Divergent Driving Mechanisms Shape the Temporal Dynamics of Benthic Prokaryotic and Eukaryotic Microbial Communities in Coastal Subtidal Zones
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area, Sampling, and Environmental Factors
2.2. DNA Extraction, PCR Amplification, and Sequencing
2.3. Sequence Analysis
2.4. Analysis of Community Assembly Mechanisms
2.5. Co-Occurrence Network Analysis
2.6. Statistical Analysis
3. Results
3.1. Temporal Dynamics of Environmental Factors
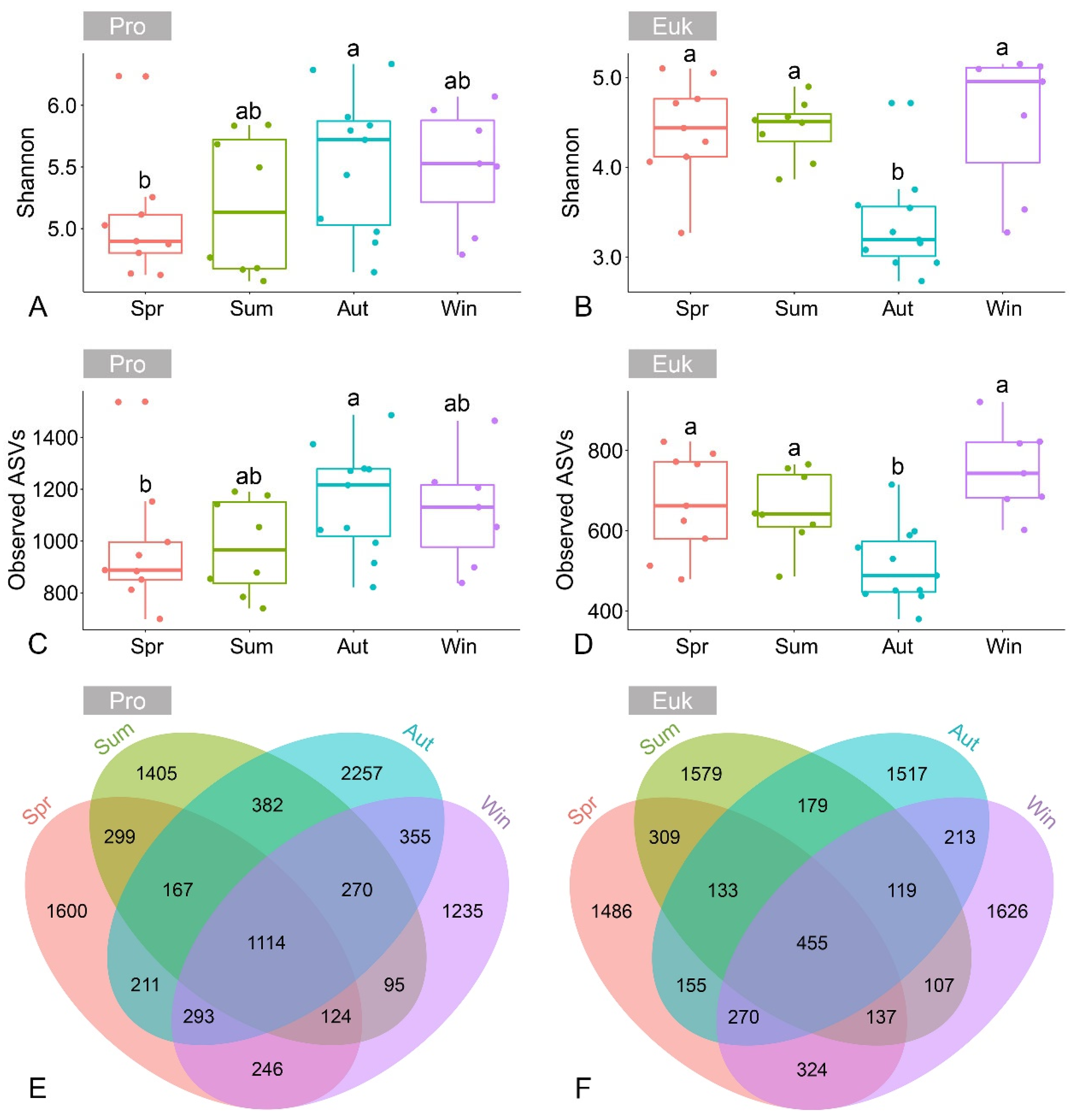
3.2. Alpha and Beta Diversity of Benthic Microbial Communities
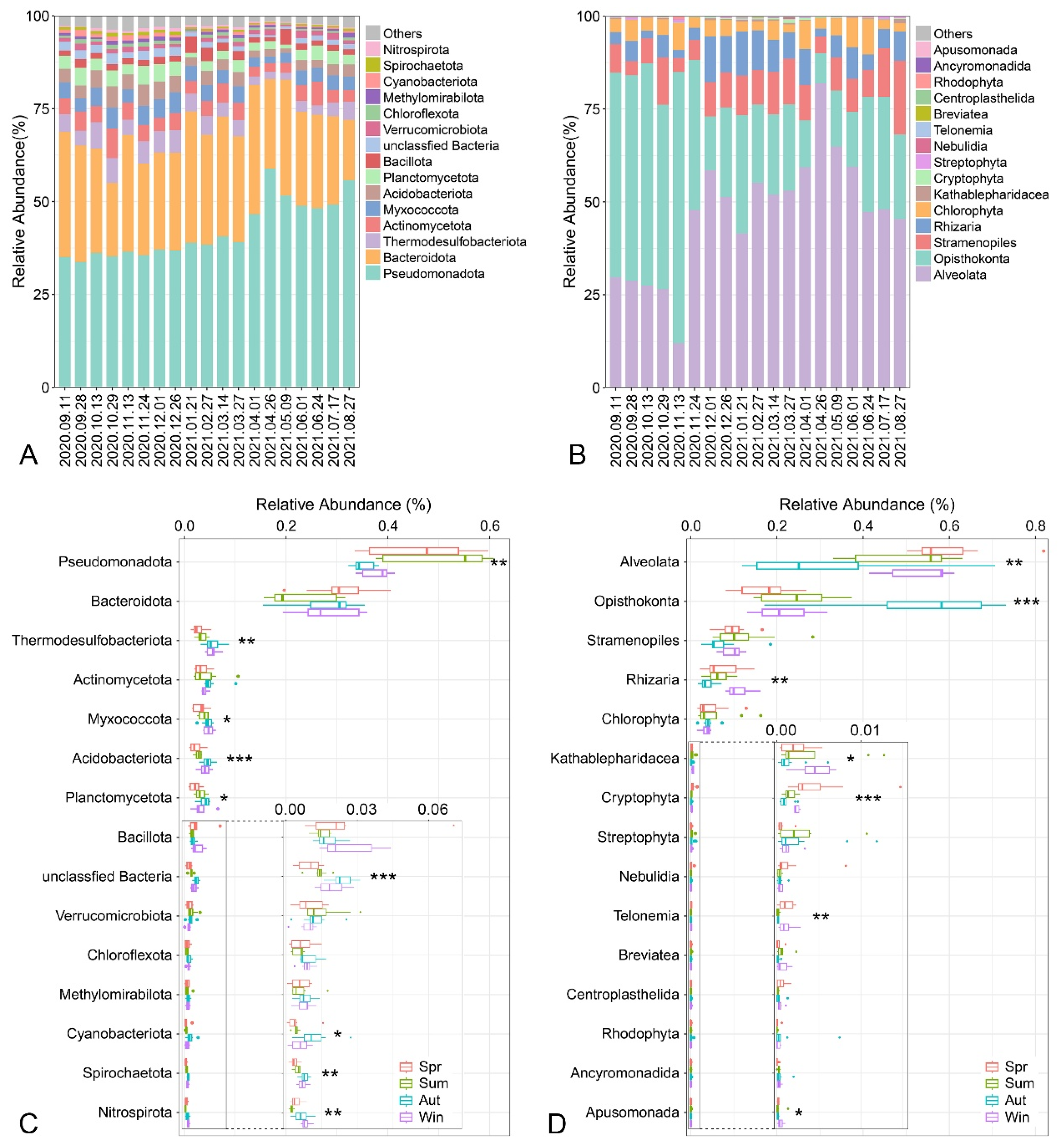
3.3. Community Composition of Benthic Microbial Communities
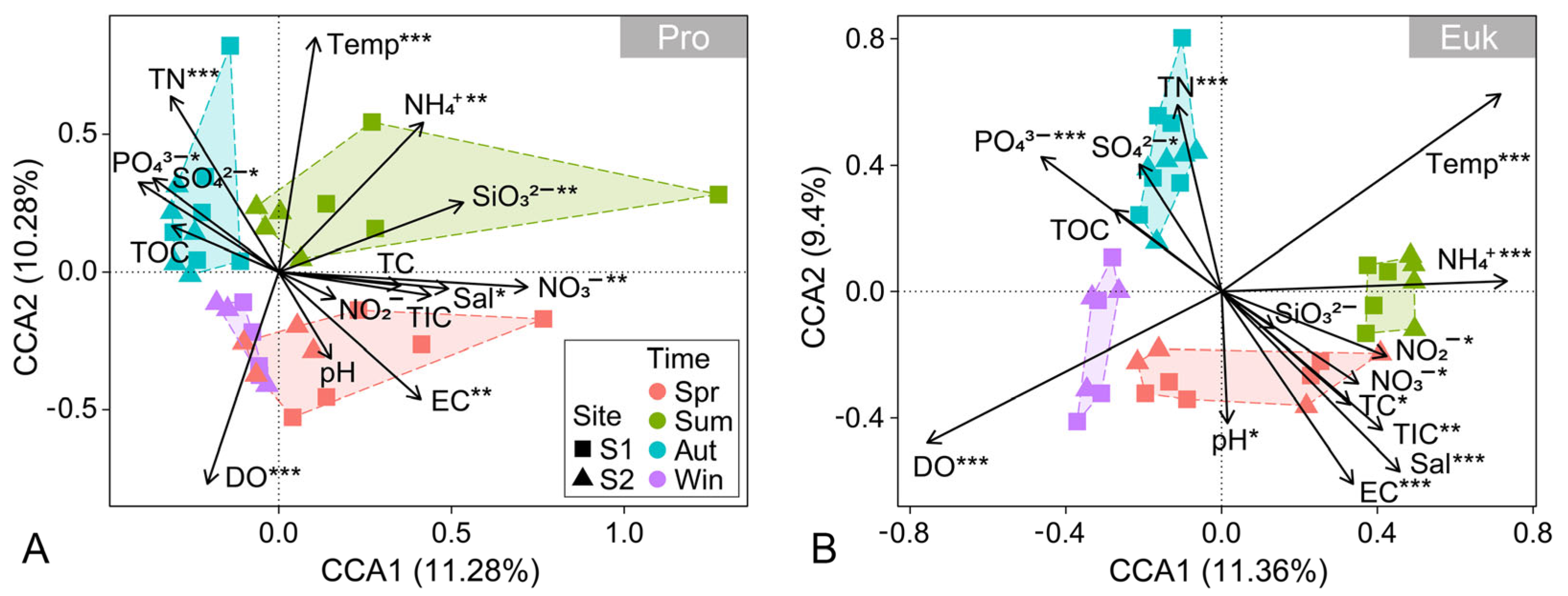
3.4. Influence of Environmental Factors on Benthic Microbial Communities
3.5. Assembly Process of Benthic Microbial Communities
3.6. Co-Occurrence Networks and Community Stability of Benthic Microbial Communities
4. Discussion
4.1. Divergent Responses of Benthic Prokaryotic and Eukaryotic Microbial Communities to Environmental Factors
4.2. The Relative Importance of Deterministic and Stochastic Processes Varied Between Benthic Prokaryotic and Eukaryotic Microbial Communities
4.3. Co-Occurrence Patterns and Seasonal Variations in the Community Stability of Benthic Prokaryotic and Eukaryotic Microbial Communities
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huettel, M.; Berg, P.; Kostka, J.E. Benthic exchange and biogeochemical cycling in permeable sediments. Annu. Rev. Mar. Sci. 2014, 6, 23–51. [Google Scholar] [CrossRef] [PubMed]
- Miksch, S.; Orellana, L.H.; Oggerin de Orube, M.; Vidal-Melgosa, S.; Solanki, V.; Hehemann, J.H.; Amann, R.; Knittel, K. Taxonomic and functional stability overrules seasonality in polar benthic microbiomes. ISME J. 2024, 18, wrad005. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.J.; Leung, P.M.; Cook, P.L.M.; Wong, W.W.; Hutchinson, T.; Eate, V.; Kessler, A.J.; Greening, C. Hydrodynamic disturbance controls microbial community assembly and biogeochemical processes in coastal sediments. ISME J. 2022, 16, 750–763. [Google Scholar] [CrossRef] [PubMed]
- Seidel, L.; Ketzer, M.; Broman, E.; Shahabi-Ghahfarokhi, S.; Rahmati-Abkenar, M.; Turner, S.; Ståhle, M.; Bergström, K.; Manoharan, L.; Ali, A.; et al. Weakened resilience of benthic microbial communities in the face of climate change. ISME Commun. 2022, 2, 21. [Google Scholar] [CrossRef]
- Moreira, V.A.; Cravo-Laureau, C.; de Carvalho, A.C.B.; Baldy, A.; Bidone, E.D.; Sabadini-Santos, E.; Duran, R. Microbial indicators along a metallic contamination gradient in tropical coastal sediments. J. Hazard. Mater. 2023, 443, 130244. [Google Scholar] [CrossRef]
- Veloso, S.; Amouroux, D.; Lanceleur, L.; Cagnon, C.; Monperrus, M.; Deborde, J.; Laureau, C.C.; Duran, R. Keystone microbial taxa organize micropollutant-related modules shaping the microbial community structure in estuarine sediments. J. Hazard. Mater. 2023, 448, 130858. [Google Scholar] [CrossRef]
- Forehead, H.; Thomson, P.; Kendrick, G.A. Shifts in composition of microbial communities of subtidal sandy sediments maximise retention of nutrients. FEMS Microbiol. Ecol. 2013, 83, 279–298. [Google Scholar] [CrossRef]
- Böer, S.I.; Hedtkamp, S.I.; van Beusekom, J.E.; Fuhrman, J.A.; Boetius, A.; Ramette, A. Time- and sediment depth-related variations in bacterial diversity and community structure in subtidal sands. ISME J. 2009, 3, 780–791. [Google Scholar] [CrossRef]
- Conte, A.; Papale, M.; Amalfitano, S.; Mikkonen, A.; Rizzo, C.; De Domenico, E.; Michaud, L.; Giudice, A.L. Bacterial community structure along the subtidal sandy sediment belt of a high Arctic fjord (Kongsfjorden, Svalbard Islands). Sci. Total Environ. 2018, 619–620, 203–211. [Google Scholar] [CrossRef]
- Lv, X.; Ma, B.; Yu, J.; Chang, S.X.; Xu, J.; Li, Y.; Wang, G.; Han, G.; Bo, G.; Chu, X. Bacterial community structure and function shift along a successional series of tidal flats in the Yellow River Delta. Sci. Rep. 2016, 6, 36550. [Google Scholar] [CrossRef]
- Stüeken, E.E.; Pellerin, A.; Thomazo, C.; Johnson, B.W.; Duncanson, S.; Schoepfer, S.D. Marine biogeochemical nitrogen cycling through Earth’s history. Nat. Rev. Earth Environ. 2024, 5, 732–747. [Google Scholar] [CrossRef]
- Ning, D.; Yuan, M.; Wu, L.; Zhang, Y.; Guo, X.; Zhou, X.; Yang, Y.; Arkin, A.P.; Firestone, M.K.; Zhou, J. A quantitative framework reveals ecological drivers of grassland microbial community assembly in response to warming. Nat. Commun. 2020, 11, 4717. [Google Scholar] [CrossRef] [PubMed]
- Dini-Andreote, F.; Stegen, J.C.; van Elsas, J.D.; Salles, J.F. Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc. Natl. Acad. Sci. USA 2015, 112, E1326–E1332. [Google Scholar] [CrossRef] [PubMed]
- Stegen, J.C.; Lin, X.; Fredrickson, J.K.; Chen, X.; Kennedy, D.W.; Murray, C.J.; Rockhold, M.L.; Konopka, A. Quantifying community assembly processes and identifying features that impose them. ISME J. 2013, 7, 2069–2079. [Google Scholar] [CrossRef]
- Zhou, J.; Ning, D. Stochastic community assembly: Does it matter in microbial ecology? Microbiol. Mol. Biol. Rev. 2017, 81, e00002–e00017. [Google Scholar] [CrossRef]
- Zhang, T.; Xu, S.; Yan, R.; Wang, R.; Gao, Y.; Kong, M.; Yi, Q.; Zhang, Y. Similar geographic patterns but distinct assembly processes of abundant and rare bacterioplankton communities in river networks of the Taihu Basin. Water Res. 2022, 211, 118057. [Google Scholar] [CrossRef]
- Qi, H.; Huang, D.; Wang, F.; Ye, M.; Jiang, X. Spatial dynamics of prokaryotic microbial communities in sediments of the Yellow Sea: Assembly process, co-occurrence relationships, and environmental implications. J. Environ. Manag. 2022, 319, 115645. [Google Scholar] [CrossRef]
- Li, Y.; Chen, J.; Wang, L.; Wang, D.; Niu, L.; Zheng, J. Hydrodynamic disturbance and nutrient accumulation co-shape the depth-dependent prokaryotic community assembly in intertidal sediments of a mountainous river estuary. J. Hydrol. 2025, 651, 132580. [Google Scholar] [CrossRef]
- Faust, K.; Raes, J. Microbial interactions: From networks to models. Nat. Rev. Microbiol. 2012, 10, 538–550. [Google Scholar] [CrossRef]
- Gao, G.; Peng, D.; Zhang, Y.; Li, Y.; Fan, K.; Tripathi, B.M.; Adams, J.M.; Chu, H. Dramatic change of bacterial assembly process and co-occurrence pattern in Spartina alterniflora salt marsh along an inundation frequency gradient. Sci. Total Environ. 2021, 755, 142546. [Google Scholar] [CrossRef]
- Yu, S.; Wang, C.; Li, H.; Zhang, X.; Wang, X.; Qu, W. Field and numerical investigations of wave effects on groundwater flow and salt transport in a sandy beach. Water Resour. Res. 2022, 58, e2022WR032077. [Google Scholar] [CrossRef]
- Dong, J.; Wang, X.; Bidegain, G.; Sun, X.; Bian, X.; Zhang, X. Assessment of the benthic ecological quality status (EcoQs) of Laizhou Bay (China) with an integrated AMBI, M−AMBI, BENTIX, BO2A and feeding evenness index. Ecol. Indic. 2023, 153, 110456. [Google Scholar] [CrossRef]
- Shu, W.; Huang, L. Microbial diversity in extreme environments. Nat. Rev. Microbiol. 2022, 20, 219–235. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Bhagwat, G.; Palanisami, T.; Liang, S.; Wan, W.; Yang, Y. Lacustrine plastisphere: Distinct succession and assembly processes of prokaryotic and eukaryotic communities and role of site, time, and polymer types. Water Res. 2024, 248, 120875. [Google Scholar] [CrossRef]
- Mu, K.; Tang, C.; Tosi, L.; Li, Y.; Zheng, X.; Donnici, S.; Sun, J.; Liu, J.; Gao, X. Coastline monitoring and prediction based on long-term remote sensing data—A case study of the eastern coast of laizhou bay, china. Remote Sens. 2024, 16, 185. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef]
- Halbach, L.; Kitzinger, K.; Hansen, M.; Littmann, S.; Benning, L.G.; Bradley, J.A.; Whitehouse, M.J.; Olofsson, M.; Mourot, R.; Tranter, M.; et al. Single-cell imaging reveals efficient nutrient uptake and growth of microalgae darkening the Greenland Ice Sheet. Nat. Commun. 2025, 16, 1521. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Guillou, L.; Bachar, D.; Audic, S.; Bass, D.; Berney, C.; Bittner, L.; Boutte, C.; Burgaud, G.; De Vargas, C.; Decelle, J.; et al. The Protist Ribosomal Reference database (PR2): A catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2012, 41, D597–D604. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, M.; Yan, Q.; Mao, C.; Shu, W.; Al-Farraj, S.A.; Song, W.; Dunthorn, M.; Jiang, Y.; Huang, J. Temporal dynamics and potential driving mechanisms of bacterioplankton communities in lake ecosystems: Insights from water diversion events. J. Hydrol. 2025, 660, 133335. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X.; Chi, Y.; Song, W.; Yan, Q.; Huang, J. Changes of the freshwater microbial community structure and assembly processes during different sample storage conditions. Microorganisms 2022, 10, 1176. [Google Scholar] [CrossRef] [PubMed]
- Ning, D.; Deng, Y.; Tiedje, J.M.; Zhou, J. A general framework for quantitatively assessing ecological stochasticity. Proc. Natl. Acad. Sci. USA 2019, 116, 16892–16898. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Jiang, Y.; Yang, Y.; He, Z.; Luo, F.; Zhou, J. Molecular ecological network analyses. BMC Bioinform. 2012, 13, 113. [Google Scholar] [CrossRef] [PubMed]
- Jiao, C.; Zhao, D.; Zhou, T.; Wu, Q.L.; Zeng, J. Habitat-specific regulation of bacterial community dynamics during phytoplankton bloom succession in a subtropical eutrophic lake. Water Res. 2023, 242, 120252. [Google Scholar] [CrossRef]
- Erdős, P.; Rényi, A. On the evolution of random graphs. Publ. Math. Inst. Hung. Acad. Sci 1960, 5, 17–60. [Google Scholar]
- Telesford, Q.K.; Joyce, K.E.; Hayasaka, S.; Burdette, J.H.; Laurienti, P.J. The ubiquity of small-world networks. Brain Connect. 2011, 1, 367–375. [Google Scholar] [CrossRef]
- Zhu, C.; Liu, W.; Li, X.; Xu, Y.; El-Serehy, H.A.; Al-Farraj, S.A.; Ma, H.; Stoeck, T.; Yi, Z. High salinity gradients and intermediate spatial scales shaped similar biogeographical and co-occurrence patterns of microeukaryotes in a tropical freshwater-saltwater ecosystem. Environ. Microbiol. 2021, 23, 4778–4796. [Google Scholar] [CrossRef]
- Olesen, J.M.; Bascompte, J.; Dupont, Y.L.; Jordano, P. The modularity of pollination networks. Proc. Natl. Acad. Sci. USA 2007, 104, 19891–19896. [Google Scholar] [CrossRef]
- Zheng, X.; Xu, K.; Naoum, J.; Lian, Y.; Wu, B.; He, Z.; Yan, Q. Deciphering microeukaryotic–bacterial co-occurrence networks in coastal aquaculture ponds. Mar. Life Sci. Technol. 2023, 5, 44–55. [Google Scholar] [CrossRef]
- Baselga, A. Partitioning abundance-based multiple-site dissimilarity into components: Balanced variation in abundance and abundance gradients. Methods Ecol. Evol. 2017, 8, 799–808. [Google Scholar] [CrossRef]
- Baselga, A.; Orme, C.D.L. betapart: An R package for the study of beta diversity. Methods Ecol. Evol. 2012, 3, 808–812. [Google Scholar] [CrossRef]
- Xun, W.; Liu, Y.; Li, W.; Ren, Y.; Xiong, W.; Xu, Z.; Zhang, N.; Miao, Y.; Shen, Q.; Zhang, R. Specialized metabolic functions of keystone taxa sustain soil microbiome stability. Microbiome 2021, 9, 35. [Google Scholar] [CrossRef] [PubMed]
- Crump, B.C.; Bowen, J.L. The microbial ecology of estuarine ecosystems. Annu. Rev. Mar. Sci. 2024, 16, 335–360. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.M.; Leigh, M.B.; Mincks, S.L. Patterns in benthic microbial community structure across environmental gradients in the Beaufort Sea shelf and slope. Front. Microbiol. 2021, 12, 581124. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, L.; Zhang, G.; Gao, H.; Chen, X.; Li, L.; Ju, F. Hydrodynamic and anthropogenic disturbances co-shape microbiota rhythmicity and community assembly within intertidal groundwater-surface water continuum. Water Res. 2023, 242, 120236. [Google Scholar] [CrossRef]
- Nyirabuhoro, P.; Liu, M.; Xiao, P.; Liu, L.; Yu, Z.; Wang, L.; Yang, J. Seasonal variability of conditionally rare taxa in the water column bacterioplankton community of subtropical reservoirs in China. Microb. Ecol. 2020, 80, 14–26. [Google Scholar] [CrossRef]
- Fuhrman, J.A.; Cram, J.A.; Needham, D.M. Marine microbial community dynamics and their ecological interpretation. Nat. Rev. Microbiol. 2015, 13, 133–146. [Google Scholar] [CrossRef]
- Cook, L.S.J.; Briscoe, A.G.; Fonseca, V.G.; Boenigk, J.; Woodward, G.; Bass, D. Microbial, holobiont, and Tree of Life eDNA/eRNA for enhanced ecological assessment. Trends Microbiol. 2025, 33, 48–65. [Google Scholar] [CrossRef]
- Guo, Z.; Lu, W.; Minpeng, S.; Liyuan, S.; Zhenlin, L.; Wenjing, C.; Xiaoyong, L.; Bo, Z.; Ha, K.J.; Zhaoyang, J. Seasonal dynamics response mechanism of benthic microbial community to artificial reef habitats. Environ. Res. 2024, 243, 117867. [Google Scholar] [CrossRef]
- Kalu, E.I.; Reyes-Prieto, A.; Barbeau, M.A. Community dynamics of microbial eukaryotes in intertidal mudflats in the hypertidal Bay of Fundy. ISME Commun. 2023, 3, 21. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Steele, J.A.; Caporaso, J.G.; Steinbrück, L.; Reeder, J.; Temperton, B.; Huse, S.; McHardy, A.C.; Knight, R.; Joint, I.; et al. Defining seasonal marine microbial community dynamics. ISME J. 2012, 6, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Hervé, V.; Morelle, J.; Lambourdière, J.; Lopez, P.J.; Claquin, P. Together throughout the year: Seasonal patterns of bacterial and eukaryotic microbial communities in a macrotidal estuary. Environ. Microbiome 2025, 20, 8. [Google Scholar] [CrossRef] [PubMed]
- Huisman, J.; Codd, G.A.; Paerl, H.W.; Ibelings, B.W.; Verspagen, J.M.H.; Visser, P.M. Cyanobacterial blooms. Nat. Rev. Microbiol. 2018, 16, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhang, P.; Li, X.; Yang, S.; Chao, X.; Liu, H.; Ba, S. Distribution patterns and community assembly processes of eukaryotic microorganisms along an altitudinal gradient in the middle reaches of the Yarlung Zangbo River. Water Res. 2023, 239, 120047. [Google Scholar] [CrossRef]
- Li, S.; Ren, K.; Yan, X.; Tsyganov, A.N.; Mazei, Y.; Smirnov, A.; Mazei, N.; Zhang, Y.; Rensing, C.; Yang, J. Linking biodiversity and ecological function through extensive microeukaryotic movement across different habitats in six urban parks. iMeta 2023, 2, e103. [Google Scholar] [CrossRef]
- Yan, J.; Zhai, F.; Gu, Y.; Liu, X.; Li, P.; Liu, Z.; Wang, Y. Drastic fluctuation in water exchange between the Yellow sea and Bohai sea caused by typhoon Lekima in August 2019: A numerical study. J. Geophys. Res. Oceans 2023, 128, e2022JC019260. [Google Scholar] [CrossRef]
- Jiang, T.; Wu, G.; Niu, P.; Cui, Z.; Bian, X.; Xie, Y.; Shi, H.; Xu, X.; Qu, K. Short-term changes in algal blooms and phytoplankton community after the passage of Super Typhoon Lekima in a temperate and inner sea (Bohai Sea) in China. Ecotoxicol. Environ. Saf. 2022, 232, 113223. [Google Scholar] [CrossRef]
- Logares, R. Decoding populations in the ocean microbiome. Microbiome 2024, 12, 67. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Z.; Wagg, C.; Castellano, M.J.; Zhang, N.; Ding, W. Soil organic carbon loss decreases biodiversity but stimulates multitrophic interactions that promote belowground metabolism. Glob. Chang. Biol. 2024, 30, e17101. [Google Scholar] [CrossRef]
- Hicks, N.; Liu, X.; Gregory, R.; Kenny, J.; Lucaci, A.; Lenzi, L.; Paterson, D.M.; Duncan, K.R. Temperature driven changes in benthic bacterial diversity influences biogeochemical cycling in coastal sediments. Front. Microbiol. 2018, 9, 1730. [Google Scholar] [CrossRef]
- Li, T.; Liu, G.; Yuan, H.; Chen, J.; Lin, X.; Li, H.; Yu, L.; Wang, C.; Li, L.; Zhuang, Y.; et al. Eukaryotic plankton community assembly and influencing factors between continental shelf and slope sites in the northern South China Sea. Environ. Res. 2023, 216, 114584. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, W.; Kou, Y.; Fang, K.; Liu, Y.; He, H.; Liu, Q. The contrasting responses of abundant and rare microbial community structures and co-occurrence networks to secondary forest succession in the subalpine region. Front. Microbiol. 2023, 14, 1177239. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.B.; Jeong, D.H.; Cho, B.C.; Park, J.S. The diversity patterns of rare to abundant microbial eukaryotes across a broad range of salinities in a solar saltern. Microb. Ecol. 2022, 84, 1103–1121. [Google Scholar] [CrossRef] [PubMed]
- Neury-Ormanni, J.; Vedrenne, J.; Morin, S. Benthic diatom growth kinetics under combined pressures of microalgal competition, predation and chemical stressors. Sci. Total Environ. 2020, 734, 139484. [Google Scholar] [CrossRef]
- Nemergut, D.R.; Schmidt, S.K.; Fukami, T.; O’Neill, S.P.; Bilinski, T.M.; Stanish, L.F.; Knelman, J.E.; Darcy, J.L.; Lynch, R.C.; Wickey, P.; et al. Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 2013, 77, 342–356. [Google Scholar] [CrossRef]
- Zheng, P.; Mao, A.; Meng, S.; Yu, F.; Zhang, S.; Lun, J.; Li, J.; Hu, Z. Assembly mechanism of microbial community under different seasons in Shantou sea area. Mar. Pollut. Bull. 2024, 205, 116550. [Google Scholar] [CrossRef]
- Zhang, Z.; Han, P.; Zheng, Y.; Jiao, S.; Dong, H.; Liang, X.; Gao, D.; Niu, Y.; Yin, G.; Liu, M.; et al. Spatiotemporal dynamics of bacterial taxonomic and functional profiles in estuarine intertidal soils of China coastal zone. Microb. Ecol. 2023, 85, 383–399. [Google Scholar] [CrossRef]
- Wei, J.; Zhang, Q.; Yin, Y.; Peng, K.; Wang, L.; Cai, Y.; Gong, Z. Limited impacts of water diversion on micro-eukaryotic community along the eastern route of China’s South-to-North Water Diversion Project. Water Res. 2024, 262, 122109. [Google Scholar] [CrossRef]
- Liu, Q.; Li, Y.; Wang, H.; Yang, G.; Kan, J.; Yang, M.; Yu, X.; Guo, C.; Wang, M.; Wang, W.; et al. Assembly and network stability of planktonic microorganisms under the influence of salinity gradient: An arctic case study from the lena river estuary to the Laptev Sea. Microbiol. Spectr. 2023, 11, e0211522. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, H.; Huang, N.; Peng, X.; Jing, H. Geographical distribution and driving force of micro-eukaryotes in the seamount sediments along the island arc of the Yap and Mariana trenches. Microbiol. Spectr. 2023, 11, e0206923. [Google Scholar] [CrossRef]
- Pearman, J.K.; Adamson, J.; Thomson-Laing, G.; Thompson, L.; Waters, S.; Vandergoes, M.J.; Howarth, J.D.; Wood, S.A. Deterministic processes drive national-scale patterns in lake surface sediment bacteria and eukaryotic assemblage composition. Limnol. Oceanogr. 2023, 68, 40–55. [Google Scholar] [CrossRef]
- Barberán, A.; Bates, S.T.; Casamayor, E.O.; Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012, 6, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Vandenkoornhuyse, P.; Li, L.; Guo, J.; Zhu, C.; Guo, S.; Ling, N.; Shen, Q. Microbial generalists and specialists differently contribute to the community diversity in farmland soils. J. Adv. Res. 2022, 40, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Thébault, E.; Fontaine, C. Stability of ecological communities and the architecture of mutualistic and trophic networks. Science 2010, 329, 853–856. [Google Scholar] [CrossRef]
- Chen, C.; Li, P.; Yin, M.; Wang, J.; Sun, Y.; Ju, W.; Liu, L.; Li, Z. Deciphering characterization of seasonal variations in microbial communities of marine ranching: Diversity, co-occurrence network patterns, and assembly processes. Mar. Pollut. Bull. 2023, 197, 115739. [Google Scholar] [CrossRef]
- Liu, S.; Yu, H.; Yu, Y.; Huang, J.; Zhou, Z.; Zeng, J.; Chen, P.; Xiao, F.; He, Z.; Yan, Q. Ecological stability of microbial communities in Lake Donghu regulated by keystone taxa. Ecol. Indic. 2022, 136, 108695. [Google Scholar] [CrossRef]
- Banerjee, S.; Schlaeppi, K.; van der Heijden, M.G.A. Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 2018, 16, 567–576. [Google Scholar] [CrossRef]
- Kerfahi, D.; Guo, Y.; Dong, K.; Wang, Q.; Adams, J.M. pH is the major predictor of soil microbial network complexity in Chinese forests along a latitudinal gradient. Catena 2024, 234, 107595. [Google Scholar] [CrossRef]
- Marinchel, N.; Casabianca, S.; Marchesini, A.; Vernesi, C.; Scardi, M.; Penna, A. Structural variability of protist assemblages in surface sediments across Italian Mediterranean marine subregions. Front. Mar. Sci. 2024, 11, 1427357. [Google Scholar] [CrossRef]
- Li, Y.; Guo, Z.; Ni, S. Dynamic stratification and biogeochemical cycling response of microbial communities in east indian ocean ridge. Chem. Eng. J. 2024, 487, 150568. [Google Scholar] [CrossRef]
- Guo, P.; Li, C.; Liu, J.; Chai, B. Predation has a significant impact on the complexity and stability of microbial food webs in subalpine lakes. Microbiol. Spectr. 2023, 11, e0241123. [Google Scholar] [CrossRef] [PubMed]
- Che, Y.; Lin, C.; Li, S.; Liu, J.; Zhu, L.; Yu, S.; Wang, N.; Li, H.; Bao, M.; Zhou, Y.; et al. Influences of hydrodynamics on microbial community assembly and organic carbon composition of resuspended sediments in shallow marginal seas. Water Res. 2024, 248, 120882. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, H.; Guo, Y.; Yan, Q.; Chen, J. Human-induced homogenization of microbial taxa and function in a subtropical river and its impacts on community stability. Water Res. 2024, 252, 121198. [Google Scholar] [CrossRef] [PubMed]
- Morriën, E.; Hannula, S.E.; Snoek, L.B.; Helmsing, N.R.; Zweers, H.; de Hollander, M.; Soto, R.L.; Bouffaud, M.-L.; Buée, M.; Dimmers, W.; et al. Soil networks become more connected and take up more carbon as nature restoration progresses. Nat. Commun. 2017, 8, 14349. [Google Scholar] [CrossRef]
- Lin, L.; Xiong, J.; Liu, L.; Wang, F.; Cao, W.; Xu, W. Microbial interactions strengthen deterministic processes during community assembly in a subtropical estuary. Sci. Total Environ. 2024, 906, 167499. [Google Scholar] [CrossRef]
- Calcagno, V.; Jarne, P.; Loreau, M.; Mouquet, N.; David, P. Diversity spurs diversification in ecological communities. Nat. Commun. 2017, 8, 15810. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, D.; Zhang, J.; Li, F.; Li, W.; Bi, L.; Li, W.; Fu, Y.; Wang, Y. Divergent Driving Mechanisms Shape the Temporal Dynamics of Benthic Prokaryotic and Eukaryotic Microbial Communities in Coastal Subtidal Zones. Microorganisms 2025, 13, 1050. https://doi.org/10.3390/microorganisms13051050
Ji D, Zhang J, Li F, Li W, Bi L, Li W, Fu Y, Wang Y. Divergent Driving Mechanisms Shape the Temporal Dynamics of Benthic Prokaryotic and Eukaryotic Microbial Communities in Coastal Subtidal Zones. Microorganisms. 2025; 13(5):1050. https://doi.org/10.3390/microorganisms13051050
Chicago/Turabian StyleJi, Daode, Jianfeng Zhang, Fan Li, Wensheng Li, Luping Bi, Wenlu Li, Yingjun Fu, and Yunfeng Wang. 2025. "Divergent Driving Mechanisms Shape the Temporal Dynamics of Benthic Prokaryotic and Eukaryotic Microbial Communities in Coastal Subtidal Zones" Microorganisms 13, no. 5: 1050. https://doi.org/10.3390/microorganisms13051050
APA StyleJi, D., Zhang, J., Li, F., Li, W., Bi, L., Li, W., Fu, Y., & Wang, Y. (2025). Divergent Driving Mechanisms Shape the Temporal Dynamics of Benthic Prokaryotic and Eukaryotic Microbial Communities in Coastal Subtidal Zones. Microorganisms, 13(5), 1050. https://doi.org/10.3390/microorganisms13051050





