV-2 by bioactive chemicals in wastewater: The suppression kinetics and their implications for wastewater-based epidemiology. Sci. Total Environ. 2024, 938, 173609. [Google Scholar] [CrossRef]
- Adams, C.; Bias, M.; Welsh, R.M.; Webb, J.; Reese, H.; Delgado, S.; Person, J.; West, R.; Shin, S.; Kirby, A. The National Wastewater Surveillance System (NWSS): From inception to widespread coverage, 2020–2022, United States. Sci. Total. Environ. 2024, 924, 171566. [Google Scholar] [CrossRef]
- Xu, X. Xiaoming Shi: Wastewater Monitoring Can Provide Early Warning for Infectious Disease Outbreaks, Building a COVID-19 Virus Monitoring Information System, with over 1300 Monitoring Points Expected by Next Year, Surpassing the United States. Available online: https://www.yicai.com/video/102264491.html (accessed on 18 May 2025).
- Luo, X.; Han, S.; Wang, Y.; Du, P.; Li, X.; Thai, P.K. Significant differences in usage of antibiotics in three Chinese cities measured by wastewater-based epidemiology. Water Res. 2024, 254, 121335. [Google Scholar] [CrossRef]
- Tao, L.; Jiong, W.; Siwen, Y.; Zhiqing, C.; Qijiong, Z.; Shangfeng, Y.; Wenjun, M.; Xiaofeng, L. Current status of public health system in Guangdong-Hong Kong-Macao Greater Bay Area and improvement suggestion. Chin. J. Epidemiol. 2023, 44, 694–698. [Google Scholar]
- Hrudey, S.E.; Silva, D.S.; Shelley, J.; Pons, W.; Isaac-Renton, J.; Chik, A.H.; Conant, B. Ethics Guidance for Environmental Scientists Engaged in Surveillance of Wastewater for SARS-CoV-2. Environ. Sci. Technol. 2021, 55, 8484–8491. [Google Scholar] [CrossRef]
- Nainani, D.; Ng, W.J.; Wuertz, S.; Thompson, J.R. Balancing public health and group privacy: Ethics, rights, and obligations for wastewater surveillance systems. Water Res. 2024, 258, 121756. [Google Scholar] [CrossRef] [PubMed]
- Ram, N.; Shuster, W.; Gable, L.; Ram, J.L. Ethical and legal wastewater surveillance. Science 2023, 379, 652. [Google Scholar] [CrossRef] [PubMed]
- Scassa, T.; Robinson, P.; Mosoff, R. The Datafication of Wastewater: Legal, Ethical and Civic Considerations. Technol. Regul. 2022, 2022, 23–35. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, S.; Chen, Y.; Zhao, J.; Prosun, T.; O’Brien, J.W.; Mueller, J.F.; Tscharke, B.J.; Coin, L.J.M.; Luby, S.P.; et al. Associations between Wastewater Microbiome and Population Smoking Rate Identified Using Wastewater-Based Epidemiology. Environ. Health 2023, 1, 394–404. [Google Scholar] [CrossRef]
- Boehm, A.B.; Wolfe, M.K.; White, B.J.; Hughes, B.; Duong, D.; Bidwell, A. More than a Tripledemic: Influenza A Virus, Respiratory Syncytial Virus, SARS-CoV-2, and Human Metapneumovirus in Wastewater during Winter 2022–2023. Environ. Sci. Technol. Lett. 2023, 10, 622–627. [Google Scholar] [CrossRef]
- Krotofil, J.; Pruc, M.; Swieczkowski, D.; Solowiej, K.; Szarpak, L. Evaluating the risk: Group A Streptococcus as a causative agent of streptococcal toxic shock syndrome and necrotizing fasciitis. Disaster Emerg. Med. J. 2024, 9, 133–135. [Google Scholar] [CrossRef]
- Prieto Riquelme, M.V.; Garner, E.; Gupta, S.; Metch, J.; Zhu, N.; Blair, M.F.; Arango-Argoty, G.; Maile-Moskowitz, A.; Li, A.D.; Flach, C.F.; et al. Demonstrating a Comprehensive Wastewater-Based Surveillance Approach That Differentiates Globally Sourced Resistomes. Environ. Sci. Technol. 2022, 56, 14982–14993. [Google Scholar] [CrossRef]
- Wang, C.; Mantilla-Calderon, D.; Xiong, Y.; Alkahtani, M.; Bashawri, Y.M.; Al Qarni, H.; Hong, P.Y. Investigation of Antibiotic Resistome in Hospital Wastewater during the COVID-19 Pandemic: Is the Initial Phase of the Pandemic Contributing to Antimicrobial Resistance? Environ. Sci. Technol. 2022, 56, 15007–15018. [Google Scholar] [CrossRef]
- Kurasawa, K.; Yoshida, M.; Nakao, M.; Muranaka, E.; Hachisu, Y.; Kishizawa, M.; Kikuchi, T.; Hase, R. A large increase in Group A streptococcus bacteremia in the 2 month short period in 2024; report from a tertiary care hospital in Chiba, Japan. J. Gen. Fam. Med. 2024, 25, 289. [Google Scholar] [CrossRef]
- Scaccia, N.; da Silva Fonseca, J.V.; Megueya, A.L.; de Aragao, G.L.; Rasolofoarison, T.; de Paula, A.V.; de Vinci Kanda Kupa, L.; Tchatchueng, J.; Makuetche, K.; Rasolojaona, T.Z.; et al. Analysis of chlorhexidine, antibiotics and bacterial community composition in water environments from Brazil, Cameroon and Madagascar during the COVID-19 pandemic. Sci. Total Environ. 2024, 932, 173016. [Google Scholar] [CrossRef]
- Wang, Z.; Cai, M.; Du, P.; Li, X. Wastewater surveillance for antibiotics and resistance genes in a river catchment: Spatiotemporal variations and the main drivers. Water Res. 2024, 251, 121090. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Gao, R.; Liang, W.; Zhou, Y.; Wang, Z.; Lan, L.; Chen, J.; Zeng, F. Feasibility of sulfated BPA and BPS as wastewater-based epidemiology biomarkers: Insights from wastewater and reported human urine analysis. Sci. Total Environ. 2024, 927, 171870. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Ceolotto, N.; Jagadeesan, K.; Standerwick, R.; Robertson, M.; Barden, R.; Kasprzyk-Hordern, B. Antimicrobials and antimicrobial resistance genes in the shadow of COVID-19 pandemic: A wastewater-based epidemiology perspective. Water Res. 2024, 257, 121665. [Google Scholar] [CrossRef]
| Method | qPCR | Amplicon Sequencing | Metagenomic Sequencing | Biosensors |
|---|---|---|---|---|
| Principle | External amplification of DNA | DNA/RNA sequencing | DNA/RNA sequencing | Specific receptors emit a clear response signal |
| Target | Specific region of viral genome | Specific region of viral genome | Whole viral genome | Specific DNA/RNA/protein |
| Time | Short | Long | Long | Short |
| Precision | High | High | High | Amplification-based: low |
| Cost | Low | Low | High | Low |
| VOC discoverable | No | Only when mutation sites are in the amplified region | Yes | No |
| Method | Applicable Conditions | Advantages | Limitations |
|---|---|---|---|
| SMA | Smoothing short-term date | Simple, stable, and easy to implement | Slow response to emergencies |
| LOESS | Long-term and nonlinear data | No need to set parameters in advance | High computational complexity |
| ANNs | Large amounts of data and time-consuming process | Extract data features through deep structures | Time-consuming and poor interpretability of the results |
| Random Forest | High-dimensional data with missing data or nonlinear relationships | Prevent overfitting | Relatively inefficient, resource-intensive, and difficult to interpret |
| Bayesian Model | Sample data are insufficient in low-dimensional spaces | Quantify the credibility of each possible outcome | Relies on the choice of priors and assumptions |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, W.; Wang, D.; Li, P.; Deng, H.; Deng, Z. Advances in Wastewater-Based Epidemiology for Pandemic Surveillance: Methodological Frameworks and Future Perspectives. Microorganisms 2025, 13, 1169. https://doi.org/10.3390/microorganisms13051169
Zhu W, Wang D, Li P, Deng H, Deng Z. Advances in Wastewater-Based Epidemiology for Pandemic Surveillance: Methodological Frameworks and Future Perspectives. Microorganisms. 2025; 13(5):1169. https://doi.org/10.3390/microorganisms13051169
Chicago/Turabian StyleZhu, Weihe, Daxi Wang, Pengsong Li, Haohao Deng, and Ziqing Deng. 2025. "Advances in Wastewater-Based Epidemiology for Pandemic Surveillance: Methodological Frameworks and Future Perspectives" Microorganisms 13, no. 5: 1169. https://doi.org/10.3390/microorganisms13051169
APA StyleZhu, W., Wang, D., Li, P., Deng, H., & Deng, Z. (2025). Advances in Wastewater-Based Epidemiology for Pandemic Surveillance: Methodological Frameworks and Future Perspectives. Microorganisms, 13(5), 1169. https://doi.org/10.3390/microorganisms13051169








