Genome Characterization of Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae Strains, Carrying Hybrid Resistance-Virulence IncHI1B/FIB Plasmids, Isolated from an Egyptian Pediatric ICU
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates
2.2. Antimicrobial Susceptibility Testing
2.3. String Test
2.4. Whole-Genome Sequencing of Clinical CR-hvKP Strains
2.5. Genomic Analysis
3. Results
3.1. Bacterial Isolation and AMR
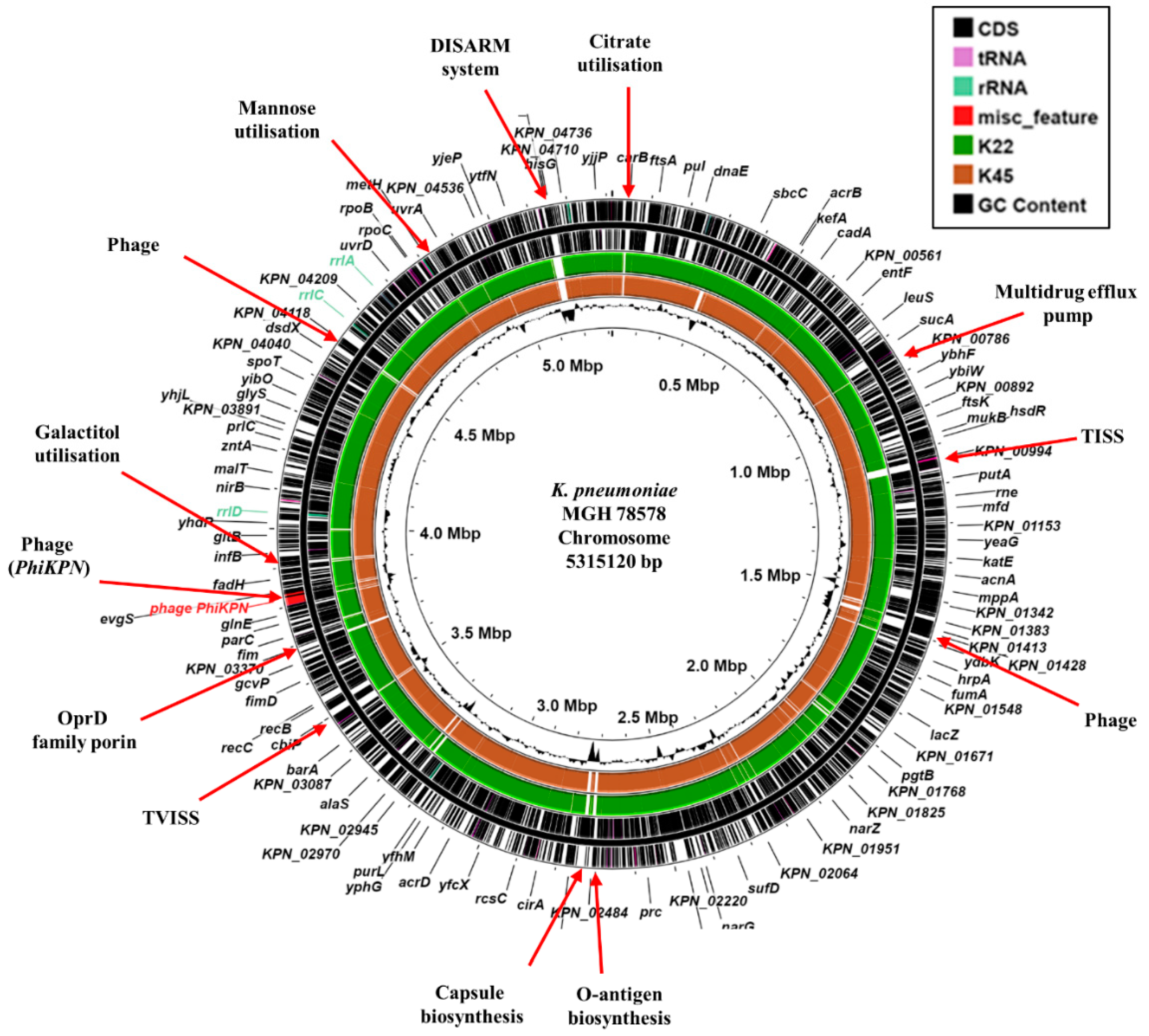
3.2. Genomic Characterization of CR-hvKP Isolates
3.3. Antimicrobial Resistance Genes
3.4. Virulence Determinants
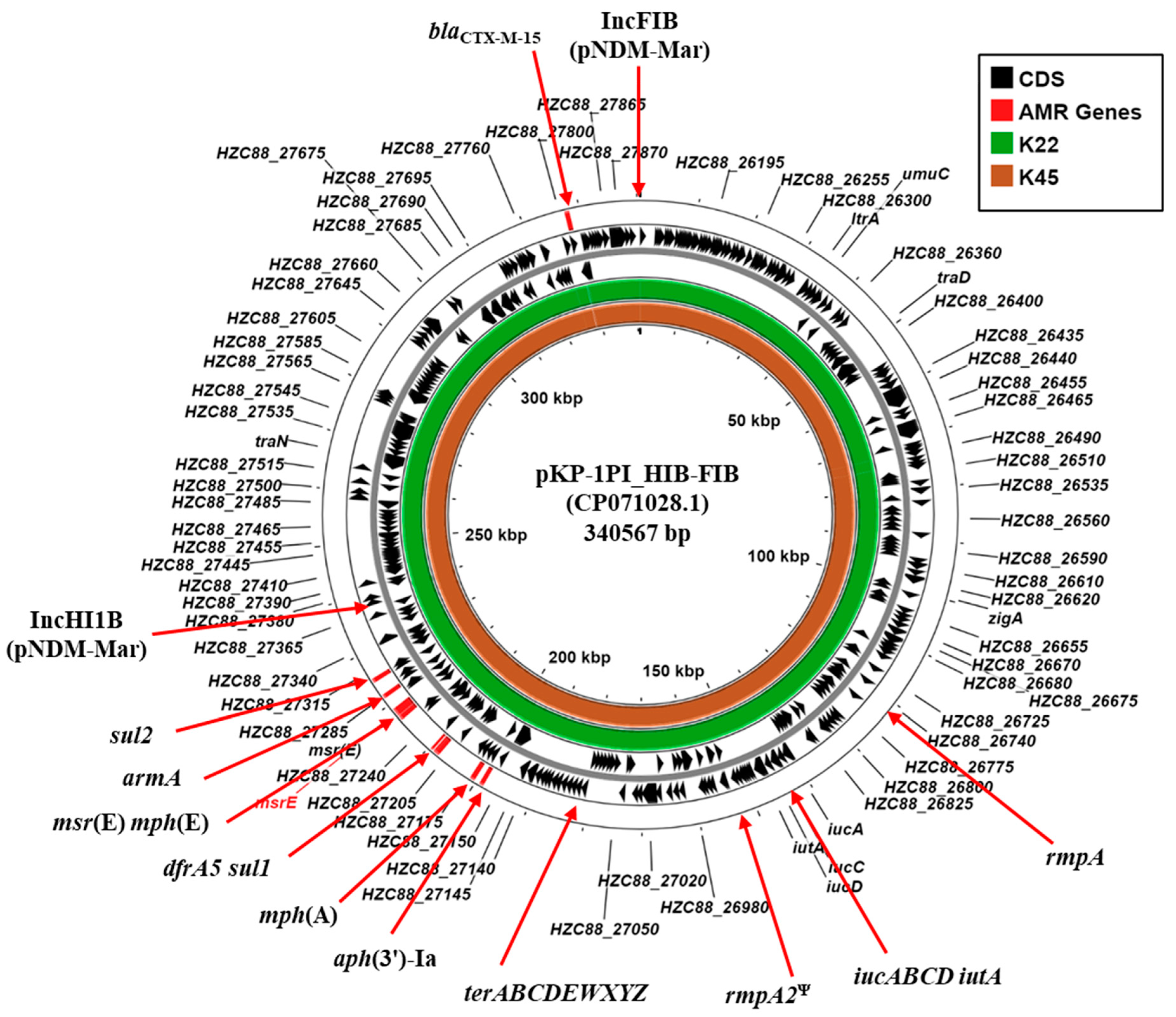
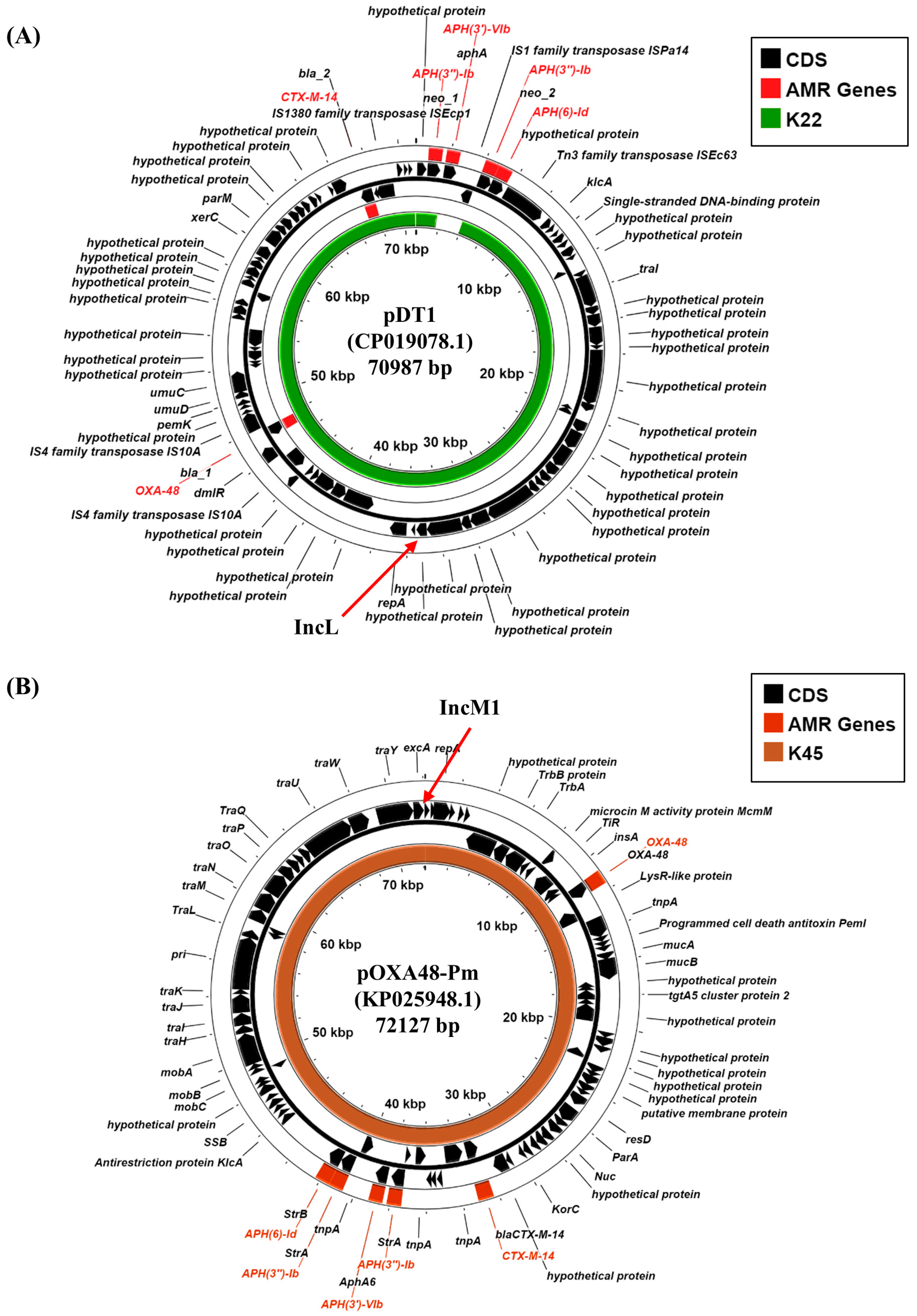
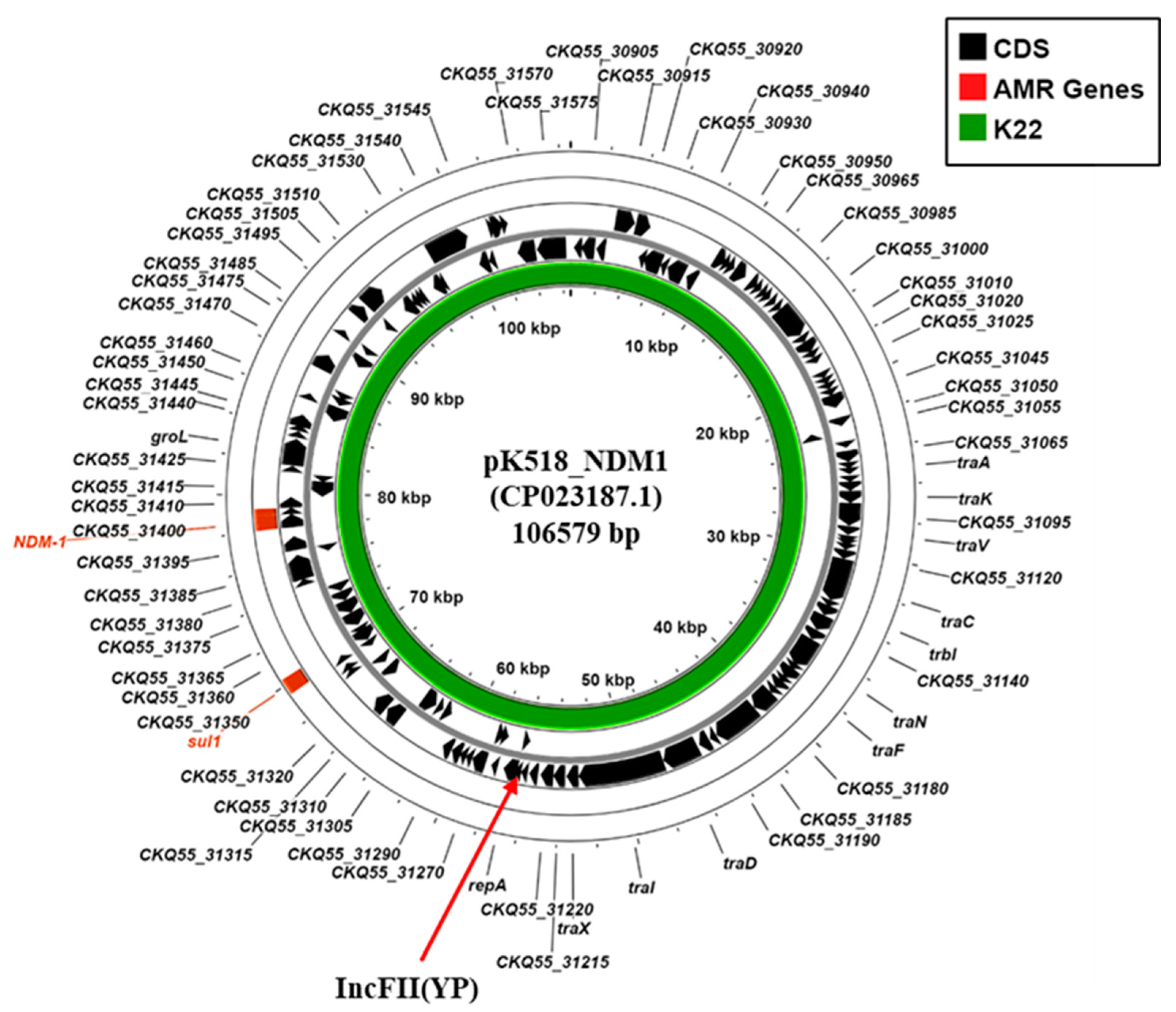
3.5. Plasmid-Associated Antimicrobial Resistance and Virulence Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shon, A.S.; Bajwa, R.P.; Russo, T.A. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: A new and dangerous breed. Virulence 2013, 4, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Russo, T.A.; Marr, C.M. Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 2019, 32. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, X.; Torres, V.V.L.; Liu, H.; Rocker, A.; Zhang, Y.; Wang, J.; Chen, L.; Bi, W.; Lin, J.; et al. An Outbreak of Carbapenem-Resistant and Hypervirulent Klebsiella pneumoniae in an Intensive Care Unit of a Major Teaching Hospital in Wenzhou, China. Front. Public Health 2019, 7, 229. [Google Scholar] [CrossRef] [PubMed]
- Gu, D.; Dong, N.; Zheng, Z.; Lin, D.; Huang, M.; Wang, L.; Chan, E.W.; Shu, L.; Yu, J.; Zhang, R.; et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: A molecular epidemiological study. Lancet Infect. Dis. 2018, 18, 37–46. [Google Scholar] [CrossRef]
- Liu, C.; Du, P.; Xiao, N.; Ji, F.; Russo, T.A.; Guo, J. Hypervirulent Klebsiella pneumoniae is emerging as an increasingly prevalent K. pneumoniae pathotype responsible for nosocomial and healthcare-associated infections in Beijing, China. Virulence 2020, 11, 1215–1224. [Google Scholar] [CrossRef]
- Lin, Y.T.; Siu, L.K.; Lin, J.C.; Chen, T.L.; Tseng, C.P.; Yeh, K.M.; Chang, F.Y.; Fung, C.P. Seroepidemiology of Klebsiella pneumoniae colonizing the intestinal tract of healthy Chinese and overseas Chinese adults in Asian countries. BMC Microbiol. 2012, 12, 13. [Google Scholar] [CrossRef]
- Choby, J.E.; Howard-Anderson, J.; Weiss, D.S. Hypervirulent Klebsiella pneumoniae—Clinical and molecular perspectives. J. Intern. Med. 2020, 287, 283–300. [Google Scholar] [CrossRef]
- Shankar, C.; Vasudevan, K.; Jacob, J.J.; Baker, S.; Isaac, B.J.; Neeravi, A.R.; Sethuvel, D.P.M.; George, B.; Veeraraghavan, B. Hybrid Plasmids Encoding Antimicrobial Resistance and Virulence Traits Among Hypervirulent Klebsiella pneumoniae ST2096 in India. Front. Cell Infect. Microbiol. 2022, 12, 875116. [Google Scholar] [CrossRef]
- Huang, Y.H.; Chou, S.H.; Liang, S.W.; Ni, C.E.; Lin, Y.T.; Huang, Y.W.; Yang, T.C. Emergence of an XDR and carbapenemase-producing hypervirulent Klebsiella pneumoniae strain in Taiwan. J. Antimicrob. Chemother. 2018, 73, 2039–2046. [Google Scholar] [CrossRef]
- He, Z.; Xu, W.; Zhao, H.; Li, W.; Dai, Y.; Lu, H.; Zhao, L.; Zhang, C.; Li, Y.; Sun, B. Epidemiological characteristics an outbreak of ST11 multidrug-resistant and hypervirulent Klebsiella pneumoniae in Anhui, China. Front. Microbiol. 2022, 13, 996753. [Google Scholar] [CrossRef]
- Sherif, M.; Palmieri, M.; Mirande, C.; El-Mahallawy, H.; Rashed, H.G.; Abd-El-Reheem, F.; El-Manakhly, A.R.; Abdel-Latif, R.A.R.; Aboulela, A.G.; Saeed, L.Y.; et al. Whole-genome sequencing of Egyptian multidrug-resistant Klebsiella pneumoniae isolates: A multi-center pilot study. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 1451–1460. [Google Scholar] [CrossRef] [PubMed]
- Elzeny, H.; Mohamed, W.; Daef, E.; El-Badawy, O.; Shaaban, L.; Osman, N.S.; Hadiya, S.; Aly, S. Detection of multiple extensively-drug resistant hypervirulent Klebsiella pneumoniae clones from patients with ventilator-associated pneumonia in Egypt. J. Med. Microbiol. 2023, 72, 001701. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, Y.; Ahmed, M.; Qin, M.; He, R.; Wu, Y.; Liang, X.; Zhong, L.L.; Chen, P.; Deng, B.; et al. Carriage of distinct bla(KPC-2) and bla(OXA-48) plasmids in a single ST11 hypervirulent Klebsiella pneumoniae isolate in Egypt. BMC Genom. 2022, 23, 20. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Yang, Y.; Yang, Y.; Yan, B.; Chen, G.; Hassan, R.M.; Zhong, L.L.; Chen, Y.; Roberts, A.P.; Wu, Y.; et al. Emergence of Hypervirulent Carbapenem-Resistant Klebsiella pneumoniae Coharboring a bla(NDM-1)-Carrying Virulent Plasmid and a bla(KPC-2)-Carrying Plasmid in an Egyptian Hospital. mSphere 2021, 6, e00088-21. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing. In CLSI Document M100, 33rd ed.; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2023. [Google Scholar]
- Yao, H.; Liu, J.; Jiang, X.; Chen, F.; Lu, X.; Zhang, J. Analysis of the Clinical Effect of Combined Drug Susceptibility to Guide Medication for Carbapenem-Resistant Klebsiella pneumoniae Patients Based on the Kirby-Bauer Disk Diffusion Method. Infect. Drug Resist. 2021, 14, 79–87. [Google Scholar] [CrossRef]
- Fang, C.T.; Chuang, Y.P.; Shun, C.T.; Chang, S.C.; Wang, J.T. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J. Exp. Med. 2004, 199, 697–705. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Rutherford, K.; Parkhill, J.; Crook, J.; Horsnell, T.; Rice, P.; Rajandream, M.A.; Barrell, B. Artemis: Sequence visualization and annotation. Bioinformatics 2000, 16, 944–945. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.J.; Rutherford, K.M.; Berriman, M.; Rajandream, M.A.; Barrell, B.G.; Parkhill, J. ACT: The Artemis Comparison Tool. Bioinformatics 2005, 21, 3422–3423. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Ponten, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Moller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef]
- Wick, R.R.; Heinz, E.; Holt, K.E.; Wyres, K.L. Kaptive Web: User-Friendly Capsule and Lipopolysaccharide Serotype Prediction for Klebsiella Genomes. J. Clin. Microbiol. 2018, 56, e00197-18. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- McClelland, M.; Sanderson, K.E.; Spieth, J.; Clifton, S.W.; Latreille, P.; Courtney, L.; Porwollik, S.; Ali, J.; Dante, M.; Du, F.; et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 2001, 413, 852–856. [Google Scholar] [CrossRef] [PubMed]
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. MMBR 2016, 80, 629–661. [Google Scholar] [CrossRef]
- Navon-Venezia, S.; Kondratyeva, K.; Carattoli, A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 2017, 41, 252–275. [Google Scholar] [CrossRef] [PubMed]
- Ofir, G.; Melamed, S.; Sberro, H.; Mukamel, Z.; Silverman, S.; Yaakov, G.; Doron, S.; Sorek, R. DISARM is a widespread bacterial defence system with broad anti-phage activities. Nat. Microbiol. 2018, 3, 90–98. [Google Scholar] [CrossRef]
- Di Pilato, V.; Henrici De Angelis, L.; Aiezza, N.; Baccani, I.; Niccolai, C.; Parisio, E.M.; Giordano, C.; Camarlinghi, G.; Barnini, S.; Forni, S.; et al. Resistome and virulome accretion in an NDM-1-producing ST147 sublineage of Klebsiella pneumoniae associated with an outbreak in Tuscany, Italy: A genotypic and phenotypic characterisation. Lancet Microbe 2022, 3, e224–e234. [Google Scholar] [CrossRef]
- Blackwell, G.A.; Doughty, E.L.; Moran, R.A. Evolution and dissemination of L and M plasmid lineages carrying antibiotic resistance genes in diverse Gram-negative bacteria. Plasmid 2021, 113, 102528. [Google Scholar] [CrossRef]
- Both, A.; Büttner, H.; Huang, J.; Perbandt, M.; Belmar Campos, C.; Christner, M.; Maurer, F.P.; Kluge, S.; König, C.; Aepfelbacher, M.; et al. Emergence of ceftazidime/avibactam non-susceptibility in an MDR Klebsiella pneumoniae isolate. J. Antimicrob. Chemother. 2017, 72, 2483–2488. [Google Scholar] [CrossRef]
- Chen, L.; Al Laham, N.; Chavda, K.D.; Mediavilla, J.R.; Jacobs, M.R.; Bonomo, R.A.; Kreiswirth, B.N. First report of an OXA-48-producing multidrug-resistant Proteus mirabilis strain from Gaza, Palestine. Antimicrob. Agents Chemother. 2015, 59, 4305–4307. [Google Scholar] [CrossRef]
- Zheng, B.; Xu, H.; Yu, X.; Lv, T.; Jiang, X.; Cheng, H.; Zhang, J.; Chen, Y.; Huang, C.; Xiao, Y. Identification and genomic characterization of a KPC-2-, NDM-1- and NDM-5-producing Klebsiella michiganensis isolate. J. Antimicrob. Chemother. 2018, 73, 536–538. [Google Scholar] [CrossRef]
- Russo, T.A.; Olson, R.; Fang, C.T.; Stoesser, N.; Miller, M.; MacDonald, U.; Hutson, A.; Barker, J.H.; La Hoz, R.M.; Johnson, J.R. Identification of Biomarkers for Differentiation of Hypervirulent Klebsiella pneumoniae from Classical K. pneumoniae. J. Clin. Microbiol. 2018, 56, e00776-18. [Google Scholar] [CrossRef] [PubMed]
- Follador, R.; Heinz, E.; Wyres, K.L.; Ellington, M.J.; Kowarik, M.; Holt, K.E.; Thomson, N.R. The diversity of Klebsiella pneumoniae surface polysaccharides. Microb. Genom. 2016, 2, e000073. [Google Scholar] [CrossRef]
- Wu, J.W.; Wang, J.T.; Lin, T.L.; Liu, Y.Z.; Wu, L.T.; Pan, Y.J. Identification of three capsule depolymerases in a bacteriophage infecting Klebsiella pneumoniae capsular types K7, K20, and K27 and therapeutic application. J. Biomed. Sci. 2023, 30, 31. [Google Scholar] [CrossRef] [PubMed]
- Hammad, H.A.; Mohamed, I.S.; El-Badawy, O.; Zakaria, A.M.; Shabaan, L.; Aly, S.A. pKpQIL-like plasmid contributes to the dissemination of bla(NDM-1) and plasmid mediated quinolone resistance determinants among multi drug resistant Klebsiella pneumoniae in Assiut university hospital, Egypt. Iran. J. Microbiol. 2023, 15, 208–218. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, Y.; Wang, M.; Li, X.; Liu, Z.; Kuang, D.; Deng, Z.; Ou, H.Y.; Qu, J. Mobilizable plasmids drive the spread of antimicrobial resistance genes and virulence genes in Klebsiella pneumoniae. Genome Med. 2023, 15, 106. [Google Scholar] [CrossRef]
- Abdel-Rahim, M.H.; El-Badawy, O.; Hadiya, S.; Daef, E.A.; Suh, S.J.; Boothe, D.M.; Aly, S.A. Patterns of Fluoroquinolone Resistance in Enterobacteriaceae Isolated from the Assiut University Hospitals, Egypt: A Comparative Study. Microb. Drug Resist. 2019, 25, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Bialek-Davenet, S.; Criscuolo, A.; Ailloud, F.; Passet, V.; Jones, L.; Delannoy-Vieillard, A.S.; Garin, B.; Le Hello, S.; Arlet, G.; Nicolas-Chanoine, M.H.; et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg. Infect. Dis. 2014, 20, 1812–1820. [Google Scholar] [CrossRef]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef]
- Mei, Y.F.; Liu, P.P.; Wan, L.G.; Liu, Y.; Wang, L.H.; Wei, D.D.; Deng, Q.; Cao, X.W. Virulence and Genomic Feature of a Virulent Klebsiella pneumoniae Sequence Type 14 Strain of Serotype K2 Harboring bla(NDM-5) in China. Front. Microbiol. 2017, 8, 335. [Google Scholar] [CrossRef]
- Al-Zahrani, I.A.; Aljabri, A.; Alhazmi, W.A.; Yasir, M.; Abujamel, T.; Alghamdi, A.K.; Azhar, E.I. Genomic analysis of extensively drug resistant (XDR) Klebsiella pneumoniae high-risk clone ST14 co-harboring bla(NDM) and bla(OXA-48) recovered from Saudi Arabia. J. Infect. Public Health 2024, 17, 669–675. [Google Scholar] [CrossRef]
- Kondo, S.; Phornsiricharoenphant, W.; Na-Rachasima, L.; Phokhaphan, P.; Ruangchai, W.; Palittapongarnpim, P.; Apisarnthanarak, A. Genomic characterization of extended-spectrum β-lactamase-producing Enterobacterales isolated from abdominal surgical patients. Epidemiol. Infect. 2024, 152, e70. [Google Scholar] [CrossRef]
- Papagiannitsis, C.C.; Giakkoupi, P.; Vatopoulos, A.C.; Tryfinopoulou, K.; Miriagou, V.; Tzouvelekis, L.S. Emergence of Klebsiella pneumoniae of a novel sequence type (ST383) producing VIM-4, KPC-2 and CMY-4 β-lactamases. Int. J. Antimicrob. Agents 2010, 36, 573–574. [Google Scholar] [CrossRef] [PubMed]
- Tsui, C.K.; Ben Abid, F.; Al Ismail, K.; McElheny, C.L.; Al Maslamani, M.; Omrani, A.S.; Doi, Y. Genomic Epidemiology of Carbapenem-Resistant Klebsiella in Qatar: Emergence and Dissemination of Hypervirulent Klebsiella pneumoniae Sequence Type 383 Strains. Antimicrob. Agents Chemother. 2023, 67, e0003023. [Google Scholar] [CrossRef] [PubMed]
- Edward, E.A.; Mohamed, N.M.; Zakaria, A.S. Whole Genome Characterization of the High-Risk Clone ST383 Klebsiella pneumoniae with a Simultaneous Carriage of bla(CTX-M-14) on IncL/M Plasmid and bla(CTX-M-15) on Convergent IncHI1B/IncFIB Plasmid from Egypt. Microorganisms 2022, 10, 1097. [Google Scholar] [CrossRef] [PubMed]
- Turton, J.F.; Payne, Z.; Coward, A.; Hopkins, K.L.; Turton, J.A.; Doumith, M.; Woodford, N. Virulence genes in isolates of Klebsiella pneumoniae from the UK during 2016, including among carbapenemase gene-positive hypervirulent K1-ST23 and ‘non-hypervirulent’ types ST147, ST15 and ST383. J. Med. Microbiol. 2018, 67, 118–128. [Google Scholar] [CrossRef]
- Turton, J.; Davies, F.; Turton, J.; Perry, C.; Payne, Z.; Pike, R. Hybrid Resistance and Virulence Plasmids in “High-Risk” Clones of Klebsiella pneumoniae, Including Those Carrying bla(NDM-5). Microorganisms 2019, 7, 326. [Google Scholar] [CrossRef]
- Starkova, P.; Lazareva, I.; Avdeeva, A.; Sulian, O.; Likholetova, D.; Ageevets, V.; Lebedeva, M.; Gostev, V.; Sopova, J.; Sidorenko, S. Emergence of Hybrid Resistance and Virulence Plasmids Harboring New Delhi Metallo-β-Lactamase in Klebsiella pneumoniae in Russia. Antibiotics 2021, 10, 691. [Google Scholar] [CrossRef]
- Falcone, M.; Tiseo, G.; Arcari, G.; Leonildi, A.; Giordano, C.; Tempini, S.; Bibbolino, G.; Mozzo, R.; Barnini, S.; Carattoli, A.; et al. Spread of hypervirulent multidrug-resistant ST147 Klebsiella pneumoniae in patients with severe COVID-19: An observational study from Italy, 2020-21. J. Antimicrob. Chemother. 2022, 77, 1140–1145. [Google Scholar] [CrossRef]
| Strain ID. | K22 | K45 |
|---|---|---|
| Date of isolation | 10 January 2015 | 12 May 2015 |
| ICU isolation | Pediatric | Pediatric |
| Type of infection | VAP | VAP |
| MIC IMP | 12 (R) | 4 (R) |
| MIC CIP | >32 (R) | 0.047 (S) |
| Resistance phenotype * | XDR 1,2,3,6,7,8 | MDR 1,2,3,7,8 |
| String test | Negative | Negative |
| Strain ID. | K22 | K45 |
|---|---|---|
| Genome size | 5,862,489 bp | 5,741,936 bp |
| Number of Contigs | 151 | 101 |
| Genes (CDs) | 5580 | 5344 |
| GC% | 56.54% | 56.65% |
| MLST | ST383 | ST14 |
| Capsular type | K30 | K2 |
| O type | O1/O2v1 Type O1 | O1/O2v1 Type O1 |
| Plasmid replicons | IncFIB (pNDM-Mar) IncHI1B (pNDM-Mar) IncFIB (pQil) IncFII (Yp) IncL, Col440II | IncFIB (pNDM-Mar) IncHI1B (pNDM-Mar) IncM Col(pHAD28) Col440I |
| Strain | K22 | K45 |
|---|---|---|
| Acquired AMR gene 1 | AmgR: aadA1, aph(3′)-Ia, aph(3″)-Ib, aph(6)-Id, armA, rmtC AmgR/FlqR: aac(6’)-Ib-cr βLR: blaCTX-M-14b, blaNDM-1, blaOXA-9, blaOXA-48, blaSHV-26 TSR: dfrA5, sul1, sul2 MclR: mph(A), mph(E), msr(E) FosR: fosA | AmgR: aph(3′)-VIb aph(3′)-Ia, aph(3″)-Ib, aph(6)-Id, armA βLR: blaCTX-M-14b, blaOXA-48, blaSHV-28 TSR: dfrA5, sul1, sul2 MclR: mph(A), mph(E), msr(E) FosR: fosA |
| Chromosomal point mutations associated with AMR | gyrA: S83F, D87N parC: S80I ompK37: I70M, I128M acrR: P161R, G164A, F172S, R173G, L195V, F197I, K201M | ompK36 N49S, L59V, L191S, F207W, A217S, N218H, D224E, L228V, E232R, T254 ompK37: I70M, I128M acrR: P161R, G164A, F172S, R173G, L195V, F197I, K201M |
| Virulence genes | Adhesion/Biofilm formation mrkABCDF, mrkHIJ, fimABCDFGH, ecpRABCDE Iron acquisition iucABCD iutA, fepA-entD, entF-fes, fepDGC, entS, fepB entCEBAH iroE, iroN Virulence regulation rmpA, rmpA2 Ψ | Adhesion/Biofilm formation mrkABCDF, mrkHIJ, fimABCDFGH, ecpRABCDE Iron acquisition iucABCD iutA, fepA-entD, entF-fes, fepDGC, entS, fepB entCEBAH iroE, iroN kfuABC Virulence regulation rmpA, rmpA2 Ψ |
| Heavy metal resistance | cusRS, cusCFBA, terABCDEWXYZ | cusRCFBA, terABCDEWXYZ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hammad, H.A.; Abdelwahab, R.; Browning, D.F.; Aly, S.A. Genome Characterization of Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae Strains, Carrying Hybrid Resistance-Virulence IncHI1B/FIB Plasmids, Isolated from an Egyptian Pediatric ICU. Microorganisms 2025, 13, 1058. https://doi.org/10.3390/microorganisms13051058
Hammad HA, Abdelwahab R, Browning DF, Aly SA. Genome Characterization of Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae Strains, Carrying Hybrid Resistance-Virulence IncHI1B/FIB Plasmids, Isolated from an Egyptian Pediatric ICU. Microorganisms. 2025; 13(5):1058. https://doi.org/10.3390/microorganisms13051058
Chicago/Turabian StyleHammad, Heba A., Radwa Abdelwahab, Douglas F. Browning, and Sherine A. Aly. 2025. "Genome Characterization of Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae Strains, Carrying Hybrid Resistance-Virulence IncHI1B/FIB Plasmids, Isolated from an Egyptian Pediatric ICU" Microorganisms 13, no. 5: 1058. https://doi.org/10.3390/microorganisms13051058
APA StyleHammad, H. A., Abdelwahab, R., Browning, D. F., & Aly, S. A. (2025). Genome Characterization of Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae Strains, Carrying Hybrid Resistance-Virulence IncHI1B/FIB Plasmids, Isolated from an Egyptian Pediatric ICU. Microorganisms, 13(5), 1058. https://doi.org/10.3390/microorganisms13051058






