Microbiome-Induced Microenvironmental Changes Before and After Breast Cancer Treatment
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction and NGS Sequencing
2.3. Metagenomic Analysis of Microbial EV Composition
2.4. Statistical Analysis
3. Results
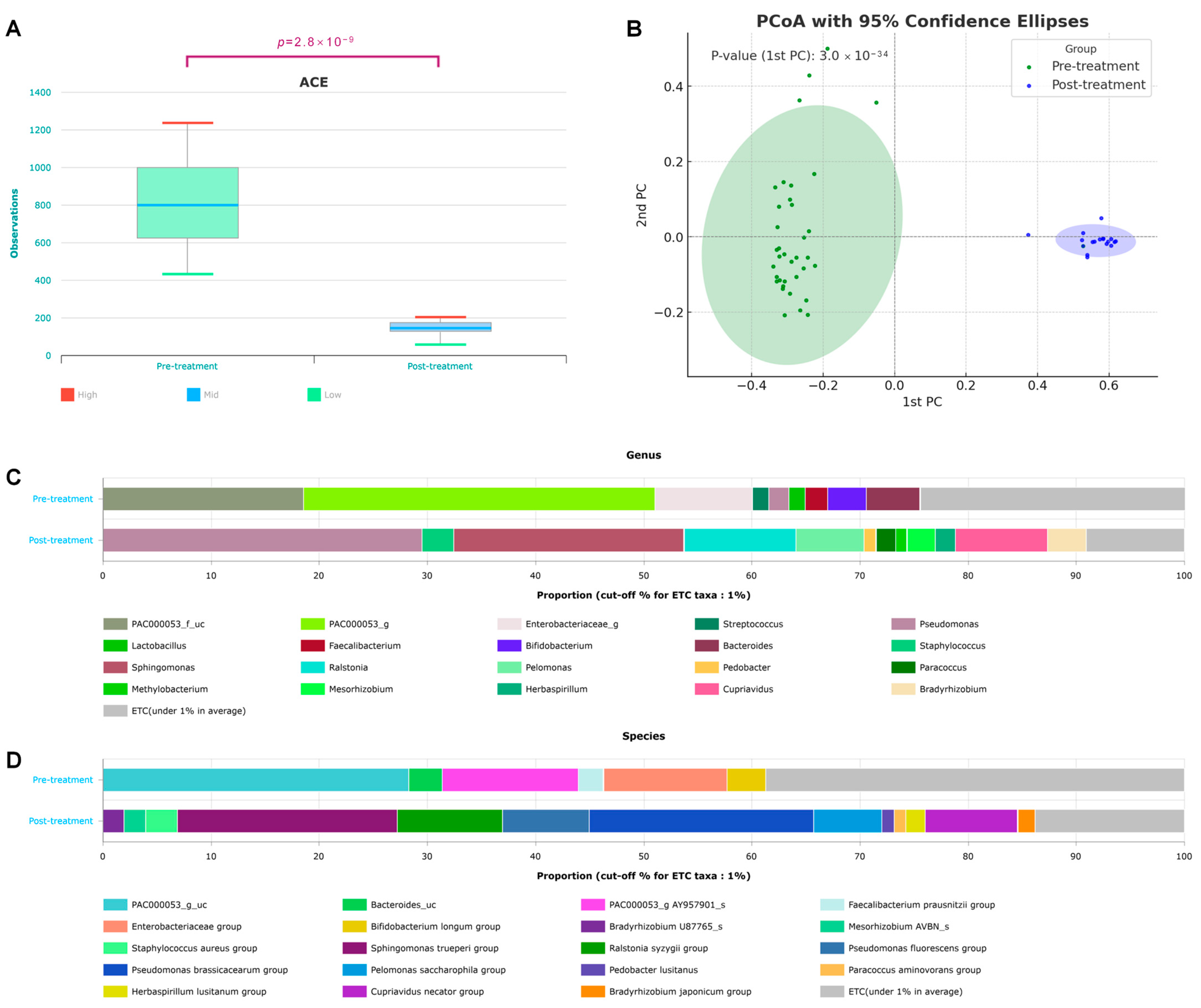
3.1. Diversity
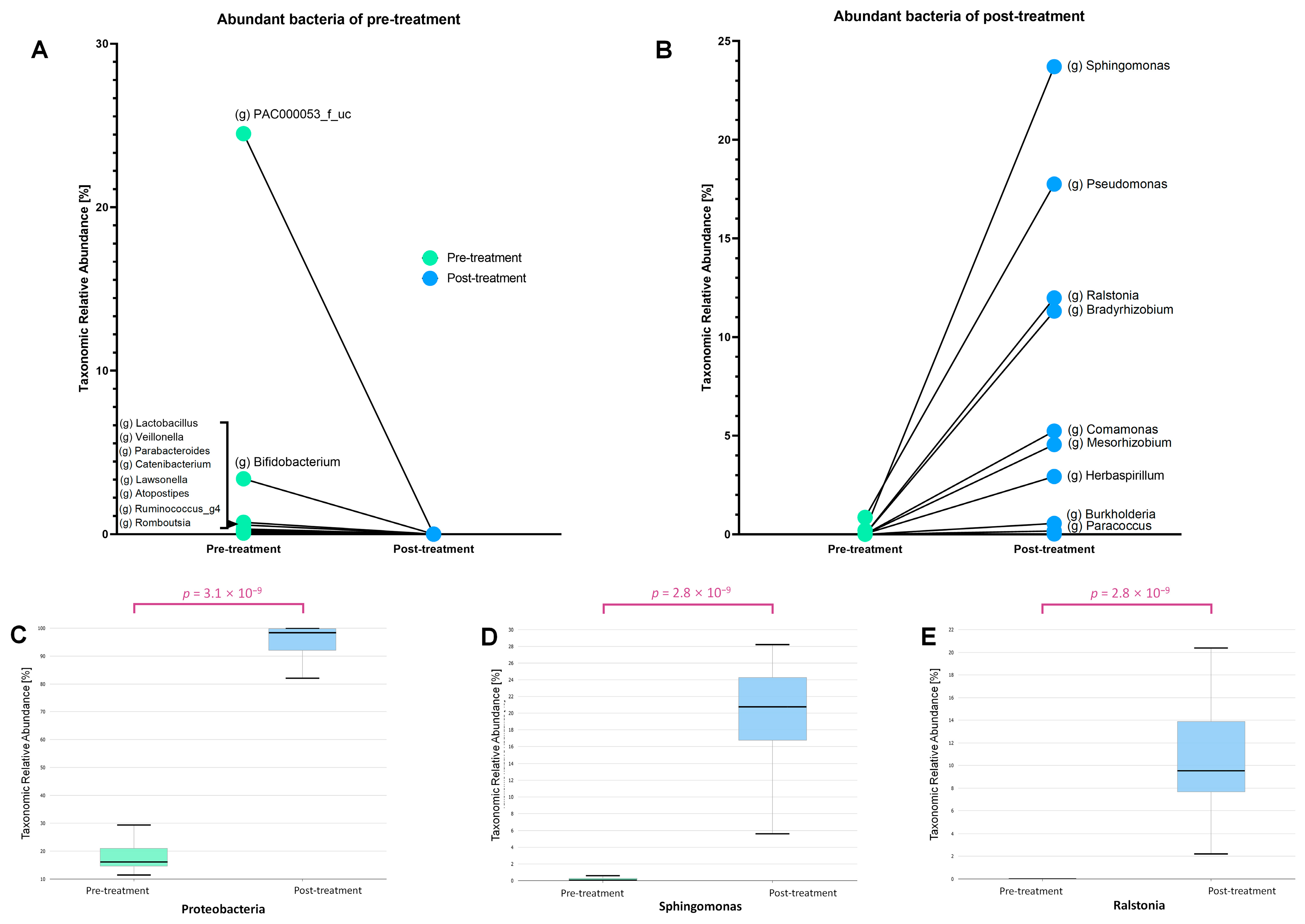
3.2. Taxonomic Shifts in the Microbiome Before and After Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rantala, J.; Seppä, K.; Eriksson, J.; Heinävaara, S.; Härkänen, T.; Jousilahti, P.; Knekt, P.; Männistö, S.; Rahkonen, O.; Malila, N.; et al. Incidence trends of early-onset breast cancer by lifestyle risk factors. BMC Cancer 2025, 25, 326. [Google Scholar] [CrossRef] [PubMed]
- Schoeler, M.; Caesar, R. Dietary lipids, gut microbiota and lipid metabolism. Rev. Endocr. Metab. Disord. 2019, 20, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, M.; Zimmermann-Kogadeeva, M.; Wegmann, R.; Goodman, A.L. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 2019, 570, 462–467. [Google Scholar] [CrossRef]
- Blayney, D.W.; Schwartzberg, L. Chemotherapy-induced neutropenia and emerging agents for prevention and treatment: A review. Cancer Treat. Rev. 2022, 109, 102427. [Google Scholar] [CrossRef]
- Shi, N.; Li, N.; Duan, X.; Niu, H. Interaction between the gut microbiome and mucosal immune system. Mil. Med. Res. 2017, 4, 14. [Google Scholar] [CrossRef]
- An, J.; Kwon, H.; Oh, S.Y.; Kim, Y.J. Association between breast cancer risk factors and blood microbiome in patients with breast cancer. Sci. Rep. 2025, 15, 6115. [Google Scholar] [CrossRef]
- Terrisse, S.; Derosa, L.; Iebba, V.; Ghiringhelli, F.; Vaz-Luis, I.; Kroemer, G.; Fidelle, M.; Christodoulidis, S.; Segata, N.; Thomas, A.M.; et al. Intestinal microbiota influences clinical outcome and side effects of early breast cancer treatment. Cell Death Differ. 2021, 28, 2778–2796. [Google Scholar] [CrossRef]
- Chronopoulos, A.; Kalluri, R. Emerging role of bacterial extracellular vesicles in cancer. Oncogene 2020, 39, 6951–6960. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Yang, J.; Kwon, H.; Lim, W.; Kim, Y.K.; Moon, B.I. Prediction of breast cancer using blood microbiome and identification of foods for breast cancer prevention. Sci. Rep. 2023, 13, 5110. [Google Scholar] [CrossRef]
- An, J.; Kwon, H.; Kim, Y.J. The Firmicutes/Bacteroidetes ratio as a risk factor of breast cancer. J. Clin. Med. 2023, 12, 2216. [Google Scholar] [CrossRef]
- Chalita, M.; Kim, Y.O.; Park, S.; Oh, H.S.; Cho, J.H.; Moon, J.; Baek, N.; Moon, C.; Lee, K.; Yang, J.; et al. EzBioCloud: A genome-driven database and platform for microbiome identification and discovery. Int. J. Syst. Evol. Microbiol. 2024, 74, 006421. [Google Scholar] [CrossRef] [PubMed]
- Kannauje, P.K.; Pandit, V.R.; Wasnik, P.N.; Venkat, N. Steroid and Sphingomonas. Ann. Afr. Med. 2022, 21, 299–300. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.A.; Bashir, M.; Rivas, M.N.; Duvall, K.; Sieling, P.A.; Pieber, T.R.; Vaishampayan, P.A.; Love, S.M.; Lee, D.J. Characterization of the microbiome of nipple aspirate fluid of breast cancer survivors. Sci. Rep. 2016, 6, 28061. [Google Scholar] [CrossRef]
- Xuan, C.; Shamonki, J.M.; Chung, A.; Dinome, M.L.; Chung, M.; Sieling, P.A.; Lee, D.J. Microbial dysbiosis is associated with human breast cancer. PLoS ONE 2014, 9, e83744. [Google Scholar] [CrossRef]
- Nandi, M.; Berry, C.; Brassinga, A.K.; Belmonte, M.F.; Fernando, W.G.; Loewen, P.C.; de Kievit, T.R. Pseudomonas brassicacearum strain DF41 kills Caenorhabditis elegans through biofilm-dependent and biofilm-independent mechanisms. Appl. Environ. Microbiol. 2016, 82, 6889–6898. [Google Scholar] [CrossRef]
- Rasimus, S.; Kolari, M.; Rita, H.; Hoornstra, D.; Salkinoja-Salonen, M. Biofilm-forming bacteria with varying tolerance to peracetic acid from a paper machine. J. Ind. Microbiol. Biotechnol. 2011, 38, 1379–1390. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Teng, Y.; Wang, X.; Xu, Y.; Li, R.; Sun, Y.; Dai, S.; Hu, W.; Wang, H.; Li, Y.; et al. Nitrogen transfer and cross-feeding between Azotobacter chroococcum and Paracoccus aminovorans promotes pyrene degradation. ISME J. 2023, 17, 2169–2181. [Google Scholar] [CrossRef]
- Lo, S.C.; Li, B.; Hung, G.C.; Lei, H.; Li, T.; Zhang, J.; Nagamine, K.; Tsai, S.; Zucker, M.J.; Olesnicky, L. Isolation and characterization of two novel bacteria Afipia cberi and Mesorhizobium hominis from blood of a patient afflicted with fatal pulmonary illness. PLoS ONE 2013, 8, e82673. [Google Scholar] [CrossRef]
- Zheng, Z.; Su, J.; Bao, X.; Wang, H.; Bian, C.; Zhao, Q.; Jiang, X. Mechanisms and applications of radiation-induced oxidative stress in regulating cancer immunotherapy. Front. Immunol. 2023, 14, 1247268. [Google Scholar] [CrossRef]
- Hanyu, M.; Fujimoto, H.; Tejima, K.; Saeki, K. Functional differences of two distinct catalases in Mesorhizobium loti MAFF303099 under free-living and symbiotic conditions. J. Bacteriol. 2009, 191, 1463–1471. [Google Scholar] [CrossRef]
- Kwiecinski, J.M.; Horswill, A.R. Staphylococcus aureus bloodstream infections: Pathogenesis and regulatory mechanisms. Curr. Opin. Microbiol. 2020, 53, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Zitvogel, L.; Daillère, R.; Roberti, M.P.; Routy, B.; Kroemer, G. Anticancer effects of the microbiome and its products. Nat. Rev. Microbiol. 2017, 15, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Xu, J.; Zhao, Y.; Zhang, P.; Zhang, J.; Yang, W.; Han, X.; Jin, H.; Zhang, W.; Wang, Y.; et al. Functional characterization of Mrr-family nuclease SLL1429 involved in MMC and phage resistance. Microbiol. Res. 2025, 296, 128123. [Google Scholar] [CrossRef] [PubMed]
- Szakács, G.; Váradi, A.; Ozvegy-Laczka, C.; Sarkadi, B. The role of ABC transporters in drug absorption, distribution, metabolism, excretion and toxicity (ADME-Tox). Drug Discov. Today 2008, 13, 379–393. [Google Scholar] [CrossRef]
- McLaughlin, H.P.; Caly, D.L.; McCarthy, Y.; Ryan, R.P.; Dow, J.M. An orphan chemotaxis sensor regulates virulence and antibiotic tolerance in the human pathogen Pseudomonas aeruginosa. PLoS ONE 2012, 7, e42205. [Google Scholar] [CrossRef]
- Sheldon, J.R.; Laakso, H.A.; Heinrichs, D.E. Iron acquisition strategies of bacterial pathogens. Microbiol. Spectr. 2016, 4, 1–32. [Google Scholar] [CrossRef]
- Nurgalieva, Z.; Liu, C.C.; Du, X.L. Chemotherapy use and risk of bone marrow suppression in a large population-based cohort of older women with breast and ovarian cancer. Med. Oncol. 2011, 28, 716–725. [Google Scholar] [CrossRef]
- Galkina, S.I.; Fedorova, N.V.; Ksenofontov, A.L.; Stadnichuk, V.I.; Baratova, L.A.; Sud’Ina, G.F. Neutrophils as a source of branched-chain, aromatic and positively charged free amino acids. Cell Adhes. Migr. 2019, 13, 98–105. [Google Scholar] [CrossRef]
- Wilkinson, A.C.; Morita, M.; Nakauchi, H.; Yamazaki, S. Branched-chain amino acid depletion conditions bone marrow for hematopoietic stem cell transplantation avoiding amino acid imbalance-associated toxicity. Exp. Hematol. 2018, 63, 12–16.e1. [Google Scholar] [CrossRef]
- Kariminik, A.; Baseri-Salehi, M.; Kheirkhah, B. Pseudomonas aeruginosa quorum sensing modulates immune responses: An updated review article. Immunol. Lett. 2017, 190, 1–6. [Google Scholar] [CrossRef]
- Ogbechi, J.; Huang, Y.S.; Clanchy, F.I.L.; Pantazi, E.; Topping, L.M.; Darlington, L.G.; Williams, R.O.; Stone, T.W. Modulation of immune cell function, IDO expression and kynurenine production by the quorum sensor 2-heptyl-3-hydroxy-4-quinolone (PQS). Front. Immunol. 2022, 13, 1001956. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]


| Pre-Treatment | Post-Treatment | ||
|---|---|---|---|
| Number of patients (%) | 36 (100%) | 9 (100%) | |
| Age (yr, mean ± SD) | 51.2 ± 6.3 | 51.2 ± 6.1 | |
| BMI (kg/m2, mean ± SD) | 22.5 ± 2.9 | 22 ± 3.0 | |
| Menopausal status | premenopausal | 16 (44.4%) | 5 (55.6%) |
| postmenopausal | 20 (55.6%) | 4 (44.4%) | |
| Estrogen receptor | positive | 26 (72.2%) | 9 (100%) |
| negative | 10 (27.8%) | 0 (0%) | |
| Progesteron receptor | positive | 24 (66.7%) | 9 (100%) |
| negative | 12 (33.3%) | 0 (0%) | |
| HER-2/new | positive | 12 (33.3%) | 1 (11.1%) |
| negative | 24 (66.7%) | 8 (88.9%) | |
| Tumor size | 0~2 | 22 (61.1%) | 1 (11.1%) |
| 2~5 | 13 (36.1%) | 8 (88.9%) | |
| 5~ | 1 (2.8%) | 0 (0%) | |
| Nodal status | positive | 9 (25%) | 2 (22.2%) |
| negative | 27 (75%) | 6 (66.7%) | |
| Stage | stage 0 | 0 (0%) | 1 (11.1%) |
| stage I | 20 (55.6%) | 2 (22.2%) | |
| stage II | 13 (36.1%) | 3 (33.3%) | |
| stage III | 3 (8.3%) | 3 (33.3%) | |
| Chemotherapy | yes | 0 (0%) | 7 (77.8%) |
| no | 36 (100%) | 2 (22.2%) | |
| Targeted therapy | yes | 0 (0%) | 1 (11.1%) |
| no | 36 (100%) | 8 (88.9%) | |
| Radiation therapy | yes | 0 (0%) | 7 (77.8%) |
| no | 36 (100%) | 2 (22.2%) | |
| Endocrine therapy | AI | 0 (0%) | 6 (66.7%) |
| tamoxifen | 0 (0%) | 3 (33.3%) |
| Ortholog | Definition | Pathway | Module | LDA Effect Size | p-Value | p-Value (FDR) | Pre-Treatment | Post-Treatment |
|---|---|---|---|---|---|---|---|---|
| K08800 | NUAK family, SNF1-like kinase | 3.62 | 2.8 × 10−9 | 1.47 × 10−8 | 0.9 | 0.05 | ||
| K07449 | similar to archaeal holliday junction resolvase and Mrr protein | 3.28 | 2.8 × 10−9 | 1.47 × 10−8 | 0.4 | 0.01 | ||
| K00934 | arginine kinase | ko00330 | 3.25 | 1.26 × 10−7 | 2.9 × 10−7 | 0.67 | 0.31 | |
| K02014 | iron complex outermembrane recepter protein | 3.24 | 3.09 × 10−9 | 1.47 × 10−8 | 0.1 | 0.46 | ||
| K03406 | methyl-accepting chemotaxis protein | ko02020, ko02030 | 3.19 | 2.8 × 10−9 | 1.47 × 10−8 | 0.06 | 0.37 | |
| K12446 | L-arabinokinase | ko00520, ko01100 | 3.13 | 6.7 × 10−9 | 2.35 × 10−8 | 0.28 | 0.01 | |
| K00059 | 3-oxoacyl-[acyl-carrier protein] reductase | ko00061, ko00333, ko00780, ko01040, ko01100, ko01130, ko01212 | M00083, M00572 | 3.1 | 2.76 × 10−9 | 1.47 × 10−8 | 0.24 | 0.49 |
| K07445 | putative DNA methylase | 3.1 | 1.59 × 10−8 | 4.7 × 10−8 | 0.29 | 0.04 | ||
| K09678 | [heparan sulfate]-glucosamine 3-sulfotransferase 4 | 3.07 | 3.09 × 10−9 | 1.47 × 10−8 | 0.01 | 0.25 | ||
| K07795 | putative tricarboxylic transport membrane protein | ko02020 | 3.04 | 1.28 × 10−8 | 3.98 × 10−8 | 0.03 | 0.25 |
| Pre-Teratment | Post-Treatment | p-Value | |
|---|---|---|---|
| WBC (×103/µL) | 6.32 | 4.91 | 0.0105 |
| ANC (×103/µL) | 5.07 | 2.7 | 0.0004 |
| Neutrophil (%) | 58.92 | 54.68 | 0.0602 |
| Lymphocyte (%) | 31.37 | 35.49 | 0.1038 |
| Monocyte (%) | 7.16 | 7.21 | 0.1368 |
| Eosinophil (%) | 2.05 | 1.92 | 0.1391 |
| Basophil (%) | 0.47 | 0.69 | 0.0313 |
| Hb (g/dL) | 14.56 | 12.46 | 0.0111 |
| Hct (%) | 41.24 | 37.27 | 0.0142 |
| RDW(CV) (%) | 26.29 | 12.67 | 0.0266 |
| Platelet (×103/µL) | 252.98 | 215.61 | 0.0021 |
| PCT (%) | 0.7 | 0.21 | 0.0001 |
| MPV (fL) | 10.03 | 9.78 | 0.2492 |
| PDW (fL) | 13.68 | 10.34 | 0.0177 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
An, J.; Kwon, H.; Kim, Y.J.; Moon, B.-I. Microbiome-Induced Microenvironmental Changes Before and After Breast Cancer Treatment. Microorganisms 2025, 13, 1057. https://doi.org/10.3390/microorganisms13051057
An J, Kwon H, Kim YJ, Moon B-I. Microbiome-Induced Microenvironmental Changes Before and After Breast Cancer Treatment. Microorganisms. 2025; 13(5):1057. https://doi.org/10.3390/microorganisms13051057
Chicago/Turabian StyleAn, Jeongshin, Hyungju Kwon, Young Ju Kim, and Byung-In Moon. 2025. "Microbiome-Induced Microenvironmental Changes Before and After Breast Cancer Treatment" Microorganisms 13, no. 5: 1057. https://doi.org/10.3390/microorganisms13051057
APA StyleAn, J., Kwon, H., Kim, Y. J., & Moon, B.-I. (2025). Microbiome-Induced Microenvironmental Changes Before and After Breast Cancer Treatment. Microorganisms, 13(5), 1057. https://doi.org/10.3390/microorganisms13051057






