Exploring the Impact of Altitude on Bacterial Communities in Informally Produced Artisanal Colonial Cheeses: Insights from 16S rRNA Gene Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Characterization of the Region and Collection of Cheese Samples
2.2. DNA Extraction and Sequencing
2.3. Read Processing and Statistical Analyses
3. Results and Discussion
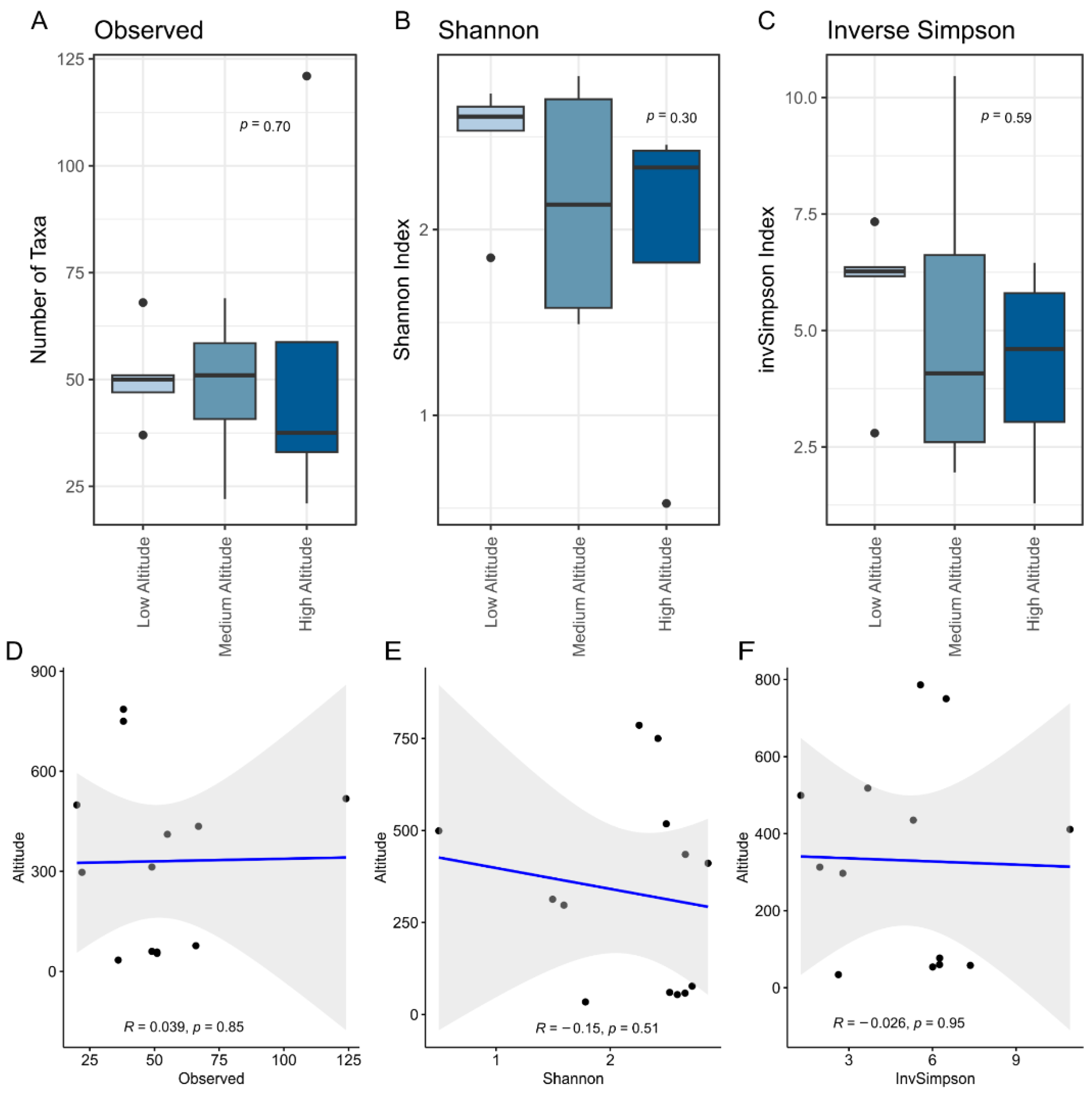
3.1. Eight Hundred Meters Are Not Enough to Change the Microbial Diversity in Artisanal Colonial Cheeses
3.2. Bacterial Taxa Are Resilient to Altitudinal Variation in Artisanal Colonial Cheeses
3.3. Physicochemical Variables Are Stable in Artisanal Colonial Cheeses Across Altitudes
3.4. Small Differences Regarding Altitude Can Be Found Only at Deep Taxonomic Levels
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- MilkPoint. Queijo Artesanal cai no Gosto do Consumidor e já Representa 20% do Mercado Nacional. Available online: https://www.milkpoint.com.br/artigos/industria-de-laticinios/queijo-artesanal-cai-no-gosto-do-consumidor-237255/ (accessed on 12 March 2025).
- Kamimura, B.A.; Magnani, M.; Luciano, W.A.; Campagnollo, F.B.; Pimentel, T.C.; Alvarenga, V.O.; Pelegrino, B.O.; Cruz, A.G.; Sant’Ana, A.S. Brazilian Artisanal Cheeses: An Overview of Their Characteristics, Main Types and Regulatory Aspects. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1636–1657. [Google Scholar] [CrossRef] [PubMed]
- Cabral, M.L.B.; de Lima, M.d.S.F.; de Araújo Fernandes, G.A.; da Costa, E.F.; Porto, A.L.F.; Cavalcanti , M.T.H. Artisan cheese: A potential source of wild lactic acid bacteria to obtain new starter cultures. J. Bioenergy Food Sci. 2016, 3, 207–215. [Google Scholar] [CrossRef]
- Margalho, L.P.; Feliciano, M.D.; Silva, C.E.; Abreu, J.S.; Piran, M.V.F.; Sant’Ana, A.S. Brazilian Artisanal Cheeses Are Rich and Diverse Sources of Nonstarter Lactic Acid Bacteria Regarding Technological, Biopreservative, and Safety Properties—Insights through Multivariate Analysis. J. Dairy Sci. 2020, 103, 7908–7926. [Google Scholar] [CrossRef] [PubMed]
- Kothe, C.I.; Mohellibi, N.; Renault, P. Revealing the Microbial Heritage of Traditional Brazilian Cheeses through Metagenomics. Food Res. Int. 2022, 157, 111265. [Google Scholar] [CrossRef] [PubMed]
- Ambrosini, L.B.; Bresolin, B.; Kroeff, D.R.; Waquil, P.D. História do Queijo Colonial do Rio Grande do Sul: Das cozinhas para o mercado. História Questões Debates 2024, 72, 270. [Google Scholar]
- Brasil Portaria MAPA n° 146. Available online: https://www.gov.br/agricultura/pt-br/assuntos/defesa-agropecuaria/suasa/regulamentos-tecnicos-de-identidade-e-qualidade-de-produtos-de-origem-animal-1/rtiq-leite-e-seus-derivados (accessed on 11 March 2025).
- Brasil Decreto N° 9.013. Available online: https://www.planalto.gov.br/ccivil_03/_ato2015-2018/2017/decreto/d9013.htm (accessed on 11 March 2025).
- De Medeiros Carvalho, M.; De Fariña, L.O.; Strongin, D.; Ferreira, C.L.L.F.; Lindner, J.D.D. Traditional Colonial-Type Cheese from the South of Brazil: A Case to Support the New Brazilian Laws for Artisanal Cheese Production from Raw Milk. J. Dairy Sci. 2019, 102, 9711–9720. [Google Scholar] [CrossRef]
- Erhardt, M.M.; Fröder, H.; Oliveira, W.d.C.; Ströher, J.A.; Savergnini, P.R.; dos Santos, V.Z.; dos Santos Richards, N.S. Physicochemical and microbiological evaluation of artisan cheeses from raw milk and verification of good practices in rural properties in the Taquari Valley-RS. Res. Soc. Dev. 2022, 11, e253111335290. [Google Scholar] [CrossRef]
- Lai, G.; Melillo, R.; Pes, M.; Addis, M.; Fadda, A.; Pirisi, A. Survival of Selected Pathogenic Bacteria during PDO Pecorino Romano Cheese Ripening. Dairy 2020, 1, 297–312. [Google Scholar] [CrossRef]
- Erhardt, M.M.; Oliveira, W.d.C.; Fröder, H.; Marques, P.H.; Oliveira, M.B.P.P.; Richards, N.S.P.d.S. Lactic Bacteria in Artisanal Cheese: Characterization through Metagenomics. Fermentation 2023, 9, 41. [Google Scholar] [CrossRef]
- de Castro Oliveira, W.; Marques, P.H.; Erhardt, M.M.; Felice, A.G.; Tristão, C.L.A.M.; Aburjaile, F.F.; Oliveira, M.B.P.P.; dos Santos Richards, N.S.P. Metagenomic Analysis and Proteins Prediction of Emerging Pathogens in Artisanal Cheese. Mol. Divers. 2025, 2025, 1–19. [Google Scholar] [CrossRef]
- de Albuquerque, T.M.N.C.; Campos, G.Z.; d’Ovidio, L.; Pinto, U.M.; Sobral, P.J.d.A.; Galvão, J.A. Unveiling Safety Concerns in Brazilian Artisanal Cheeses: A Call for Enhanced Ripening Protocols and Microbiological Assessments. Foods 2024, 13, 1644. [Google Scholar] [CrossRef] [PubMed]
- Rio Grande do Sul Instrução Normativa n° 002/2023. Aprova o Regulamento Técnico de Identidade e Qualidade do Queijo Colonial Artesanal, conforme o Anexo desta Instrução Normativa. Available online: https://www.diariooficial.rs.gov.br/materia?id=837948 (accessed on 11 March 2025).
- Hendy, J.; Rest, M.; Aldenderfer, M.; Warinner, C. Cultures of Fermentation: Living with Microbes: An Introduction to Supplement 24. Curr. Anthropol. 2021, 62, S197–S206. [Google Scholar] [CrossRef]
- Gobbetti, M.; Di Cagno, R.; Calasso, M.; Neviani, E.; Fox, P.F.; De Angelis, M. Drivers That Establish and Assembly the Lactic Acid Bacteria Biota in Cheeses. Trends Food Sci. Technol. 2018, 78, 244–254. [Google Scholar] [CrossRef]
- McClure, S.B.; Magill, C.; Podrug, E.; Moore, A.M.T.; Harper, T.K.; Culleton, B.J.; Kennett, D.J.; Freeman, K.H. Fatty Acid Specific δ13C Values Reveal Earliest Mediterranean Cheese Production 7,200 Years Ago. PLoS ONE 2018, 13, e0202807. [Google Scholar] [CrossRef]
- Fox, P.F.; McSweeney, P.L.H.; Cogan, T.M.; Guinee, T.P. Cheese: Chemistry, Physics and Microbiology, Volume 1: General Aspects; Elsevier: Amsterdam, The Netherlands, 2004; ISBN 978-0-08-050093-5. [Google Scholar]
- Randazzo, C.L.; Caggia, C.; Neviani, E. Cheese Ripening: Quality, Safety and Health Aspects; Nova Science Publishers Inc.: Hauppauge, NY, USA, 2013; ISBN 978-1-62417-032-4. [Google Scholar]
- Neviani, E.; Gatti, M.; Gardini, F.; Levante, A. Microbiota of Cheese Ecosystems: A Perspective on Cheesemaking. Foods 2025, 14, 830. [Google Scholar] [CrossRef]
- Melkonian, C.; Zorrilla, F.; Kjærbølling, I.; Blasche, S.; Machado, D.; Junge, M.; Sørensen, K.I.; Andersen, L.T.; Patil, K.R.; Zeidan, A.A. Microbial Interactions Shape Cheese Flavour Formation. Nat. Commun. 2023, 14, 8348. [Google Scholar] [CrossRef]
- Serrano, S.; Morais, S.; Semedo-Lemsaddek, T. Tradition Unveiled: A Comprehensive Review of Microbiological Studies on Portuguese Traditional Cheeses, Merging Conventional and OMICs Analyses. Front. Ind. Microbiol. 2024, 2, 1420042. [Google Scholar] [CrossRef]
- Ströher, J.A.; Sant’Anna, V.; Oliveira, W.d.C.; Padilha, R.L. The Effect of Temperature on Physicochemical and Microbiological Aspects of Serrano Artisanal Cheese Ripening. Braz. Arch. Biol. Technol. 2023, 66, e23220530. [Google Scholar] [CrossRef]
- Korena, K.; Krzyzankova, M.; Florianova, M.; Karasova, D.; Babak, V.; Strakova, N.; Juricova, H. Microbial Succession in the Cheese Ripening Process—Competition of the Starter Cultures and the Microbiota of the Cheese Plant Environment. Microorganisms 2023, 11, 1735. [Google Scholar] [CrossRef]
- Correddu, F.; Murgia, M.A.; Mangia, N.P.; Lunesu, M.F.; Cesarani, A.; Deiana, P.; Pulina, G.; Nudda, A. Effect of Altitude of Flock Location, Season of Milk Production and Ripening Time on the Fatty Acid Profile of Pecorino Sardo Cheese. Int. Dairy J. 2021, 113, 104895. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, W.; Cao, Z.; Yang, H.; Wang, Y.; Li, S. Effects of Altitude on the Gut Microbiome and Metabolomics of Sanhe Heifers. Front. Microbiol. 2023, 14, 1076011. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; He, C.; Xu, Y.; Zhang, C.; Li, C.; Jing, X.; Wang, M.; Yang, Y.; Suo, L.; Kalds, P.; et al. Taxonomic and Functional Adaption of the Gastrointestinal Microbiome of Goats Kept at High Altitude (4800 m) under Intensive or Extensive Rearing Conditions. FEMS Microbiol. Ecol. 2021, 97, fiab009. [Google Scholar] [CrossRef] [PubMed]
- Rossi, A.; Marroni, F.; Renoldi, N.; Di Filippo, G.; Gover, E.; Marino, M.; Innocente, N. An Integrated Approach to Explore the Microbial Biodiversity of Natural Milk Cultures for Cheesemaking. J. Dairy Sci. 2024, 107, 4288–4297. [Google Scholar] [CrossRef]
- Wang, Y.; Qian, P.-Y. Conservative Fragments in Bacterial 16S rRNA Genes and Primer Design for 16S Ribosomal DNA Amplicons in Metagenomic Studies. PLoS ONE 2009, 4, e7401. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-High-Throughput Microbial Community Analysis on the Illumina HiSeq and MiSeq Platforms. ISME J. 2012, 2012, 1621–1624. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Development Core Team: Vienna, Austria, 2022. [Google Scholar]
- Wickham, H. Ggplot2. WIREs Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Dinno, A. Nonparametric Pairwise Multiple Comparisons in Independent Groups Using Dunn’s Test. Stata J. 2015, 15, 292–300. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Liu, C.; Cui, Y.; Li, X.; Yao, M. Microeco: An R Package for Data Mining in Microbial Community Ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef] [PubMed]
- Puth, M.-T.; Neuhäuser, M.; Ruxton, G.D. Effective Use of Spearman’s and Kendall’s Correlation Coefficients for Association between Two Measured Traits. Anim. Behav. 2015, 102, 77–84. [Google Scholar] [CrossRef]
- Gloor, G.B.; Macklaim, J.M.; Pawlowsky-Glahn, V.; Egozcue, J.J. Microbiome Datasets Are Compositional: And This Is Not Optional. Front. Microbiol. 2017, 8, 2224. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package; R Package Vegan, Vers. 2.2-1; World Agroforestry Centre: Nairobi, Kenya, 2015. [Google Scholar]
- Fernandes, A.D.; Macklaim, J.M.; Linn, T.G.; Reid, G.; Gloor, G.B. ANOVA-Like Differential Expression (ALDEx) Analysis for Mixed Population RNA-Seq. PLoS ONE 2013, 8, e67019. [Google Scholar] [CrossRef]
- Ercolini, D. Secrets of the Cheese Microbiome. Nat. Food 2020, 1, 466–467. [Google Scholar] [CrossRef]
- De Pasquale, I.; Calasso, M.; Mancini, L.; Ercolini, D.; La Storia, A.; De Angelis, M.; Di Cagno, R.; Gobbetti, M. Causal Relationship between Microbial Ecology Dynamics and Proteolysis during Manufacture and Ripening of Protected Designation of Origin (PDO) Cheese Canestrato Pugliese. Appl. Environ. Microbiol. 2014, 80, 4085–4094. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Gesang, Q.; Luo, J.; Wu, X.; Rebi, A.; You, Y.; Zhou, J. Drivers of Plant Diversification along an Altitudinal Gradient in the Alpine Desert Grassland, Northern Tibetan Plateau. Glob. Ecol. Conserv. 2024, 53, e02987. [Google Scholar] [CrossRef]
- De Filippis, F.; Genovese, A.; Ferranti, P.; Gilbert, J.A.; Ercolini, D. Metatranscriptomics Reveals Temperature-Driven Functional Changes in Microbiome Impacting Cheese Maturation Rate. Sci. Rep. 2016, 6, 21871. [Google Scholar] [CrossRef]
- Ströher, J.A.; Oliveira, W.d.C.; de Freitas, A.S.; Salazar, M.M.; Flôres, S.H.; Malheiros, P.d.S. Microbial Dynamics and Volatile Compound Profiles in Artisanal Kefir During Storage. Fermentation 2025, 11, 105. [Google Scholar] [CrossRef]
- Ströher, J.A.; Silva, D.M.C.; NUNES, I.D.S. Qualidade Microbiológica Do Queijo Artesanal Colonial (QAC): Impacto Das Boas Práticas de Fabricação (BPF)-Uma Revisão Sistemática. Nutr. Rev. Nutr. E Vigilância Em Saúde 2023, 10, e11943. [Google Scholar] [CrossRef]
- Bava, L.; Zucali, M.; Tamburini, A.; Morandi, S.; Brasca, M. Effect of Different Farming Practices on Lactic Acid Bacteria Content in Cow Milk. Animals 2021, 11, 522. [Google Scholar] [CrossRef] [PubMed]
- Leclercq-Perlat, M.-N.; Hélias, A.; Corrieu, G. Short Communication: Little Change Takes Place in Camembert-Type Cheese Water Activities throughout Ripening in Terms of Relative Humidity and Salt. J. Dairy Sci. 2013, 96, 7521–7525. [Google Scholar] [CrossRef] [PubMed]
- Erhardt, M.M.; Fröder, H.; de Castro Oliveira, W.; Ströher, J.A.; Savergnini, P.R.; dos Santos, V.Z.; dos Santos Richards, N.S.P. Avaliação Físico-Química e Microbiológica de Queijos Artesanais a Partir de Leite Cru e Verificação de Boas Práticas Em Propriedades Rurais No Vale Do Taquari-RS. Res. Soc. Dev. 2022, 11, e253111335290. [Google Scholar] [CrossRef]
- Benincá, T.; Santos, V.Z.d.; Sant’Anna, V.; Berreta, M.d.S.R. Correlação entre dados microbiológicos e físicoquímicos com as boas práticas de fabricação de queijos coloniais produzidos no Sul do Brasil. Cad. Ciência Tecnol. 2022, 39, 27176. [Google Scholar] [CrossRef]
- Neves, L.F.; Fonseca, H.C.; Oliveira, M.L.P.; de Souza, C.N.; Durães, G.L.L.S.; Duarte, E.R.; de Souza, M.R. Perfil físico-químico de queijos artesanais do Norte de Minas Gerais. Phys.-Chem. Profile Artis. Cheeses North Minas Gerais 2021, 23, 1–10. [Google Scholar] [CrossRef]
- Jung, M.Y.; Lee, C.; Seo, M.-J.; Roh, S.W.; Lee, S.H. Characterization of a Potential Probiotic Bacterium Lactococcus Raffinolactis WiKim0068 Isolated from Fermented Vegetable Using Genomic and in Vitro Analyses. BMC Microbiol. 2020, 20, 136. [Google Scholar] [CrossRef]
- Yoon, J.-H.; Lee, C.-H.; Kang, S.-J.; Oh, T.-K. Psychrobacter celer sp. Nov., Isolated from Sea Water of the South Sea in Korea. Int. J. Syst. Evol. Microbiol. 2005, 55, 1885–1890. [Google Scholar] [CrossRef]
- Huang, F.; Zhao, Y.; Hou, Y.; Yang, Y.; Yue, B.; Zhang, X. Unraveling the Antimicrobial Potential of Lactiplantibacillus Plantarum Strains TE0907 and TE1809 Sourced from Bufo Gargarizans: Advancing the Frontier of Probiotic-Based Therapeutics. Front. Microbiol. 2024, 15, 1347830. [Google Scholar] [CrossRef]
- Afshari, R.; Pillidge, C.J.; Dias, D.A.; Osborn, A.M.; Gill, H. Cheesomics: The Future Pathway to Understanding Cheese Flavour and Quality. Crit. Rev. Food Sci. Nutr. 2020, 60, 33–47. [Google Scholar] [CrossRef]
- Nugroho, A.D.W.; Kleerebezem, M.; Bachmann, H. Growth, Dormancy and Lysis: The Complex Relation of Starter Culture Physiology and Cheese Flavour Formation. Curr. Opin. Food Sci. 2021, 39, 22–30. [Google Scholar] [CrossRef]
- Qiao, H.; Chen, L.; Yang, J.; Zhi, W.; Chen, R.; Lu, T.; Tan, H.; Sheng, Z. Effect of Lactic Acid Bacteria on Bacterial Community Structure and Characteristics of Sugarcane Juice. Foods 2022, 11, 3134. [Google Scholar] [CrossRef] [PubMed]
- Nam, J.H.; Cho, Y.S.; Rackerby, B.; Goddik, L.; Park, S.H. Shifts of Microbiota during Cheese Production: Impact on Production and Quality. Appl. Microbiol. Biotechnol. 2021, 105, 2307–2318. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, L.; Claret, A.; Verbeke, W.; Sulmont-Rossé, C.; Hersleth, M. Chapter 6—Innovation in Traditional Food Products: Does It Make Sense? In Innovation Strategies in the Food Industry, 2nd ed.; Galanakis, C.M., Ed.; Academic Press: Cambridge, MA, USA, 2022; pp. 87–95. ISBN 978-0-323-85203-6. [Google Scholar]
- Azevedo, G.D.; de Castro Oliveira, W.; Ströher, J.A.; Oliveira, M.B.P.P.; dos Santos Richards, N.S.P.; Sant’Anna, V. Exploring the Marination of Colonial Cheese into Wine and Vinegar through Ripening: Physicochemical Characteristics, Sensorial Attributes and Emotions Evoked. Int. J. Gastron. Food Sci. 2024, 38, 101066. [Google Scholar] [CrossRef]
- André, J.a.M.; Balbinot, L.; Casaril, K.B.P.B. Selection of lactic acid bacteria with probiotic potential from colonial cheeses. Arq. Bras. Med. Vet. Zootec. 2025, 77. [Google Scholar] [CrossRef]
- Myazaki, N.L.; Cardoso, M.F.; Verruck, S. Metataxonomic Analysis of Bacterial and Fungal Communities in Colonial Cheese. Food Sci. Technol. 2024, 44. [Google Scholar] [CrossRef]



| Sample | Altitude (m) | Classification | Raw Milk | Cheese |
|---|---|---|---|---|
| LJ | 34 | Low | Yes | Colonial type |
| AM | 54 | Low | Yes | Colonial type |
| EN | 58 | Low | Yes | Colonial type |
| MU | 77 | Low | Yes | Colonial type |
| RC | 60 | Low | Yes | Colonial type |
| RV | 297 | Medium | Yes | Colonial type |
| NB | 313 | Medium | Yes | Colonial type |
| AG | 411 | Medium | Yes | Colonial type |
| PT | 435 | Medium | Yes | Colonial type |
| DR | 499 | High | Yes | Colonial type |
| VC | 518 | High | Yes | Colonial type |
| AV | 750 | High | Yes | Colonial type |
| IL | 786 | High | Yes | Colonial type |
| Taxonomic Level | Taxa | Rab a Low Altitude | Rab a High Altitude | Overlap b | Effect c | p-Value d |
|---|---|---|---|---|---|---|
| Phylum | Actinobacteriota | 1.80 | −2.75 | 0.11 | 1.01 | 0.04 |
| Class | - | - | - | - | - | - |
| Order | - | - | - | - | - | - |
| Family | - | - | - | - | - | - |
| Genus | Lactiplantibacillus | −3.56 | 3.98 | 0.07 | −1.59 | 0.06 |
| Species | Psychrobacter celer | 4.47 | −4.59 | 0.10 | 1.14 | 0.06 |
| Lactococcus raffinolactis | 8.81 | −4.36 | 0.14 | 1.09 | 0.06 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveira, W.d.C.; de Freitas, A.S.; Ströher, J.A.; Richards, N.S.P.d.S.; Oliveira, M.B.P.P.; Erhardt, M.M. Exploring the Impact of Altitude on Bacterial Communities in Informally Produced Artisanal Colonial Cheeses: Insights from 16S rRNA Gene Sequencing. Microorganisms 2025, 13, 1116. https://doi.org/10.3390/microorganisms13051116
Oliveira WdC, de Freitas AS, Ströher JA, Richards NSPdS, Oliveira MBPP, Erhardt MM. Exploring the Impact of Altitude on Bacterial Communities in Informally Produced Artisanal Colonial Cheeses: Insights from 16S rRNA Gene Sequencing. Microorganisms. 2025; 13(5):1116. https://doi.org/10.3390/microorganisms13051116
Chicago/Turabian StyleOliveira, Wemerson de Castro, Anderson Santos de Freitas, Jeferson Aloísio Ströher, Neila Silvia Pereira dos Santos Richards, Maria Beatriz Prior Pinto Oliveira, and Magnolia Martins Erhardt. 2025. "Exploring the Impact of Altitude on Bacterial Communities in Informally Produced Artisanal Colonial Cheeses: Insights from 16S rRNA Gene Sequencing" Microorganisms 13, no. 5: 1116. https://doi.org/10.3390/microorganisms13051116
APA StyleOliveira, W. d. C., de Freitas, A. S., Ströher, J. A., Richards, N. S. P. d. S., Oliveira, M. B. P. P., & Erhardt, M. M. (2025). Exploring the Impact of Altitude on Bacterial Communities in Informally Produced Artisanal Colonial Cheeses: Insights from 16S rRNA Gene Sequencing. Microorganisms, 13(5), 1116. https://doi.org/10.3390/microorganisms13051116








