Non-Secretor Status Due to FUT2 Stop Mutation Is Associated with Reduced Rotavirus Infections but Not with Other Enteric Pathogens in Rwandan Children
Abstract
1. Introduction
2. Materials and Methods
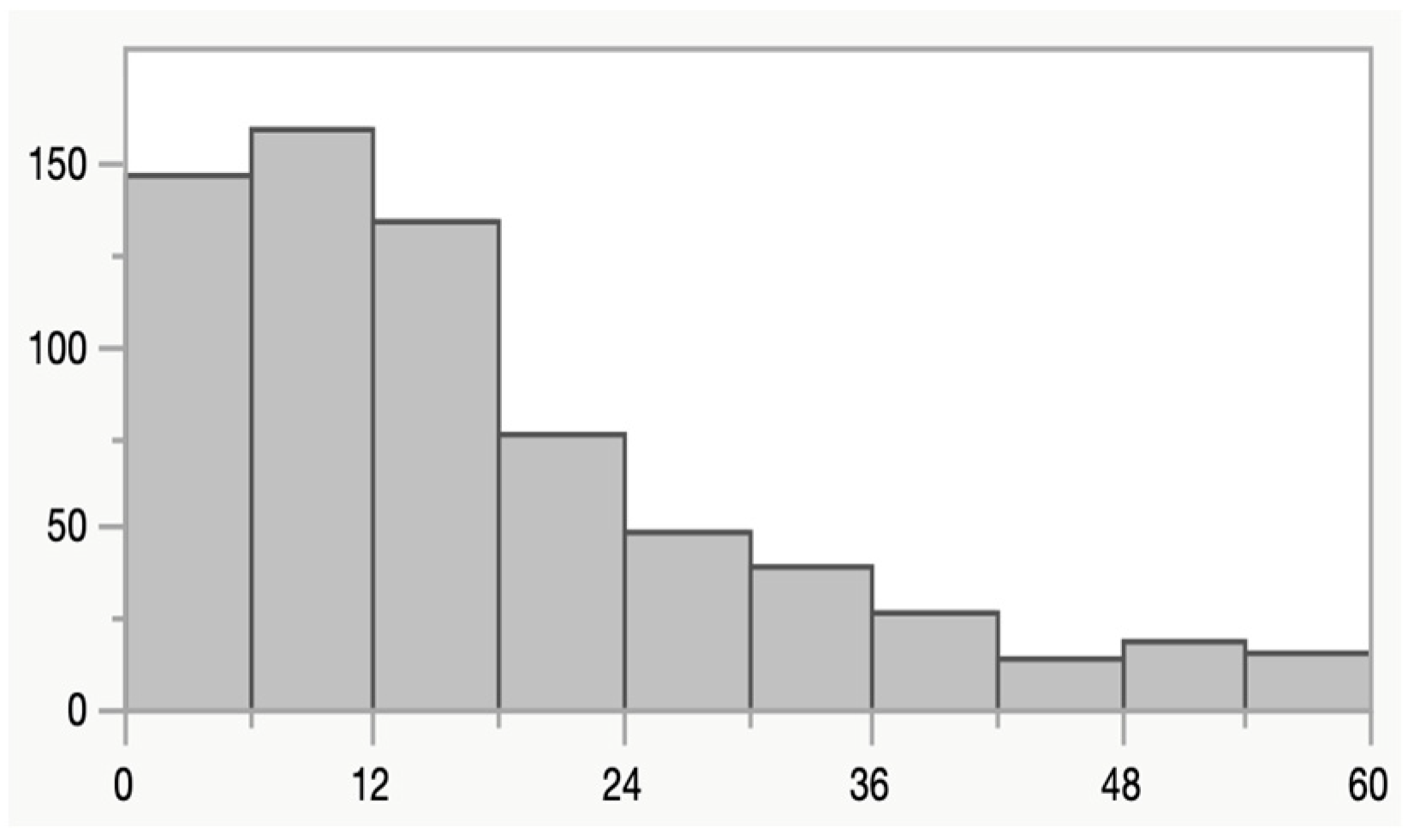
2.1. Study Design, Study Area and Characteristics of Participants
2.2. PCR for Pathogen Detection
2.3. FUT2 SNP Genotyping
2.4. Data Analysis
2.5. Ethical Approval
3. Results
3.1. Pathogen Association with Diarrhea and FUT2 Genotype
3.2. Rotavirus P Types and Secretor Status
3.3. Norovirus GII and Secretor Status
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kotloff, K.L. The Burden and Etiology of Diarrheal Illness in Developing Countries. Pediatr. Clin. N. Am. 2017, 64, 799–814. [Google Scholar] [CrossRef] [PubMed]
- Vila-Guilera, J.; Parikh, P.; Chaturvedi, H.; Ciric, L.; Lakhanpaul, M. Towards transformative WASH: An integrated case study exploring environmental, sociocultural, economic and institutional risk factors contributing to infant enteric infections in rural tribal India. BMC Public Health 2021, 21, 1331. [Google Scholar] [CrossRef]
- Baker, K.K.; Mumma, J.A.O.; Simiyu, S.; Sewell, D.; Tsai, K.; Anderson, J.D.; MacDougall, A.; Dreibelbis, R.; Cumming, O. Environmental and behavioural exposure pathways associated with diarrhoea and enteric pathogen detection in 5-month-old, periurban Kenyan infants: A cross-sectional study. BMJ Open 2022, 12, e059878. [Google Scholar] [CrossRef] [PubMed]
- Kwok, A.J.; Mentzer, A.; Knight, J.C. Host genetics and infectious disease: New tools, insights and translational opportunities. Nat. Rev. Genet. 2021, 22, 137–153. [Google Scholar] [CrossRef]
- Hu, M.; Zhang, X.; Li, J.; Chen, L.; He, X.; Sui, T. Fucosyltransferase 2: A Genetic Risk Factor for Intestinal Diseases. Front. Microbiol. 2022, 13, 940196. [Google Scholar] [CrossRef] [PubMed]
- Nordgren, J.; Svensson, L. Genetic Susceptibility to Human Norovirus Infection: An Update. Viruses 2019, 11, 226. [Google Scholar] [CrossRef]
- Sharma, S.; Hagbom, M.; Svensson, L.; Nordgren, J. The Impact of Human Genetic Polymorphisms on Rotavirus Susceptibility, Epidemiology, and Vaccine Take. Viruses 2020, 12, 324. [Google Scholar] [CrossRef]
- Lopman, B.A.; Trivedi, T.; Vicuna, Y.; Costantini, V.; Collins, N.; Gregoricus, N.; Parashar, U.; Sandoval, C.; Broncano, N.; Vaca, M.; et al. Norovirus Infection and Disease in an Ecuadorian Birth Cohort: Association of Certain Norovirus Genotypes With Host FUT2 Secretor Status. J. Infect. Dis. 2015, 211, 1813–1821. [Google Scholar] [CrossRef]
- MacDonald, J.; Groome, M.J.; Mans, J.; Page, N. FUT2 Secretor Status Influences Susceptibility to VP4 Strain-Specific Rotavirus Infections in South African Children. Pathogens 2020, 9, 795. [Google Scholar] [CrossRef]
- Nordgren, J.; Sharma, S.; Bucardo, F.; Nasir, W.; Gunaydin, G.; Ouermi, D.; Nitiema, L.W.; Becker-Dreps, S.; Simpore, J.; Hammarstrom, L.; et al. Both Lewis and secretor status mediate susceptibility to rotavirus infections in a rotavirus genotype-dependent manner. Clin. Infect. Dis. 2014, 59, 1567–1573. [Google Scholar] [CrossRef]
- Bucardo, F.; Kindberg, E.; Paniagua, M.; Grahn, A.; Larson, G.; Vildevall, M.; Svensson, L. Genetic susceptibility to symptomatic norovirus infection in Nicaragua. J. Med. Virol. 2009, 81, 728–735. [Google Scholar] [CrossRef]
- Pena-Gil, N.; Santiso-Bellon, C.; Gozalbo-Rovira, R.; Buesa, J.; Monedero, V.; Rodriguez-Diaz, J. The Role of Host Glycobiology and Gut Microbiota in Rotavirus and Norovirus Infection, an Update. Int. J. Mol. Sci. 2021, 22, 13473. [Google Scholar] [CrossRef] [PubMed]
- Munyemana, J.B.; Kabayiza, J.C.; Nilsson, S.; Andersson, M.E.; Lindh, M. Shigella and Enterotoxigenic Escherichia coli Have Replaced Rotavirus as Main Causes of Childhood Diarrhea in Rwanda After 10 Years of Rotavirus Vaccination. J. Infect. Dis. 2024, 230, e1176–e1180. [Google Scholar] [CrossRef] [PubMed]
- Widström, J.; Andersson, M.E.; Westin, J.; Wahllöf, M.; Lindh, M.; Rydell, G.E. Complex norovirus transmission dynamics at hospital wards revealed by deep sequencing. J. Clin. Microbiol. 2023, 61, e0060823. [Google Scholar] [CrossRef]
- Andersson, M.; Lindh, M. Rotavirus genotype shifts among Swedish children and adults—Application of a real-time PCR genotyping. J. Clin. Virol. 2017, 96, 1–6. [Google Scholar] [CrossRef]
- Cannon, J.L.; Bonifacio, J.; Bucardo, F.; Buesa, J.; Bruggink, L.; Chan, M.C.; Fumian, T.M.; Giri, S.; Gonzalez, M.D.; Hewitt, J.; et al. Global Trends in Norovirus Genotype Distribution among Children with Acute Gastroenteritis. Emerg. Infect. Dis. 2021, 27, 1438–1445. [Google Scholar] [CrossRef] [PubMed]
- van Beek, J.; de Graaf, M.; Al-Hello, H.; Allen, D.J.; Ambert-Balay, K.; Botteldoorn, N.; Brytting, M.; Buesa, J.; Cabrerizo, M.; Chan, M.; et al. Molecular surveillance of norovirus, 2005–2016: An epidemiological analysis of data collected from the NoroNet network. Lancet Infect. Dis. 2018, 18, 545–553. [Google Scholar] [CrossRef]
- Rossouw, E.; Brauer, M.; Meyer, P.; du Plessis, N.M.; Avenant, T.; Mans, J. Virus Etiology, Diversity and Clinical Characteristics in South African Children Hospitalised with Gastroenteritis. Viruses 2021, 13, 215. [Google Scholar] [CrossRef]
- Imbert-Marcille, B.M.; Barbe, L.; Dupe, M.; Le Moullac-Vaidye, B.; Besse, B.; Peltier, C.; Ruvoen-Clouet, N.; Le Pendu, J. A FUT2 gene common polymorphism determines resistance to rotavirus A of the P[8] genotype. J. Infect. Dis. 2014, 209, 1227–1230. [Google Scholar] [CrossRef]
- Huang, P.; Xia, M.; Tan, M.; Zhong, W.; Wei, C.; Wang, L.; Morrow, A.; Jiang, X. Spike protein VP8* of human rotavirus recognizes histo-blood group antigens in a type-specific manner. J. Virol. 2012, 86, 4833–4843. [Google Scholar] [CrossRef]
- Van Trang, N.; Vu, H.T.; Le, N.T.; Huang, P.; Jiang, X.; Anh, D.D. Association between norovirus and rotavirus infection and histo-blood group antigen types in Vietnamese children. J. Clin. Microbiol. 2014, 52, 1366–1374. [Google Scholar] [CrossRef] [PubMed]
- Ayouni, S.; Sdiri-Loulizi, K.; de Rougemont, A.; Estienney, M.; Ambert-Balay, K.; Aho, S.; Hamami, S.; Aouni, M.; Neji-Guediche, M.; Pothier, P.; et al. Rotavirus P[8] Infections in Persons with Secretor and Nonsecretor Phenotypes, Tunisia. Emerg. Infect. Dis. 2015, 21, 2055–2058. [Google Scholar] [CrossRef]
- Khachou, A.; Le Moullac-Vaidye, B.; Peltier, C.; Breiman, A.; Imbert-Marcille, B.M.; Ruvoen-Clouet, N.; Aouni, M.; Mastouri, M.; Chouchane, S.; Le Pendu, J. Host-Range Shift Between Emerging P[8]-4 Rotavirus and Common P[8] and P[4] Strains. J. Infect. Dis. 2020, 222, 836–839. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Farkas, T.; Marionneau, S.; Zhong, W.; Ruvoen-Clouet, N.; Morrow, A.L.; Altaye, M.; Pickering, L.K.; Newburg, D.S.; LePendu, J.; et al. Noroviruses bind to human ABO, Lewis, and secretor histo-blood group antigens: Identification of 4 distinct strain-specific patterns. J. Infect. Dis. 2003, 188, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Marionneau, S.; Ruvoen, N.; Le Moullac-Vaidye, B.; Clement, M.; Cailleau-Thomas, A.; Ruiz-Palacois, G.; Huang, P.; Jiang, X.; Le Pendu, J. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 2002, 122, 1967–1977. [Google Scholar] [CrossRef]
- Thorven, M.; Grahn, A.; Hedlund, K.O.; Johansson, H.; Wahlfrid, C.; Larson, G.; Svensson, L. A homozygous nonsense mutation (428G-->A) in the human secretor (FUT2) gene provides resistance to symptomatic norovirus (GGII) infections. J. Virol. 2005, 79, 15351–15355. [Google Scholar] [CrossRef]
- Tan, M.; Jin, M.; Xie, H.; Duan, Z.; Jiang, X.; Fang, Z. Outbreak studies of a GII-3 and a GII-4 norovirus revealed an association between HBGA phenotypes and viral infection. J. Med. Virol. 2008, 80, 1296–1301. [Google Scholar] [CrossRef]
- Lindesmith, L.; Moe, C.; Lependu, J.; Frelinger, J.A.; Treanor, J.; Baric, R.S. Cellular and humoral immunity following Snow Mountain virus challenge. J. Virol. 2005, 79, 2900–2909. [Google Scholar] [CrossRef]
- Le Pendu, J.; Rydell, G.E.; Nasir, W.; Larson, G. Human norovirus receptors. In Viral Gastroenteritis: Molecular Epidemiology and Pathogenesis; Svensson, L., Desselberger, U., Greenberg, H.B., Estes, M.K., Eds.; Elsevier: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Brazil, J.C.; Parkos, C.A. Finding the sweet spot: Glycosylation mediated regulation of intestinal inflammation. Mucosal Immunol. 2022, 15, 211–222. [Google Scholar] [CrossRef]
- Ruiz-Palacios, G.M.; Cervantes, L.E.; Ramos, P.; Chavez-Munguia, B.; Newburg, D.S. Campylobacter jejuni binds intestinal H(O) antigen (Fuc alpha 1, 2Gal beta 1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J. Biol. Chem. 2003, 278, 14112–14120. [Google Scholar] [CrossRef]
- Ahmed, T.; Lundgren, A.; Arifuzzaman, M.; Qadri, F.; Teneberg, S.; Svennerholm, A.M. Children with the Le(a+b−) blood group have increased susceptibility to diarrhea caused by enterotoxigenic Escherichia coli expressing colonization factor I group fimbriae. Infect. Immun. 2009, 77, 2059–2064. [Google Scholar] [CrossRef] [PubMed]
- Mottram, L.; Wiklund, G.; Larson, G.; Qadri, F.; Svennerholm, A.M. FUT2 non-secretor status is associated with altered susceptibility to symptomatic enterotoxigenic Escherichia coli infection in Bangladeshis. Sci. Rep. 2017, 7, 10649. [Google Scholar] [CrossRef] [PubMed]
| FUT2+ (n = 536) | FUT2− (n = 132) | OR | p | |
|---|---|---|---|---|
| Adenovirus 40/41 (n = 52) | 7.1% (38) | 11% (14) | 0.64 | 0.20 |
| Astrovirus (n = 30) | 3.8% (25) | 4.7% (5) | 1.24 | 0.81 |
| Norovirus GI (n = 24) | 0.6% (21) | 1.3% (3) | 1.75 | 0.45 |
| Norovirus GII (n = 69) | 10% (52) | 13% (17) | 0.72 | 0.27 |
| Rotavirus (n = 74) | 13% (67) | 5.3% (7) | 2.55 | 0.019 |
| Sapovirus (n = 78) | 12% (62) | 12% (16) | 0.95 | 0.88 |
| Campylobacter (n = 52) | 7.8% (42) | 7.6% (10) | 1.04 | 1.0 |
| ETEC-eltB (n = 158) | 23% (125) | 25% (33) | 1.08 | 1.0 |
| ETEC-estA (n = 81) | 12% (67) | 11% (14) | 1.20 | 0.66 |
| Salmonella (n = 38) | 4.8% (26) | 9.1% (12) | 0.51 | 0.09 |
| Shigella (n = 99) | 15% (83) | 12% (16) | 1.32 | 0.41 |
| Cryptosporidium (n = 22) | 3.7% (20) | 1.5% (2) | 2.52 | 0.28 |
| FUT2 + | FUT2 − | Pint. | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Diarrhea | No Diarrhea | OR | p | Diarrhea | No Diarrhea | OR | p | ||
| (n = 384) | (n = 152) | (n = 86) | (n = 46) | ||||||
| Adenovirus 40/41 (n = 52) | 7.6% (29) | 5.9% (9) | 1.30 | 0.58 | 10% (9) | 11% (5) | 0.96 | 1.0 | 0.67 |
| Astrovirus (n = 30) | 5.2% (20) | 3.3% (5) | 1.62 | 0.50 | 4.6% (4) | 2.2% (1) | 2.20 | 0.66 | 0.80 |
| Norovirus GI (n = 24) | 4.2% (16) | 3.3% (5) | 1.28 | 0.81 | 2.3% (2) | 2.2% (1) | 1.07 | 1.0 | 0.90 |
| Norovirus GII (n = 69) | 11% (42) | 6.6% (10) | 1.74 | 0.15 | 14% (12) | 11% (5) | 1.33 | 0.79 | 0.69 |
| Rotavirus (n = 74) | 16% (60) | 4.6% (7) | 3.84 | 0.0003 | 5.8% (5) | 4.4% (2) | 1.35 | 1.0 | 0.30 |
| Sapovirus (n = 78) | 12% (45) | 11% (17) | 1.05 | 1.0 | 13% (11) | 11% (5) | 1.20 | 1.0 | 0.84 |
| Campylobacter (n = 52) | 9.6% (37) | 3.3% (5) | 3.13 | 0.012 | 10% (9) | 2.2% (1) | 5.25 | 0.16 | 0.65 |
| ETEC-eltB (n = 158) | 29% (110) | 10% (15) | 3.67 | <0.0001 | 29% (25) | 17% (8) | 1.95 | 0.21 | 0.25 |
| ETEC-estA (n = 81) | 16% (67) | 4.6% (7) | 3.83 | 0.0003 | 14% (12) | 4.4% (2) | 3.57 | 0.14 | 0.94 |
| Salmonella (n = 38) | 5.7% (22) | 2.6% (4) | 2.25 | 0.18 | 8.1% (7) | 11% (5) | 0.73 | 0.75 | 0.17 |
| Shigella (n = 99) | 20% (75) | 5.3% (8) | 4.37 | <0.0001 | 16% (14) | 4.4% (2) | 4.28 | 0.052 | 0.98 |
| Cryptosporidium (n = 22) | 4.9% (19) | 0.7% (1) | 7.86 | 0.02 | 2.3% (2) | 0% (0) | 0.54 | 0.62 | |
| FUT2+ | FUT2− | |||||
|---|---|---|---|---|---|---|
| All | with Diarrhea | All | with Diarrhea | OR | p | |
| Adenovirus 40/41 (n = 52) | 38 | 29 (76%) | 14 | 9 (64%) | 1.79 | 0.48 |
| Astrovirus (n = 30) | 25 | 20 (80%) | 5 | 4 (80%) | 1.00 | 1.00 |
| Norovirus GI (n = 24) | 21 | 16 (76%) | 3 | 2 (67%) | 1.60 | 1.00 |
| Norovirus GII (n = 69) | 52 | 42 (81% | 17 | 12 (71%) | 1.75 | 0.50 |
| Rotavirus (n = 74) | 67 | 60 (90%) | 7 | 5 (71%) | 3.43 | 0.20 |
| Sapovirus (n = 78) | 62 | 45 (73%) | 16 | 11 (69%) | 1.20 | 0.76 |
| Campylobacter (n = 52) | 42 | 37 (88%) | 10 | 9 (90%) | 0.82 | 1.00 |
| ETEC-eltB (n = 158) | 125 | 110 (88%) | 33 | 25 (76%) | 2.35 | 0.10 |
| ETEC-estA (n = 81) | 67 | 60 (90%) | 14 | 12 (86%) | 1.43 | 0.65 |
| Salmonella (n = 38) | 26 | 22 (85%) | 12 | 7 (58%) | 3.93 | 0.11 |
| Shigella (n = 99) | 83 | 75 (90%) | 16 | 14 (88%) | 1.34 | 0.66 |
| Cryptosporidium (n = 22) | 20 | 19 (95%) | 2 | 2 (100%) | 0.00 | 1.00 |
| FUT2+ (n = 536) | FUT2− (n = 132) | ||||
|---|---|---|---|---|---|
| n | Diarrhea | No Diarrhea | Diarrhea | No Diarrhea | |
| Rotavirus | 74 | 60 | 7 | 5 | 2 |
| P[8] | 50 | 42 | 5 | 2 | 1 |
| P[4] | 7 | 7 | 0 | 0 | 0 |
| P[6] | 3 | 2 | 0 | 1 | 0 |
| P[9] | 1 | 1 | 0 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Munyemana, J.B.; Kabayiza, J.C.; Seruyange, E.; Nilsson, S.; Rydell, G.E.; Martner, A.; Andersson, M.E.; Lindh, M. Non-Secretor Status Due to FUT2 Stop Mutation Is Associated with Reduced Rotavirus Infections but Not with Other Enteric Pathogens in Rwandan Children. Microorganisms 2025, 13, 1071. https://doi.org/10.3390/microorganisms13051071
Munyemana JB, Kabayiza JC, Seruyange E, Nilsson S, Rydell GE, Martner A, Andersson ME, Lindh M. Non-Secretor Status Due to FUT2 Stop Mutation Is Associated with Reduced Rotavirus Infections but Not with Other Enteric Pathogens in Rwandan Children. Microorganisms. 2025; 13(5):1071. https://doi.org/10.3390/microorganisms13051071
Chicago/Turabian StyleMunyemana, Jean Bosco, Jean Claude Kabayiza, Eric Seruyange, Staffan Nilsson, Gustaf E. Rydell, Anna Martner, Maria E. Andersson, and Magnus Lindh. 2025. "Non-Secretor Status Due to FUT2 Stop Mutation Is Associated with Reduced Rotavirus Infections but Not with Other Enteric Pathogens in Rwandan Children" Microorganisms 13, no. 5: 1071. https://doi.org/10.3390/microorganisms13051071
APA StyleMunyemana, J. B., Kabayiza, J. C., Seruyange, E., Nilsson, S., Rydell, G. E., Martner, A., Andersson, M. E., & Lindh, M. (2025). Non-Secretor Status Due to FUT2 Stop Mutation Is Associated with Reduced Rotavirus Infections but Not with Other Enteric Pathogens in Rwandan Children. Microorganisms, 13(5), 1071. https://doi.org/10.3390/microorganisms13051071







