Abstract
This systematic review aimed to assess the prevalence of Escherichia coli O157:H7 using a One Health approach, integrating data from human, animal, and environmental sources across Africa. Following PRISMA guidelines, studies reporting on E. coli O157:H7 in human, animal, and environment samples from African countries were retrieved from PubMed, Scopus, Web of Science, and Google Scholar. All data were analyzed using a binary random-effects model by the DerSimonian–Laird method at a 95% confidence interval. Out of 1757 publications generated, 56 from 9 countries including Ethiopia (17/56), South Africa (13/56), Nigeria (10/56), Egypt (9/56), Ghana (2/56), Tanzania (2/56), Benin (1/56), Namibia (1/56), and Senegal (1/56) were included. The pooled prevalence of E. coli O157:H7 was 4.7%, with the highest prevalence observed among animal samples (5.4%) followed by the environmental and human samples (3.4 and 2.8%, respectively). The pooled prevalence of antibiotic resistance was observed to be 96.5%, 82.8%, 76.8%, 70.7%, 62.1%, 50.4%, and 40.2% for cefoxitin, ampicillin, cefuroxime, nitrofurantoin, amikacin, amoxiclav, and ciprofloxacin, respectively. This distribution highlights the interconnectedness between animals, the environment, and human populations in the transmission and persistence of this pathogen and the need to implement a suitable and appropriate One Health pathogenic and antimicrobial resistance surveillance system in the African region.
1. Introduction
Escherichia coli (E. coli) is one of the many bacteria that live in the intestines of healthy humans and most warm-blooded animals. Most strains are harmless and help with digestion, but some strains can cause severe illness. E. coli O157:H7 serotype is a recognized food and waterborne pathogen that causes severe intestinal infection in humans [1,2]. It is the most common strain that causes acute hemorrhagic diarrhea, which may progress to hemolytic uremic syndrome, with systemic complications occurring more frequently in children [3]. E. coli O157:H7 can be differentiated from other E. coli by the production of a potent toxin, the Shiga toxin, which damages the lining of the intestinal wall leading to bloody diarrhea [4]. In Africa, Shiga toxin-producing E. coli (STEC) infections are estimated at 10,200 cases annually, with an incidence rate of 1.4 cases per 100,000 person per year [5]. Among these, E. coli O157:H7, a key STEC serotype, contributes 10% of reported infections [5,6].
Transmission occurs via the fecal–oral route after the consumption of contaminated, undercooked liquids, and foods or through person-to-person via fecal shedding [1]. Studies have identified cattle and other ruminants as the main reservoirs of E. coli O157:H7, with the pathogen being isolated from animals, food products, clinical samples, and environmental sources across all continents [5].
Reports have indicated prevalence of E. coli O157:H7 in cattle, sheep, goats, beef, meat products, chicken, dairy products, milk, fruits, and vegetables from several countries including Egypt, Algeria, and Libya [7]. In Ethiopia, the prevalence of E. coli O157:H7 was found to be 4% (95% CI = 3–5%) in foods of animal origin [8]. Similarly, in the North West Province of South Africa, the prevalence of E. coli O157:H7 was 9.5% from human, cattle, and pig samples [9]. According to community-based prevalence studies, the E. coli O157:H7 strain is responsible for 20 and 15.3% of E. coli infections in Nigeria [10] and Ethiopia [11], respectively.
Recently, several systematic reviews in Africa have focused on the epidemiology of E. coli using the One Health approach [12,13,14,15]. However, to the best of our knowledge, there is limited pooled data on the prevalence of serotype E. coli O157:H7 in any systematic review using the One Health approach. Nevertheless, the few data reported have been those described in only a few African countries [16,17,18,19,20]. This systematic review was undertaken, to report on the pooled prevalence of E. coli O157:H7 using a One Health perspective across humans, animals, and the environment. Specifically, we aimed to (i) evaluate the pooled prevalence of E. coli O157:H7, (ii) identify the most favorable host to the spread of E. coli O157:H7 among these three components, and (iii) determine the pooled prevalence of the antibiotic resistance profile of the bacteria.
2. Materials and Methods
2.1. Study Design
This systematic review was conducted from 7 September 2024 to 28 October 2024 following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (Supplementary Table S1). Studies focused on African countries and published between 1 January 2002 and 28 October 2024 were eligible for inclusion. Literature searches were conducted in PubMed, Scopus, Web of Science, and Google Scholar using Boolean operators to combine specific keywords such as Escherichia coli O157:H7, One Health, clinical and environmental samples, food products, and water sources, along with individual African country names (Supplementary Table S2). Only articles published in English and French were considered for inclusion.
2.2. Study Eligibility Criteria
All search results from the different databases were exported to Excel software (v.13) and compiled. Rayyan was utilized to eliminate duplicates and to categorize and consolidate the results (available at https://www.rayyan.com/, accessed on 7 September 2024), while Endnote X9 software (v.20) was used to manage the collected publications and citations.
Inclusion criteria: Full-text research articles reporting on E. coli O157:H7 isolated in African countries if they investigated its presence in human–animal–environment, animal–human, human–environment, or animal–environment contexts. Studies were also required to provide details on the study population, sample source, number of isolates, and methods used to detect E. coli O157:H7. The literature search spanned all publications in the last 22 years (1 January 2002 to 28 October 2024) and was conducted by NSS and TOA.
Exclusion criteria: Articles were excluded if they contained incomplete information or focused solely on clinical, animal/livestock, or environmental samples without a broader context. Additionally, review articles, conference proceedings, duplicate publications, abstracts, posters, short communications, letters, and studies from non-African countries were not considered.
2.3. Screening and Data Extraction
Full texts of the selected publications were screened using Rayyan AI (https://www.rayyan.com/, accessed on 17 September 2024). The titles and abstracts relevant to the study question were carefully reviewed. Next, the selected papers at this stage further underwent full-text reviews, and only studies meeting the inclusion criteria were included in the review. Data were systematically collected and organized in an MS Excel spreadsheet by two reviewers, NSS and TOA. Both reviewers performed data extraction separately, and their results were compared for consistency. Extracted data included the study period, publication year, country, sampling population (specific animal, human, or environmental source/host), reservoirs studied (human–animal–environment, animal–human, animal–environment, human–environment), methods for E. coli O157:H7 detection, number of isolates from each source (animal, human, environment), prevalence of isolates, and multi-drug resistance pattern.
2.4. Study Quality
The Joanna Briggs Institute (JBI) checklist for prevalence studies was used to assess the quality of the studies [21] and was carried out by two independent reviewers (NSS and TOA). The JBI checklist contains nine questions that were weighted as follows: 1 for a YES response or 0 for a NO response (Supplementary Table S3).
2.5. Data Analysis
The extracted data were used for descriptive statistics. Further analysis was carried out in multiple steps. Excel 2013 was used for data entry, while the meta-analysis, forest plots, and funnel plots of E. coli O157:H7 as well as the estimation of the country effect were conducted using the comprehensive meta-analysis. All data generated were analyzed using the binary random-effects model. This model is based on the DerSimonian–Laird method with a 95% confidence interval. The random-effects model was used to calculate the pooled prevalence of each sample source. The inverse variance index (I2) was used to quantify the heterogeneity across the studies and estimate the random-effects model. An I2 value of >75% indicated considerable heterogeneity. Statistical significance was denoted as a p-value < 0.05 [22]. Egger’s test was used to validate the asymmetry of the funnel plot.
3. Results
3.1. Characteristics of the Included Research Articles
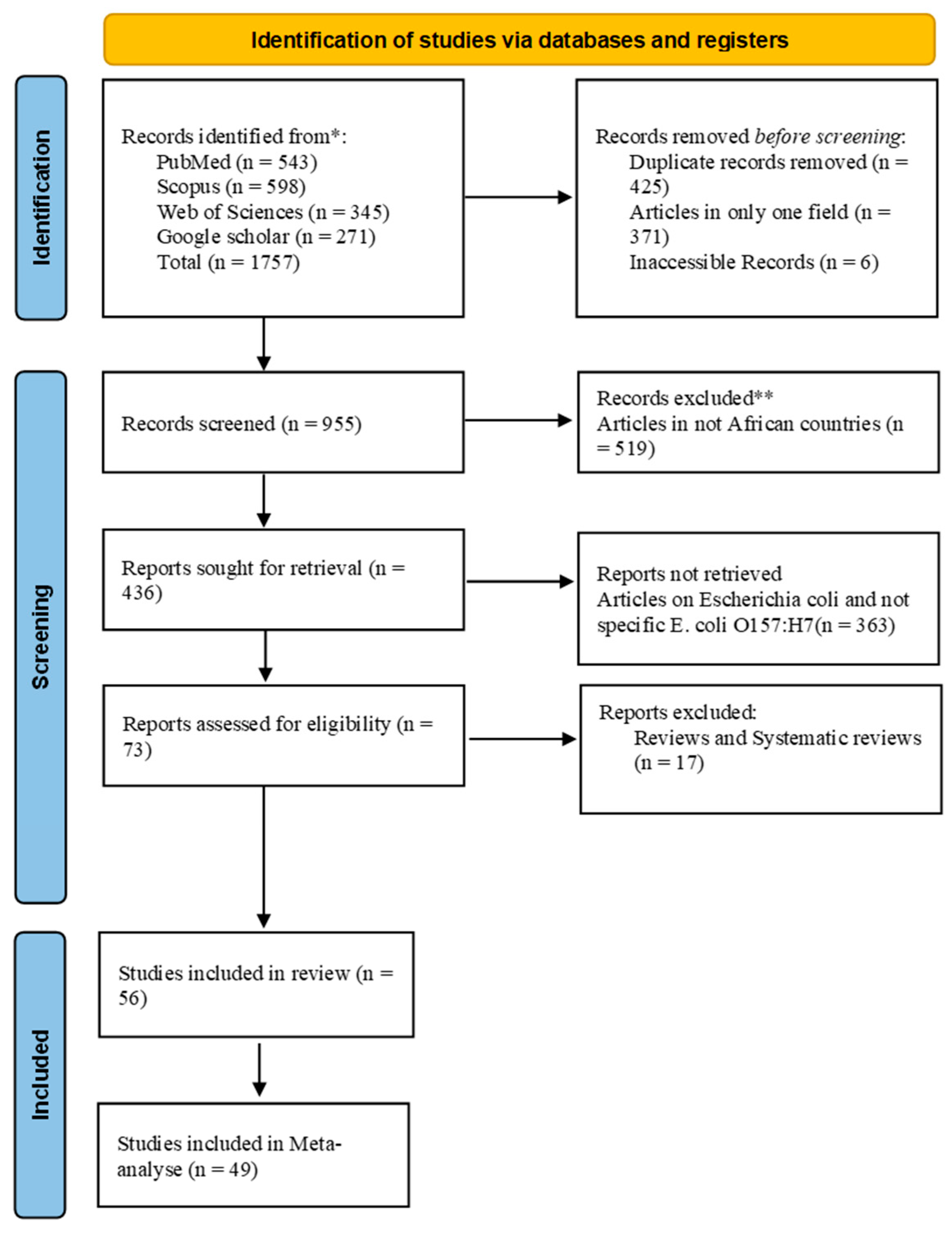
A total of 1757 articles were generated in our initial search including 598 publications (34%) from Scopus, 543 (30.9%) from PubMed, 343 (19.5%) from Web of Science, and 271 (15.4%) from Google Scholar. After screening, a total of 56 (3.2%) articles were included in the review (Figure 1 and Table 1).
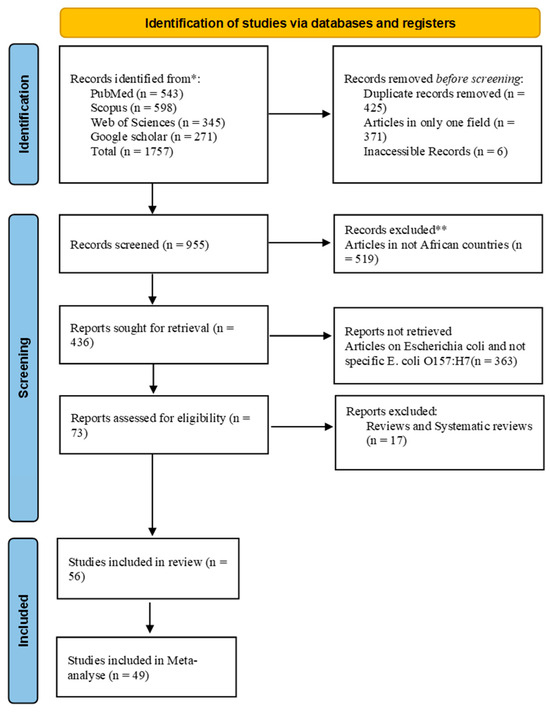
Figure 1.
Flowchart (PRISMA flow diagram) of the systematic literature search, identification, screening, and article selection. * first screening; ** second screening (deep screening).

Table 1.
Study characteristics.
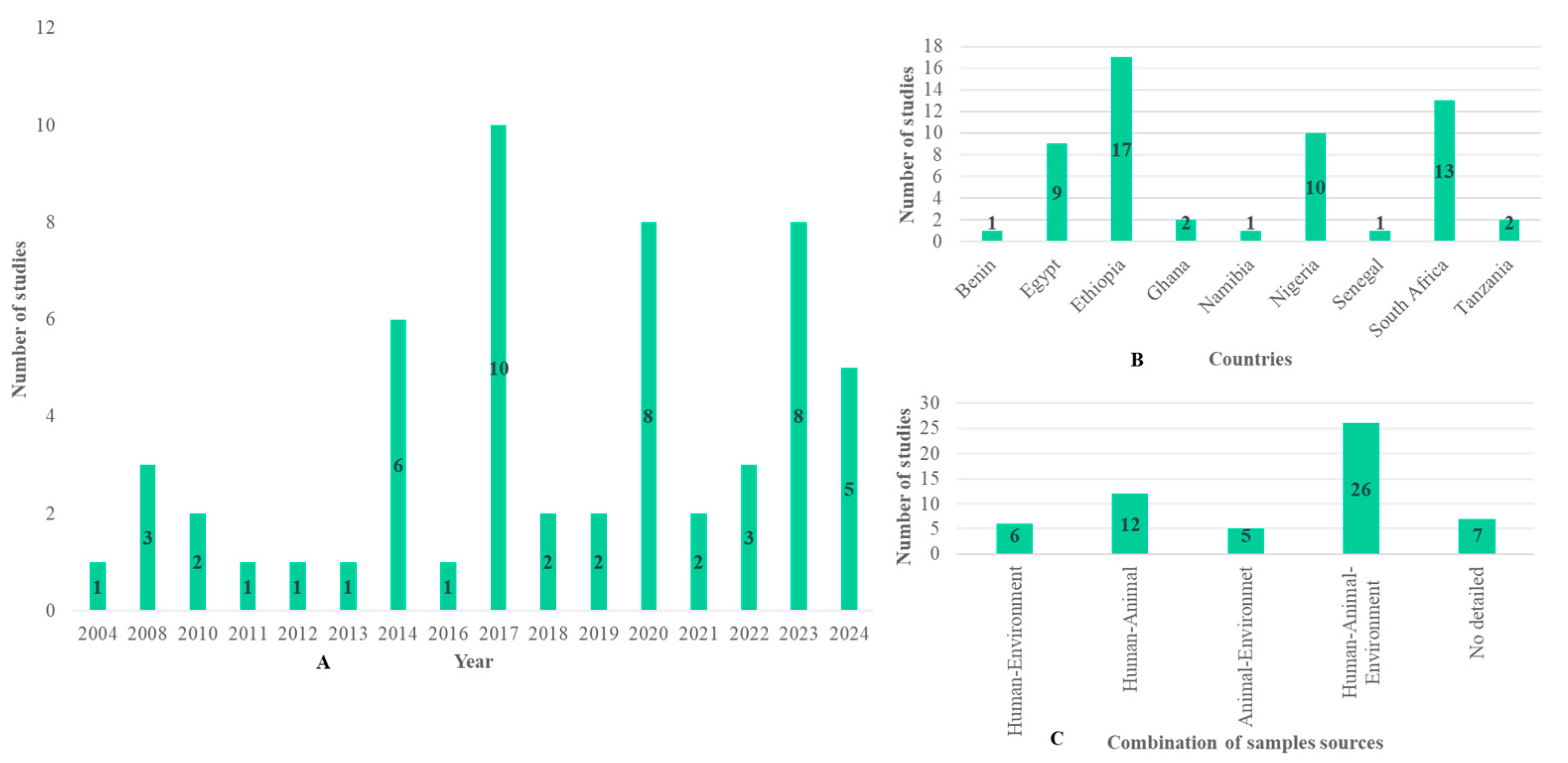
Figure 2A shows the evolution of the number of publications according to year. Papers included in this review were found between 2004 and 2024. Out of the publications included in our study, 6/56 were published in 2014, 10/56 in 2017, 8/56 in 2020 and 2023, and 5/56 in 2024. Only a few articles were found in the other years, and the evolution of publications did not follow a normal curve. These 56 articles were found in only 9 countries including Ethiopia (17/56), South Africa (13/56), Nigeria (10/56), Egypt (9/56), Ghana (2/56), Tanzania (2/56), Benin (1/56), Namibia (1/56), and Senegal (1/56) (Figure 2B). Out of the 56 publications, 7 were excluded due to the absence of data on the sample population. Only 49 detailed information about the sample origin, sample number, isolate number, and methods. These 49 publications were used for the meta-analysis, of which 26 were from human–animal–environment samples, 12 from human–animal samples, 6 from human–environment samples, and 5 from animal–environment samples (Figure 2C).
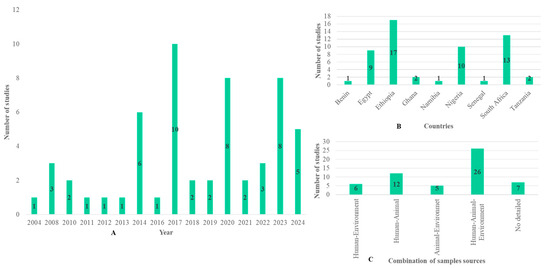
Figure 2.
Summary of the selected studies showing the number of studies (A) per year, (B) per country, and (C) per combination of sample sources.
3.2. Distribution of Samples, the Sampling Sources, and E. coli O157:H7 Identification Methods Used
Samples from human studies were obtained from stools (n = 19), urine (n = 1), and workers’ hand swabs (n = 7). Some studies did not specify the type of sample and mentioned only that there were human samples (n = 14). For the animal studies, most studies reported isolates from cattle (n = 16), meat and meat products (n = 15), beef (n = 14), pork (n = 8), and milk and dairy products (n = 7). A few studies reported on poultry (n = 4), chicken (n = 4), animal feces (n = 4), fish (n = 2), and mouton (n = 1). Some studies did not specify the type of samples and mentioned only that the samples were from animals (n = 3). Samples from the environment were obtained from water sources (n = 26), knives (n = 15), cutting boards (n = 6), manure or soil (n = 5), tables (n = 4), floors (n = 3), vegetables (n = 3), sinks (n = 3), vehicles for meat transport (n = 2), utensils (n = 2), door handles (n = 2), equipment (n = 2), walls (n = 2), bedrail (n = 1), cupboard (n = 1), stamp (n = 1), stretcher (n = 1), swivels (n = 1), and toilet seat (n = 1). Some studies did not specify the type of samples and only mentioned environmental samples (n = 3). The identification methods utilized in these studies included culture-based techniques (100%, n = 56), standard biochemical tests (91.1%, n = 51), PCR (53.6%, n = 30), serological (agglutination) test (16.1%, n = 9), serotyping using E. coli O157:H7 anti-sera (10.7%, n = 6), sequencing (8.9%, n = 5), pulse field gel electrophoresis (PFGE) (3.6%, n = 2), and immunochromatographic separation (1.8%, n = 1). In terms of antibiotic susceptibility testing, the Kirby–Bauer disk diffusion method was the only method used in all publications that described the antimicrobial susceptibility test (100%, n = 28).
3.3. Meta-Analysis
3.3.1. Distribution of E. coli O157:H7 Prevalence by Country
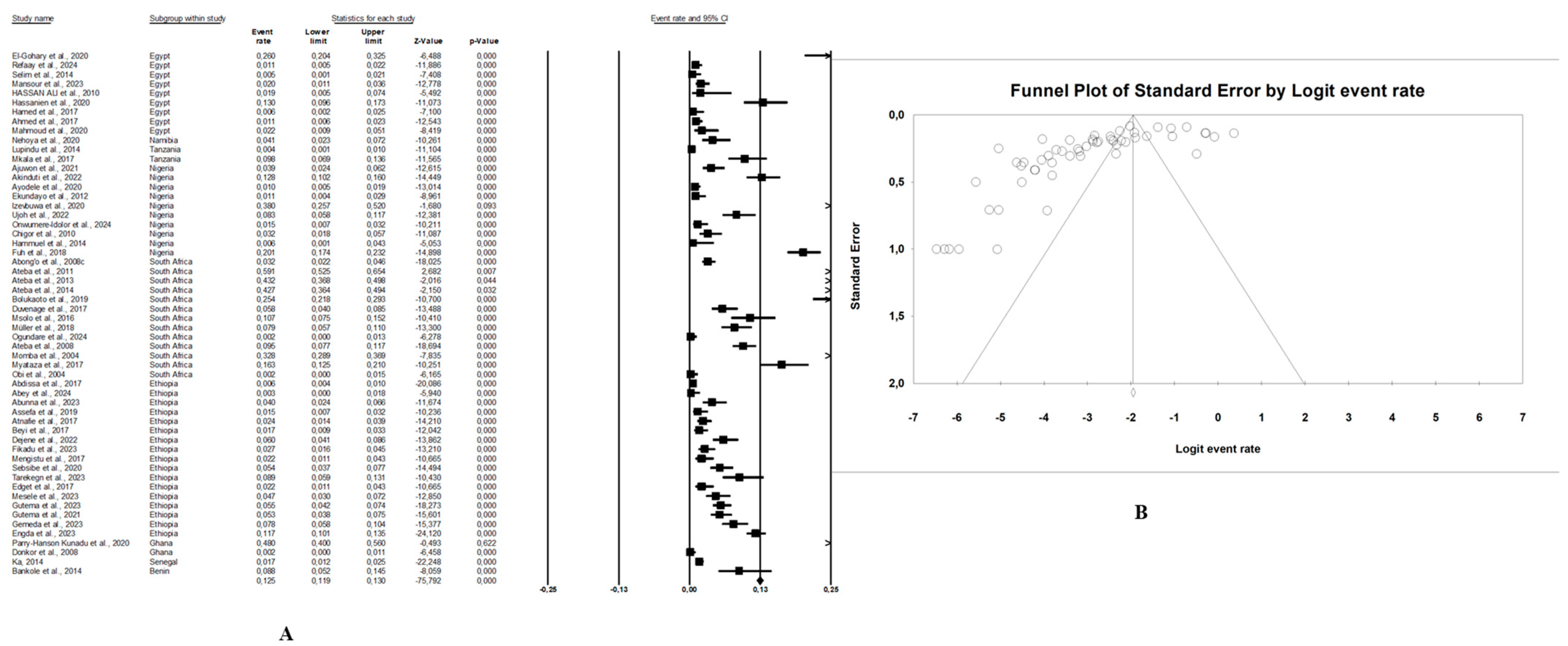
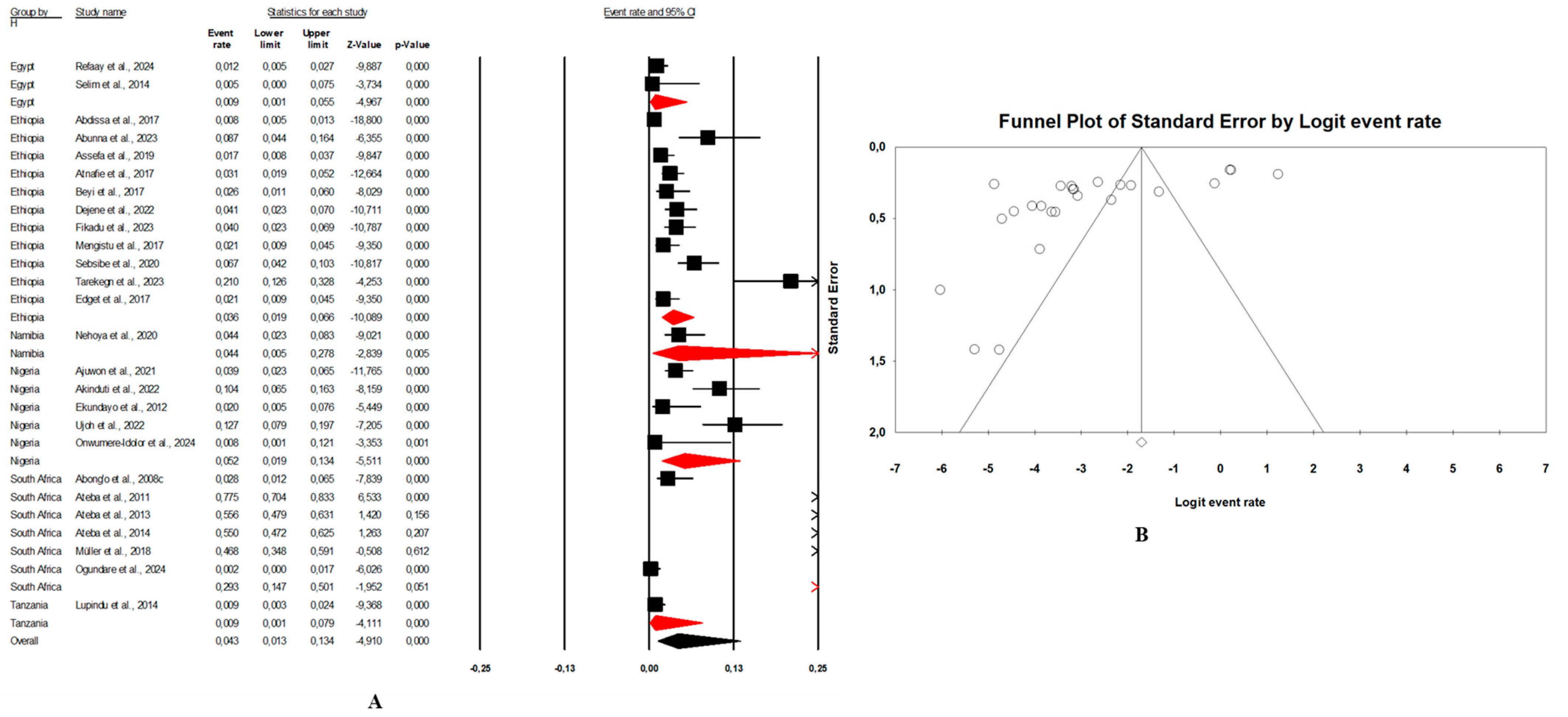
In this review, the pooled prevalence of E. coli O157:H7 in Africa in the reviewed studies was 4.7% (95% CI: 3.4–6.5, I2 = 97.6%, p < 0.05) (Figure 3A). The highest prevalence was seen in animal samples with pooled prevalence of 5.4% (95% CI: 3.4–8.6, I2 = 96.9%, p < 0.05), followed by the environmental samples at 3.4% (95% CI: 2.3–5.0, I2 = 82.9%, p < 0.05), and human samples at 2.8% (95% CI: 1.8–4.2, I2 = 91.9%, p < 0.05). The presence of publication bias, represented by an asymmetrical funnel plot, was statistically confirmed with the random-effects model (p < 0.0001) (Figure 3B). The asymmetrical distribution of effect estimates, shown by a funnel plot of the study distribution, allowed us to examine the data according to the countries. Therefore, the pooled prevalence of E. coli O157:H7 was 12.6% (n = 13; 95% CI: 7.1–21.2; I2 = 98.1%; p < 0.05) in South Africa, 6.0% (n = 17, 95% CI: 5.5–6.5, I2 = 93.6%; p < 0.05) in Ethiopia, 4.8% (n = 10; 95% CI: 2.4–9.4; I2 = 96.2%; p < 0.05) in Nigeria, and 2.4% (n = 9; 95% CI: 0.8–6.8; I2 = 96.2%; p < 0.05) in Egypt. Notably, the prevalence of E. coli O157:H7 was 9.1% (n = 2) in Ghana, 8.8% (n = 1) in Benin, 4.1% (n = 1) in Namibia, 2.5% (n = 2) in Tanzania, and 1.7% (n = 1) in Senegal.
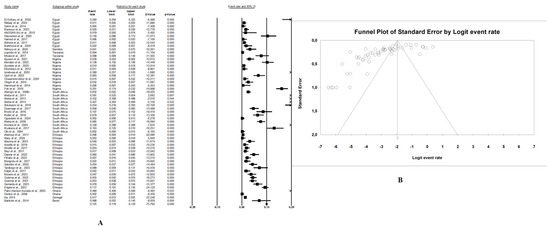
Figure 3.
(A) Forest plot with the adjusted average prevalence of E. coli O157:H7 from the One Health perspective in Africa. Legend: Random effects mode (95% CI: 3.4–6.5, I2 = 97.6%, p < 0.05). X-axis is the proportion of countries reported in individual studies as listed along the Y-axis, with the range of proportion in the 95% confidence interval (CI). I2 = heterogenicity, p = p-value. The estimate of prevalence was calculated by pooling 56 studies using the random-effects model. (B) Funnel plot with the adjusted average prevalence of E. coli O157:H7 from the One Health perspective in Africa. Legend: The graph displays the standard error of the estimate (prevalence) on the Y-axis, while the X-axis represents the transformed proportions (prevalence), with individual studies represented by small circles. The 95% confidence interval is indicated by solid lines on the graph.
3.3.2. Distribution of E. coli O157:H7 Prevalence According to Combination of Sample Sources
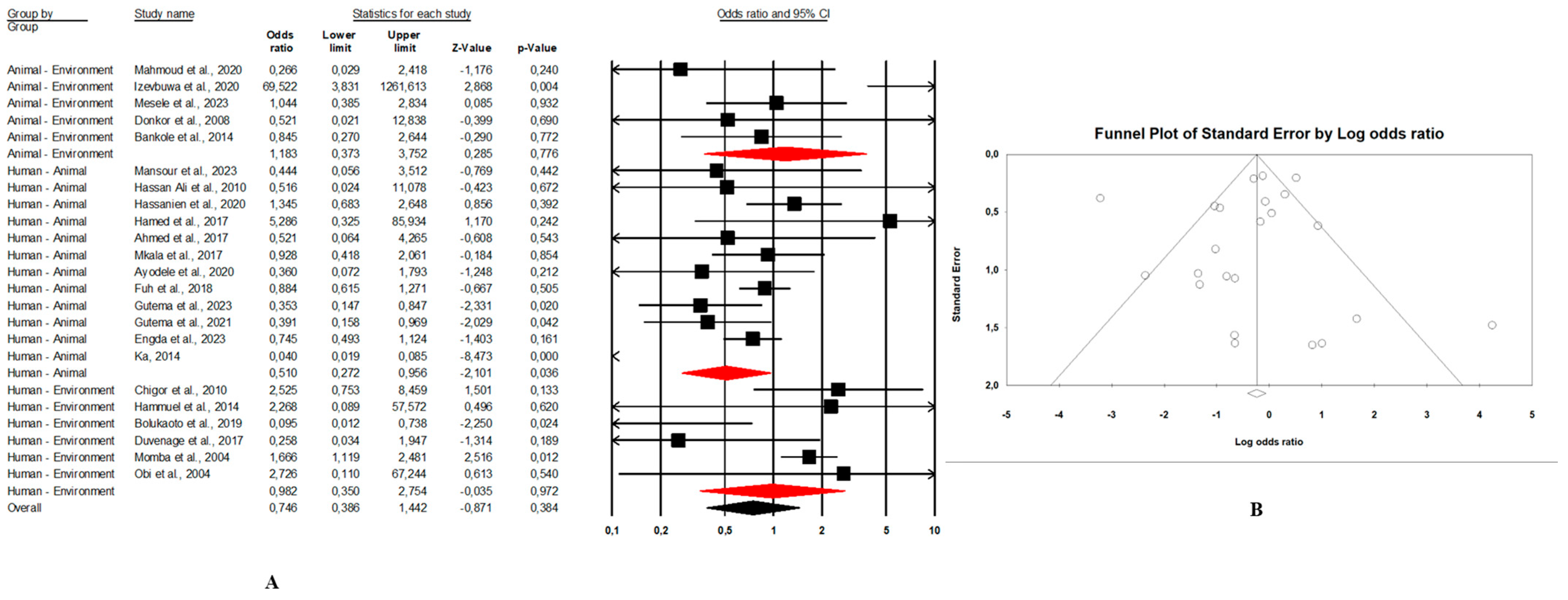
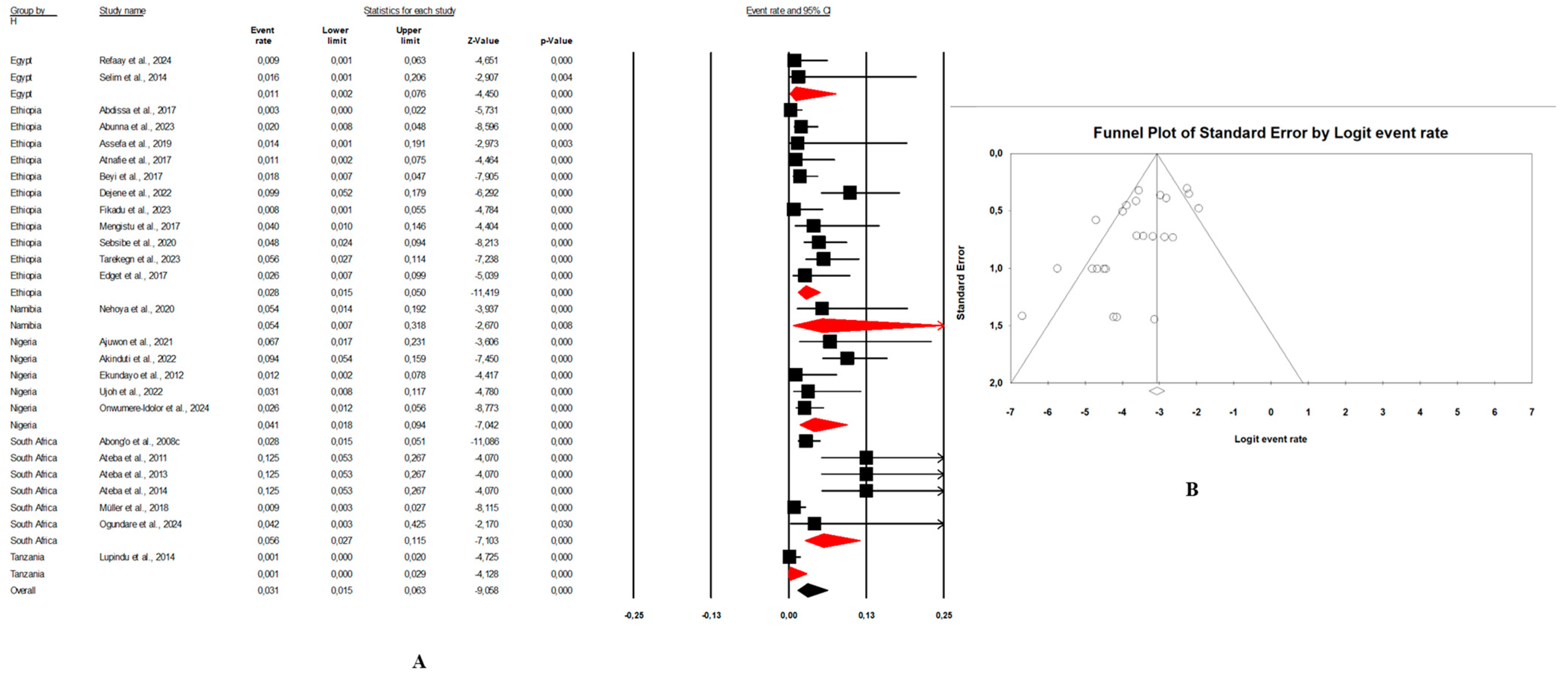
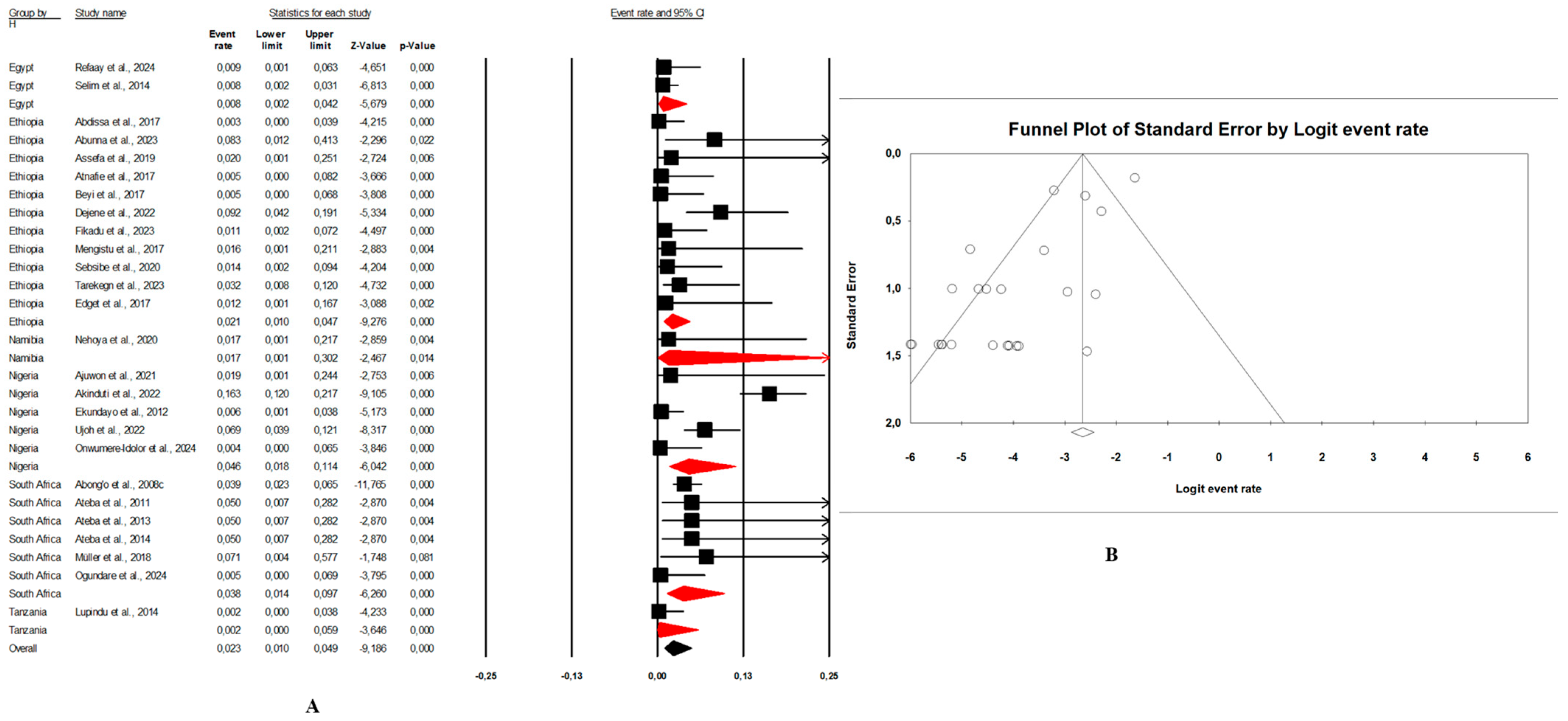
- Combination human–animal–environment sources: Figure 4A shows the distribution of the pooled prevalence of E. coli O157:H7 from the 26 articles reviewed having human–animal–environment samples. According to the random effects analysis, this prevalence was 3.7% (95% CI: 2.0–6.8; I2 = 98.1%; p < 0.05). Figure 4B shows a funnel plot showing the distribution bias in effect estimates among studies examining the prevalence of E. coli O157:H7 in Africa. Among these 26 articles, the pooled prevalence of E. coli O157:H7 was 4.3% (95% CI: 1.3–13.4; I2 = 97.8%; p < 0.05) from animal samples (Figure 5A,B), followed by 3.1% (95% CI: 1.5–6.3; I2 = 66.4%; p < 0.05) from environment samples (Figure 6A,B), and 2.3% (95% CI: 1.0–4.9; I2 = 73.6%; p < 0.05) from human samples (Figure 7A,B). These publications were found in 6 countries from which the pooled prevalence of E. coli O157:H7 was 13.8% (n = 6; 95% CI: 4.4–35.8; I2 = 98.8%; p < 0.05) in South Africa, 4.1% (n = 5; 95% CI: 1.8–8.9; I2 = 93.3%; p < 0.05) in Nigeria, 2.8% (n = 11, 95% CI: 1.7–4.5; I2 = 96.2%; p < 0.05) in Ethiopia, 0.9% (n = 2; 95% CI: 0.5–1.8; I2 = 0%; p < 0.05) in Egypt, 4.1% (n = 1) in Namibia, and 0.4% (n = 1) in Tanzania.
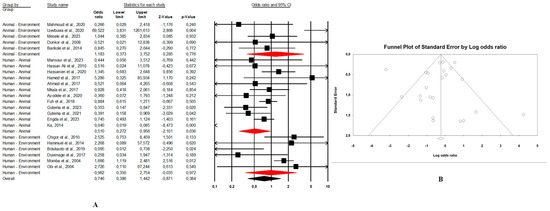 Figure 4. (A) Forest plot with the adjusted average prevalence of E. coli O157:H7 from human–animal–environment samples. (B) Funnel plot of E. coli O157:H7 from the human–animal–environment samples. Legend: Random effects mode (95% CI: 2.0–6.8; I2 = 98.1%; p < 0.05). X-axis is the proportion of E. coli O157:H7 reported in individual studies as listed along the Y-axis, with the range of proportion in the 95% confidence interval (CI). I2 = heterogenicity, p = p-value. The estimate of prevalence was calculated by pooling 26 studies that reported E. coli O157:H7 using the random-effects model.
Figure 4. (A) Forest plot with the adjusted average prevalence of E. coli O157:H7 from human–animal–environment samples. (B) Funnel plot of E. coli O157:H7 from the human–animal–environment samples. Legend: Random effects mode (95% CI: 2.0–6.8; I2 = 98.1%; p < 0.05). X-axis is the proportion of E. coli O157:H7 reported in individual studies as listed along the Y-axis, with the range of proportion in the 95% confidence interval (CI). I2 = heterogenicity, p = p-value. The estimate of prevalence was calculated by pooling 26 studies that reported E. coli O157:H7 using the random-effects model.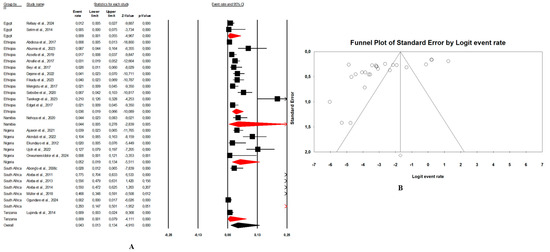 Figure 5. (A) Forest plot with the adjusted average prevalence of E. coli O157:H7 from animal samples. (B) Funnel plot of E. coli O157:H7 from animal samples. Legend: Random effects mode (95% CI: 1.3–13.4; I2 = 97.8%; p < 0.05). X-axis is the proportion of E. coli O157:H7 reported in individual studies as listed along the Y-axis, with the range of proportion in the 95% confidence interval (CI). I2 = heterogenicity, p = p-value. The estimate of prevalence was calculated by pooling all studies that reported E. coli O157:H7 from animal samples using the random-effects model.
Figure 5. (A) Forest plot with the adjusted average prevalence of E. coli O157:H7 from animal samples. (B) Funnel plot of E. coli O157:H7 from animal samples. Legend: Random effects mode (95% CI: 1.3–13.4; I2 = 97.8%; p < 0.05). X-axis is the proportion of E. coli O157:H7 reported in individual studies as listed along the Y-axis, with the range of proportion in the 95% confidence interval (CI). I2 = heterogenicity, p = p-value. The estimate of prevalence was calculated by pooling all studies that reported E. coli O157:H7 from animal samples using the random-effects model.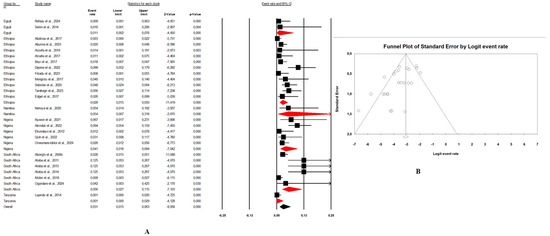 Figure 6. (A) Forest plot with the adjusted average prevalence of E. coli O157:H7 from environment samples. (B) Funnel plot of E. coli O157:H7 from environment samples. Legend: Random effects mode (95% CI: 1.5–6.3; I2 = 66.4%; p < 0.05). X-axis is the proportion of E. coli O157:H7 reported in individual studies as listed along the Y-axis, with the range of proportion in the 95% confidence interval (CI). I2 = heterogenicity, p = p-value. The estimate of prevalence was calculated by pooling all studies that reported E. coli O157:H7 from environment samples using the random-effects model.
Figure 6. (A) Forest plot with the adjusted average prevalence of E. coli O157:H7 from environment samples. (B) Funnel plot of E. coli O157:H7 from environment samples. Legend: Random effects mode (95% CI: 1.5–6.3; I2 = 66.4%; p < 0.05). X-axis is the proportion of E. coli O157:H7 reported in individual studies as listed along the Y-axis, with the range of proportion in the 95% confidence interval (CI). I2 = heterogenicity, p = p-value. The estimate of prevalence was calculated by pooling all studies that reported E. coli O157:H7 from environment samples using the random-effects model.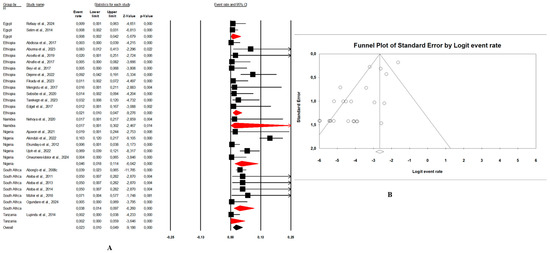 Figure 7. (A) Forest plot with the adjusted average prevalence of E. coli O157:H7 from human samples. (B) Funnel plot of E. coli O157:H7 from human samples. Legend: Random effects mode (95% CI: 1.0–4.9; I2 = 73.6%; p < 0.05). X-axis is the proportion of E. coli O157:H7 reported in individual studies as listed along the Y-axis, with the range of proportion in the 95% confidence interval (CI). I2 = heterogenicity, p = p-value. The estimate of prevalence was calculated by pooling all studies that reported E. coli O157:H7 from human samples using the random-effects model.
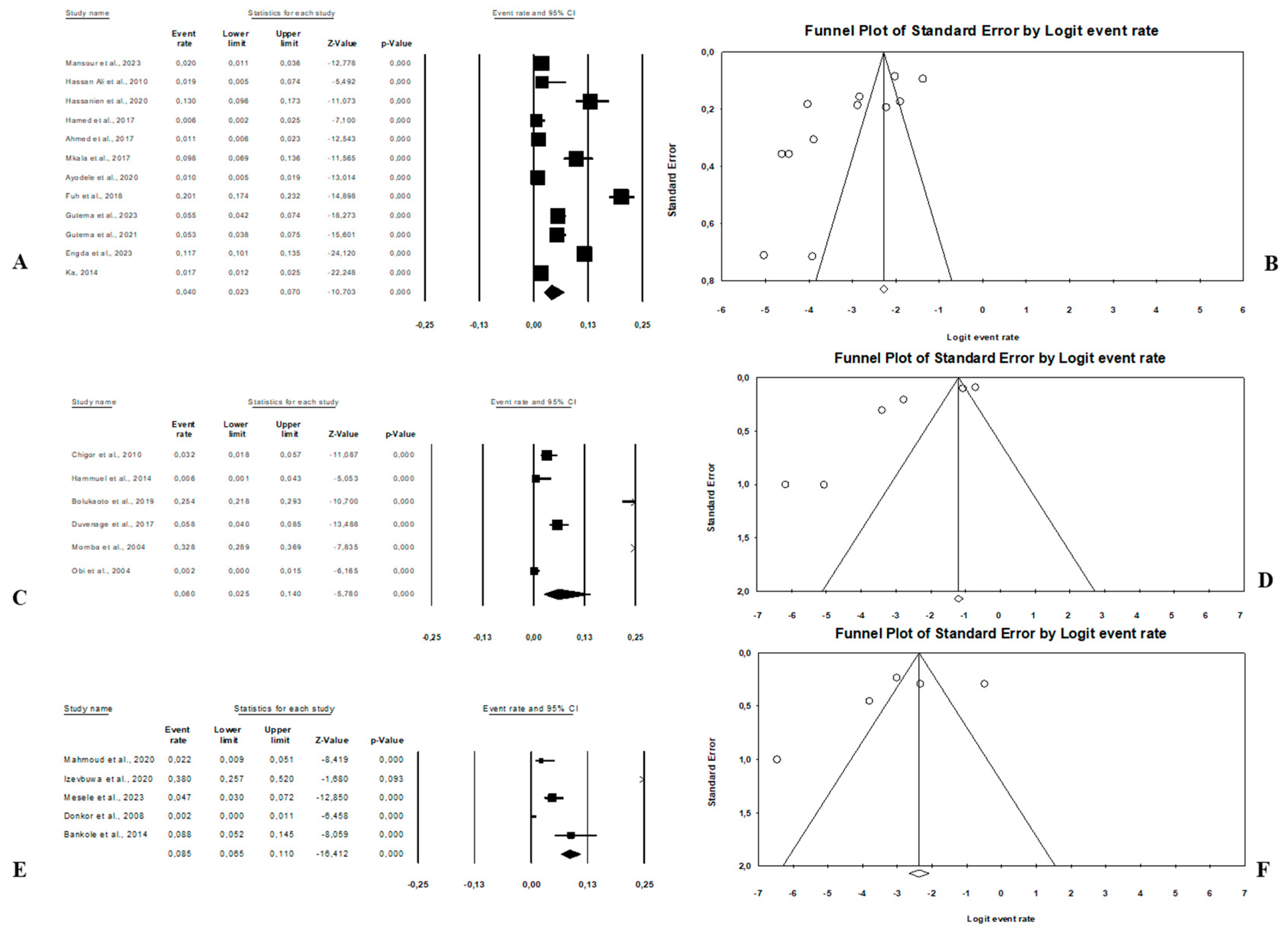
Figure 7. (A) Forest plot with the adjusted average prevalence of E. coli O157:H7 from human samples. (B) Funnel plot of E. coli O157:H7 from human samples. Legend: Random effects mode (95% CI: 1.0–4.9; I2 = 73.6%; p < 0.05). X-axis is the proportion of E. coli O157:H7 reported in individual studies as listed along the Y-axis, with the range of proportion in the 95% confidence interval (CI). I2 = heterogenicity, p = p-value. The estimate of prevalence was calculated by pooling all studies that reported E. coli O157:H7 from human samples using the random-effects model. - Combination of human–animal sources: In this review, 12 studies described concrete information on E. coli O157:H7 from human and animal sources. The average pooled prevalence was 4.0% (95% CI: 2.3–7.0; I2 = 96.9%; p < 0.05) (Figure 8A). The asymmetrical distribution of the effect estimates, shown by a funnel plot of the study distribution, allowed us to examine the data according to the countries and sample sources (Figure 8B). Specifically, the pooled prevalence of E. coli O157:H7 was 3.6% (95% CI: 1.8–6.9; I2 = 94.1%; p < 0.05) from the human samples and 6.7% (95% CI: 3.5–12.3; I2 = 95.6%; p < 0.05) from the animal samples. In this review, these publications were from 5 countries including Egypt (n = 5; 2.2% (95% CI: 0.6–8.1; I2 = 94.6%; p < 0.05), Ethiopia (n = 3; 7.1% (95% CI: 3.9–12.6; I2 = 93.9%; p < 0.05), Nigeria (n = 2; 4.8% (95% CI: 0.2–54.9; I2 = 98.7%; p < 0.05), Tanzania (n = 1; 9.8%), and Senegal (n = 1; 1.7%).
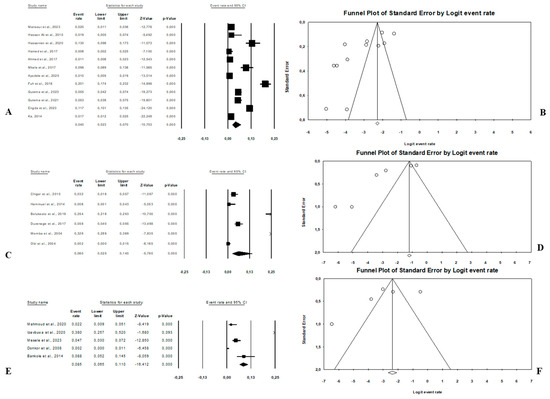 Figure 8. Forest plot and funnel plot with the adjusted average prevalence of E. coli O157:H7. (A,B) Human–animal samples; (C,D) human–environment samples; (E,F) animal–environment samples.
Figure 8. Forest plot and funnel plot with the adjusted average prevalence of E. coli O157:H7. (A,B) Human–animal samples; (C,D) human–environment samples; (E,F) animal–environment samples. - Combination of human–environment sources: Our study highlighted 6 publications in this area, of which the pooled prevalence of E. coli O157:H7 was 6.0% (95% CI: 2.5–14.0; I2 = 97.2%; p < 0.05) (Figure 8C). The asymmetrical distribution of the effect estimates, which is shown by a funnel plot of the study distribution, led us to further examine the data according to countries and sample sources (Figure 8D). These research articles were described in 2 countries such as in South Africa (n = 4), where the pooled prevalence of E. coli O157:H7 was 10.9% (95% CI: 4.6–23.6; I2 = 97.3%; p < 0.05), and in Nigeria (n = 2), it was 1.9% (95% CI: 0.4–8.2; I2 = 60.6%; p > 0.05). The pooled prevalence of E. coli O157:H7 from the human samples was 2.7% (95% CI: 0.4–15.8; I2 = 94.3%; p < 0.05), and it was 3.7% (95% CI: 1.2–10.7; I2 = 94.2%; p < 0.05) from the environment samples.
- Combination of animal–environment sources: For this combination, the pooled prevalence of E. coli O157:H7 was 8.5% (95% CI: 6.5–11.0; I2 = 94.7%; p < 0.05) in the 5 publications reviewed (Figure 8E). Figure 8F is a funnel plot showing the distribution bias in the effect estimates among studies that examined the prevalence of E. coli O157:H7 in the animal–environment samples. Specifically, the pooled prevalence of E. coli O157:H7 from the animal samples in this combination was 5.7% (95% CI: 1.0–26.9; I2 = 94.1%; p < 0.05), and it was 3.6% (95% CI: 1.4–8.9; I2 = 69.9%; p < 0.05) in the environment samples. The prevalence of E. coli O157:H7 was 38% (n = 1) in Nigeria, 8.8% (n = 1) in Benin, 4.7% (n = 1) in Ethiopia, 2.2% (n = 1) in Egypt, and 0.02% (n = 1) in Ghana.
3.3.3. Pooled Antimicrobial Resistance (AMR) Prevalence of E. coli O157:H7 Across the Human, Animal, and Environmental Studies in Africa
Out of the 56 studies included in this systematic review, only 28 studies described the antimicrobial resistance (AMR) patterns. Among these 28 publications, 23 presented the antibiotic resistance levels of E. coli O157:H7 to 15 antibiotics with all details, while the other 5 publications presented the antibiotic resistance level of E. coli generally. Out of twenty-three studies, fourteen were carried out on a combination of human–animal–environment sources, nine from human–animal sources, two from the human–environment area, and one from an animal-environment source. Considering the type of investigation (phenotypic or genotypic and phenotypic), all studies used the phenotypic investigation method, which mostly relies on the standard microbiological culture (Table 1). Only one publication from Nigeria used genotypic method to detect antimicrobial resistance gene; extended spectrum beta-lactamase (ESBL) genes were detected in E. coli O157:H7, indicating the presence of blaSHV, blaCTX-M, and blaTEM in one (12.5%), one (12.5%), and three (37.5%) isolates, respectively [38]. All the 23 publications reported the presence of antibiotic resistance in E. coli O157:H7.
According to the random-effect analysis, the pooled prevalence was 77.4% for ampicillin (95% CI: 59.4–88.9; I2 = 75.0%; p < 0.05), 73.2% for tetracycline (95% CI: 55.3–85.7; I2 = 86.1; p < 0.05), 67.9% for trimethoprim/sulfamethoxazole (95% CI: 31.1–90.9; I2 = 82.4%; p = 0.342), 66.9% for gentamicin (95% CI: 56.6–75.8; I2 = 26.9%; p < 0.05), 63.5% for cefoxitin (95% CI: 6.4–97.8; I2 = 88.7%; p = 0.737), 53.4% for nitrofurantoin (95% CI: 37.8–68.4; I2 = 59.6%; p = 0.674), and 51.7% for cefuroxime (95% CI: 15.4–86.3; I2 = 71.9%; p = 0.941). However, the pooled prevalence was observed to be low in amikacin, 45.8% (95% CI: 26.9–65.9; I2 = 65.6%; p = 0.688), ceftazidime, 44.5% (95% CI: 9.3–86.3; I2 = 79.8%; p = 0.833), ceftriaxone, 43.5% (95% CI: 25.6–63.4; I2 = 70.0%; p = 0.528), amoxiclav, 43.1% (95% CI: 28.3–59.3; I2 = 81.3%; p = 0.408), ciprofloxacin, 35.9% (95% CI: 16.7–61.0; I2 = 91.6%; p = 0.269), nalidixic-acid, 25.5% (95% CI: 9.7–52.2; I2 = 0%; p = 0.07), and chloramphenicol, 23.9% (95% CI: 7.7–54.2; I2 = 93.3%; p = 0.09). There was a significant difference observed in all antibiotic-resistant E. coli O157:H7 described in this systematic review, except for ampicillin and tetracycline. This difference observed was not consistent; it sometimes showed a high prevalence in human studies, was sometimes high in animal studies, and at times high in environmental studies (Table 2). The observed antibiotic resistance in these studies was found to be the highest in animal studies compared to the human and environmental studies. The pooled antibiotic resistance prevalences were 96.5% for cefoxitin, 82.8% for ampicillin, 76.8% for cefuroxime, 70.7% for nitrofurantoin, 62.1% for amikacin, 50.4% for amoxiclav, and 40.2% for ciprofloxacin. Moreover, the pooled prevalence in trimethoprim/sulfamethoxazole was 82.9%, while it was 80.7% for tetracycline, and 48.7%, 45.7%, and 26.4% for ceftazidime, ceftriaxone, and chloramphenicol, respectively, in the human studies. Of note, imipenem was only described in human studies, of which the prevalence was 83.3%, while only gentamicin was reported in the environmental studies with a pooled prevalence of 72.6%. In the animal isolates, the resistance levels were higher in cefoxitin than in other antibiotics with 96.5% as the pooled prevalence. For human isolates, the pooled prevalence of E. coli O157:H7 was 83.3% to nalidixic-acid and imipenem.

Table 2.
Subgroup meta-analysis of the pooled antimicrobial resistance (AMR) prevalence of E. coli O157:H7 across the human, animal, and environment studies.
3.4. Risk of Bias
In our review, the included studies reported high heterogeneity, as indicated by the I2 = 97.6% and Cochrane Q test (Q = 2317.28370, p < 0.0001). Visual inspection of the funnel plot showed a slight asymmetrical distribution. The intercept of the Eggers regression model was 1.33121 (95% CI: −11.01740–5.67955) with a t statistic of 6.27 and a p-value of 0.000. This finding suggests that potential publication bias in the included studies was unlikely (Figure 3B).
4. Discussion
This study was a systematic review of E. coli O157:H7 from the One Health approach, focusing simultaneously on animals, humans, and their environment. Our investigations showed that Ethiopia, South Africa, Nigeria, and Egypt were the African countries with the highest number of publications on E. coli O157:H7. Many factors could be attributed to this fact. According to Untaman et al. [74], the contribution of the number of publications in these countries could be due to the number of journal outlets, the high number of research institutions or universities, and the research specialization observed in these countries. Additionally, these countries are English-speaking nations, and the English language is most often used for scientific publications [75]. Recently, a systematic review and meta-analysis carried out on E. coli isolates from water in Africa showed that South Africa, Ethiopia, and Nigeria had the highest number of studies [14]. According to these authors, the higher socio-economic status of these countries in the region, and thus their ability to invest well in research and facilities, could explain this fact [14]. Moreover, not reporting data on E. coli O157:H7 from the One Health perspective in some African countries does not explain the absence of the bacterium, as there may be a high chance that this organism exists in these countries. Lupindua et al. showed that the lack of reports on E. coli O157:H7 isolation in some African countries could be due to the poor and insufficient diagnostic facilities even in some national reference laboratories, especially in rural settings where infections may be undiagnosed [5]. Most studies also used conventional culture methods for E. coli O157:H7 detection. This conventional method may be a limitation to the identification of E. coli O157:H7, since the evolution of molecular methods offers more robust and diverse methods for the detection of E. coli O157:H7 [76].
The data obtained from the research articles in this study did not present an exponential evolution in the publication years. In our investigation, most articles were published in 2017, 2020, and 2023. However, some systematic reviews in Africa have shown that the evolution of publications by year followed a normal curve [12,14,75,77].
To the best of our knowledge, this study is the first comprehensive systematic review and meta-analysis on the prevalence of E. coli O157:H7 simultaneously isolated from human, animal, and environment samples. This will contribute to the surveillance of this pathogenic bacteria and aid in designing preventive measures that will reduce the associated mortality of children under five years due to diarrhea in different regions of Africa. Over the last ten years, many studies have reported the incidence of E. coli O157:H7 in Africa [78,79,80,81,82]. However, data using the One Health perspective have been limited in the region.
It was observed that the pooled prevalence of E. coli O157:H7 from articles using the One Health approach in Africa was 4.7%. The random effects analysis showed that the pooled prevalence of the bacteria was 3.7%, 4%, 6%, and 8.5% in the human–animal–environment, human–animal, human–environment, and animal–environment publications, respectively. These results highlight the possibility of this bacteria being able to spread in different interfaces (human, animal, and environment). This could be linked to many factors, notably human activities. In terms of the sample source of E. coli O157:H7, the animal samples had the highest pooled prevalence of 5.4%. In Brazil, the pooled prevalence of E. coli O157:H7 in bovine meat and meat products was 1% lower than the prevalence in our findings [83]. According to Assefa and Bihon (2018), the pooled prevalence of E. coli O157:H7 in foods of animal origin in Ethiopia was 4% [84]. In Tunisia, E. coli O157:H7 isolated from cattle had a prevalence of 4.2% [85]. These results are slightly similar to the results of our findings. According to some reports, cattle are the primary reservoirs of E. coli O157:H7, and the consumption of beef and beef products have been identified as major sources of foodborne transmission [55]. Cattle could also be considered as the origin for E. coli O157:H7 spreading in environment, as they shed the pathogen normally through their feces. There is a high chance that these pathogens could contaminate vegetables, as farmers often use cow dung as natural fertilizers or irrigate vegetables with water that has been contaminated with cattle feces [7]. Our investigation showed that there was a variation in the prevalence of E. coli O157:H7 in foods and animal products from one region to another. This could be the result of geographical variations in slaughter hall conditions and handling practices (transportation trucks, carcass, and carcass with contact surfaces) [86].
From the environmental samples, our systematic review report the pooled prevalence of E. coli O157:H7 at 3.4%. Some environmental-based studies have reported near approximate prevalence; this is the case in Ethiopia and the United Kingdom, where the overall prevalence of E. coli O157:H7 was reported as 4.7% and 4.3%, respectively [87]. This bacterium is often considered as a pathogen frequently isolated from waters and wastewater. In African countries, vegetables are rinsed using various water sources including rivers and ponds close to the garden or selling site, increasing the risk of contamination with E. coli O157:H7 and other pathogens [88].
Our investigation showed that the pooled prevalence of E. coli O157:H7 from human samples was 2.8%. Generally, E. coli O157:H7 is most commonly implicated in human infections, especially food poisoning. Its transmission to humans mainly occurs through the consumption of contaminated foods such as raw or undercooked ground meat, raw milk, raw vegetables, and contaminated sprouted seeds [89]. The low prevalence of E. coli 0157:H7 reported in this review from humans could be due to the fact that many countries now observe an amelioration of sanitation, and the populace have started adopting better hygienic measures such as practices linked to food preparation and sanitation conditions.
In addition, the systematic review focused on 28 studies that highlighted the antibiotic resistance in E. coli 0157:H7 isolates. The pooled prevalence of antibiotic resistance in E. coli O157:H7 varied from one antibiotic to another and the different sample sources. For most of the antibiotics tested, resistance was observed more in the isolates from animal samples than from the environmental and human samples. Numerous studies that have reported antibiotic resistance in E. coli using the One Health approach have described similar results to our findings. This has been notably reported in Vietnam [90], Africa [14,91,92], and all over the world [93]. This fact is not surprising as the National Agency for Sanitary Safety of Food, the Environment, and Work (ANSES) has reported that farmers in many developing countries freely give antibiotics to their animals as soon as they are sick, without first checking whether they are indeed suffering from bacterial infections. These are often even used routinely on healthy animals to prevent infections or as growth promoters [94]. Therefore, animal and environmental samples, especially wastewater, are considered as potential sources of antibiotic resistance genes and multidrug resistance bacteria [88]. Moreover, the higher antibiotic resistance observed from animal and environmental samples calls for a lot of sensitization regarding the prudent use of antibiotics in these sectors, knowing that antibiotic-resistant bacteria reach humans indirectly through the food chain [95]. This could be through the consumption of contaminated food or food-derived products [95]. Therefore, there is a need for continuous and strong surveillance of bacterial infections from these three compartments (human, environment, and animal). Furthermore, infection reporting, technical staff training, the acquisition of laboratory equipment, implementation of common standard operating procedures, and the sharing of AMR data and expertise are needed in Africa to guide the required new approaches for the control and treatment of bacterial infections.
Only one publication in this systematic review used the genotypic method to detect antimicrobial resistance genes, out of which ESBL genes were detected in E. coli O157:H7. Therefore, it is recommended that future research should investigate the presence of genes associated with antibiotic resistance in E. coli 0157:H7 isolated from the One Health perspective. The results will provide a useful database on the molecular epidemiology of antibiotic-resistant E. coli O157:H7 in the context of One Health in Africa. This will help policymakers and scientists to develop ways to combat the spread of antibiotic resistance using the One Health approach.
There were, however, a number of limitations in this systematic review that should be considered. For instance, our study protocol was not registered on the standard PROSPERO platform like other studies. In addition, the available data were not representative and varied from one country to another. Furthermore, many of the reviewed articles had small sample sizes, potentially creating a bias in our statistical analyses.
5. Conclusions
In this systematic review, most of the E. coli 0157:H7 isolates were recovered from animals, followed by environmental and human samples, with antibiotic resistance being more common in animal-derived isolates. The systematic review emphasizes the interconnectedness between animals, the environment, and human populations in the transmission and persistence of this pathogen and the need to implement a suitable and appropriate One Health pathogenic and antimicrobial resistance surveillance system in the African region. A more coordinated approach, including standardized procedures, improved laboratory capacity, and better data sharing, will strengthen efforts to monitor pathogen spread and antimicrobial resistance under a One Health framework.
Our systematic review revealed that the available data on E. coli O157:H7 from a One Health approach in Africa are not representative and vary from one country to another.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms13040902/s1. Reference [96] is cited in the Supplementary Materials.
Author Contributions
Conception and design: N.S.S. and E.S.D.; Development of data screening form: N.S.S.; Data screening: N.S.S. and T.O.A.; Data analysis and interpretation: N.S.S., T.O.A. and P.B.T.-Q.; Draft preparation and revisions: N.S.S., T.O.A., P.B.T.-Q. and E.S.D.; Funding acquisition: E.S.D. All authors have read and agreed to the published version of the manuscript.
Funding
This review paper was supported by the Fogarty International Center of the National Institutes of Health through the Research and Capacity Building in Antimicrobial Resistance in West Africa (RECABAW) Training Program hosted at the Department of Medical Microbiology, University of Ghana Medical School (Award Number: D43TW012487). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed are included in this review.
Acknowledgments
The authors acknowledge and thank all the staff at the Department of Medical Microbiology, Medical School, University of Ghana.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Ameer, M.A.; Wasey, A.; Salen, P. Escherichia coli (E. coli 0157 H7); StatPearls Publishing: Treasure Island, FL, USA, 2018. [Google Scholar]
- Karmali, M.A.; Gannon, V.; Sargeant, J.M. Verocytotoxin-producing Escherichia coli (VTEC). Vet. Microbiol. 2010, 140, 360–370. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, G.L.; Hollingsworth, J.; Morris, J.G., Jr. Emerging foodborne pathogens: Escherichia coli O157: H7 as a model of entry of a new pathogen into the food supply of the developed world. Epidemiol. Rev. 1996, 18, 29–51. [Google Scholar] [CrossRef] [PubMed]
- Dunn, J.R.; Keen, J.E.; Moreland, D.; Thompson, R.A.J. Prevalence of Escherichia coli O157: H7 in white-tailed deer from Louisiana. J. Wildl. Dis. 2004, 40, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Lupindua, A.M. Epidemiology of Shiga toxin-producing Escherichia coli O157: H7 in Africa in review. S. Afr. J Infect. Dis. 2018, 33, 24–30. [Google Scholar] [CrossRef]
- Majowicz, S.E.; Scallan, E.; Jones-Bitton, A.; Sargeant, J.M.; Stapleton, J.; Angulo, F.J.; Yeung, D.H.; Kirk, M.D. Global incidence of human Shiga toxin–producing Escherichia coli infections and deaths: A systematic review and knowledge synthesis. Foodborne Pathog. Dis. 2014, 11, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Jaradat, Z.W.; Abulaila, S.; Al-Rousan, E.; Ababneh, Q.O. Prevalence of Escherichia coli O157: H7 in foods in the MENA region between years 2000 and 2022: A review. Arab. J. Basic Appl. Sci. 2024, 31, 104–120. [Google Scholar] [CrossRef]
- Assefa, A.; Regassa, F.; Ayana, D.; Amenu, K.; Abunna, F. Prevalence and antibiotic susceptibility pattern of Escherichia coli O157: H7 isolated from harvested fish at Lake Hayq and Tekeze dam, Northern Ethiopia. Heliyon 2019, 5, e02996. [Google Scholar] [CrossRef]
- Ateba, C.N.; Bezuidenhout, C.C. Characterisation of Escherichia coli O157 strains from humans, cattle and pigs in the North-West Province, South Africa. Int. J. Food Microbiol. 2008, 128, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Natal, I.; Saez-Nieto, J.A.; Medina-Pascual, M.J.; Albersmeier, A.; Valdezate, S.; Guerra-Laso, J.; Rodríguez, H.; Marrodán, T.; Parras, T.; Tauch, A. Dermabacter hominis: A usually daptomycin-resistant gram-positive organism infrequently isolated from human clinical samples. New Microbes New Infect. 2013, 1, 35–40. [Google Scholar] [CrossRef]
- Getaneh, D.K.; Hordofa, L.O.; Ayana, D.A.; Tessema, T.S.; Regassa, L.D. Prevalence of Escherichia coli O157: H7 and associated factors in under-five children in Eastern Ethiopia. PLoS ONE 2021, 16, e0246024. [Google Scholar] [CrossRef]
- Escher, N.A.; Muhummed, A.M.; Hattendorf, J.; Vonaesch, P.; Zinsstag, J.; Health, I. Systematic review and meta-analysis of integrated studies on antimicrobial resistance genes in Africa—A One Health perspective. Trop. Med. Int. Health 2021, 26, 1153–1163. [Google Scholar] [CrossRef]
- Gemeda, B.A.; Assefa, A.; Jaleta, M.B.; Amenu, K.; Wieland, B.J. Antimicrobial resistance in Ethiopia: A systematic review and meta-analysis of prevalence in foods, food handlers, animals, and the environment. One Health 2021, 13, 100286. [Google Scholar] [CrossRef] [PubMed]
- Ramatla, T.; Ramaili, T.; Lekota, K.E.; Ndou, R.; Mphuti, N.; Bezuidenhout, C.; Thekisoe, O. A systematic review and meta-analysis on prevalence and antimicrobial resistance profile of Escherichia coli isolated from water in africa (2000–2021). Heliyon 2023, 9, e16123. [Google Scholar] [CrossRef]
- Rizzo, D.M.; Lichtveld, M.; Mazet, J.A.; Togami, E.; Miller, S.A. Plant health and its effects on food safety and security in a One Health framework: Four case studies. One Health Outlook 2021, 3, 6. [Google Scholar] [CrossRef]
- Abey, S.L.; Teka, M.; Bitew, A.B.; Molla, W.; Ejo, M.; Dagnaw, G.G.; Adugna, T.; Nigatu, S.; Mengistu, B.A.; Kinde, M.Z. Detection and antibiogram profile of diarrheagenic Escherichia coli isolated from two abattoir settings in northwest Ethiopia: A one health perspective. One Health Outlook 2024, 6, 8. [Google Scholar] [CrossRef]
- Akinduti, A.P.; Ayodele, O.; Motayo, B.O.; Obafemi, Y.D.; Isibor, P.O.; Aboderin, O.W. Cluster analysis and geospatial mapping of antibiotic resistant Escherichia coli O157 in southwest Nigerian communities. One Health 2022, 15, 100447. [Google Scholar] [CrossRef] [PubMed]
- Bolukaoto, J.Y.; Kock, M.M.; Strydom, K.-A.; Mbelle, N.M.; Ehlers, M.M. Molecular characteristics and genotypic diversity of enterohaemorrhagic Escherichia coli O157: H7 isolates in Gauteng region, South Africa. Sci. Total Environ. 2019, 692, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Mansour, A.M.; Shehab, S.A.; Nossair, M.A.; Ayyad, A.S.; Tawfik, R.G.; El-Lami, S.A.; Eskander, M.J. Molecular Characterization of Shiga Toxin-producing Escherichia coli Isolated from Some Food Products as well as Human Stool in Alexandria, Egypt. Sci. Total Environ. 2023, 13, 1056–1062. [Google Scholar]
- Nehoya, K.N.; Hamatui, N.; Shilangale, R.P.; Onywera, H.; Kennedy, J.; Mwapagha, L.M. Characterization of Shiga toxin-producing Escherichia coli in raw beef from informal and commercial abattoirs. PLoS ONE 2020, 15, e0243828. [Google Scholar] [CrossRef] [PubMed]
- Munn, Z.; Moola, S.; Riitano, D.; Lisy, K. The development of a critical appraisal tool for use in systematic reviews addressing questions of prevalence. Int. J. Health Policy Manag. 2014, 3, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.P.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring inconsistency in meta-analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef]
- El-Gohary, F.A.; Abdel-Hafez, L.J.M.; Zakaria, A.I.; Shata, R.R.; Tahoun, A.; El-Mleeh, A.; Abo Elfadl, E.A.; Elmahallawy, E.K. Enhanced antibacterial activity of silver nanoparticles combined with hydrogen peroxide against multidrug-resistant pathogens isolated from dairy farms and beef slaughterhouses in Egypt. Infect. Drug Resist. 2020, 13, 3485–3499. [Google Scholar] [CrossRef]
- Refaay, T.; Elshafiee, E.; Mansour, H.A.; Sabry, M.A. Occurrence, Antimicrobial Susceptibility and Phylogroups of Escherichia coli O157:H7 Isolated from Food Outlets in Some Touristic Cities in Egypt. Pak. J. Zool. 2024, 56, 503–512. [Google Scholar] [CrossRef]
- Selim, S.A.; Ahmed, S.F.; Aziz, M.H.A.; Zakaria, A.M.; Klena, J.D.; Pangallo, D. Prevalence and characterization of Shiga-toxin O157: H7 and non-O157: H7 enterohemorrhagic Escherichia coli isolated from different sources. Biotechnol. Equip. 2013, 27, 3834–3842. [Google Scholar] [CrossRef]
- Ali, S.F.H.; Hassanein, R.; El-Malek, A.; Elsayh, K.I. Incidence and characterization of E. coli O157:H7 isolated from minced beef, chicken meats and human stools in Assiut City. Assiut Vet. Med. J. 2010, 56, 1–13. [Google Scholar] [CrossRef]
- Hassanien, A.A.; Shaker, E. Investigation of the effect of chitosan and silver nanoparticles on the antibiotic resistance of Escherichia coli O157: H7 isolated from some milk products and diarrheal patients in Sohag City, Egypt. Vet. World 2020, 13, 1647–1653. [Google Scholar] [CrossRef]
- Hamed, O.M.; Sabry, M.A.; Hassanain, N.A.; Hamza, E.; Hegazi, A.G.; Salman, M.B. Occurrence of virulent and antibiotic-resistant Shiga toxin-producing Escherichia coli in some food products and human stool in Egypt. Vet. World 2017, 10, 1233–1240. [Google Scholar] [CrossRef]
- Ahmed, H.; MacLeod, E.T.; El Bayomi, R.M.; Mohsen, R.A.; Nassar, A.H. Molecular characterization of Escherichia coli O157: H7 and non-O157 shiga toxin producing E. coli from retail meat and humans. Zagazig. Vet. J. 2017, 45, 250–261. [Google Scholar] [CrossRef]
- Mahmoud, M.; Zaki, R.; Abd-Elhafeez, H. An epifluorescence-based technique accelerates risk assessment of aggregated bacterial communities in carcass and environment. Environ. Pollut. 2020, 260, 113950. [Google Scholar] [CrossRef]
- Lupindu, A.M.; Olsen, J.E.; Ngowi, H.A.; Msoffe, P.L.; Mtambo, M.M.; Scheutz, F.; Dalsgaard, A. Occurrence and characterization of Shiga toxin-producing Escherichia coli o157: h7 and other non-sorbitol–fermenting e. coli in cattle and humans in urban areas of Morogoro, Tanzania. Vector-Borne Zoonotic Dis. 2014, 14, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Mkala, R.S.; Azizi, K.A. Prevalence and Antimicrobial Resistance Patterns of Extended Spectrum Beta Lactamase Producing Enterohemorrhagic Escherichia coli Strain O157: H7 from Cattle and Humans in Moshi, Northern Tanzania. Microbiol. Res. J. Int. 2017, 19, 1–10. [Google Scholar] [CrossRef]
- Ajuwon, B.I.; Babatunde, S.K.; Kolawole, O.M.; Ajiboye, A.E.; Lawal, A.H. Prevalence and antibiotic resistance of Escherichia coli O157: H7 in beef at a commercial slaughterhouse in Moro, Kwara State, Nigeria. Access Microbiol. 2021, 3, 000289. [Google Scholar] [CrossRef] [PubMed]
- Ayodele, O.A.; Deji-Agboola, A.M.; Akinduti, P.A.; Faneye, A.O. Phylo-diversity of prevalent human E. coli O157: H7 with strains from retailed meat and fish in selected markets in Ibadan Nigeria. J Immunoass. Immunochem. 2020, 41, 117–131. [Google Scholar] [CrossRef]
- Ekundayo, A.; Isibor, J. Screening of human and non-human specimens for Escherichia coli O157: H7 and Typhoid organisms in Benin City, Nigeria. J. Med. Res. 2012, 1, 052–056. [Google Scholar]
- Izevbuwa, O.E.; Okhuebor, S.O. Occurrence of l O157 H7 from meat products sold in Obinze abattoir, IMO State, Nigeria. Int. J. Appl. Biol. 2020, 4, 49–58. [Google Scholar]
- Ujoh, N.A.; Amala, S.E.; Nwokah, E.G.; Wachukwu, C.K. Detection of Escherichia coli O157: H7 and Non-O157 Strains in Meat, Human Stool, Abattoir Wastewater and Hygiene Practices among Meat Peddlers in Port Harcourt, Nigeria. Int. J. Pathog. Res. 2022, 9, 31–40. [Google Scholar] [CrossRef]
- Onwumere-Idolor, O.S.; Kperegbeyi, J.I.; Imonikebe, U.G.; Okoli, C.E.; Ajibo, F.E.; Njoga, E.O. Epidemiology of multidrug-resistant zoonotic E. coli from beef processing and retail points in Delta State, Nigeria: Public health implications. Prev. Vet. Med. 2024, 224, 106132. [Google Scholar] [CrossRef]
- Chigor, V.N.; Umoh, V.J.; Smith, S.I.; Igbinosa, E.O.; Okoh, A. Multidrug resistance and plasmid patterns of Escherichia coli O157 and other E. coli isolated from diarrhoeal stools and surface waters from some selected sources in Zaria, Nigeria. Int. J. Environ. Res. Public Health 2010, 7, 3831–3841. [Google Scholar] [CrossRef] [PubMed]
- Hammuel, C.; Jatau, E.D.; Whong, C.M. Prevalence and antibiogram pattern of some nosocomial pathogens isolated from Hospital Environment in Zaria, Nigeria. Aceh Int. J. Sci. Technol. 2014, 3, 131–139. [Google Scholar] [CrossRef]
- Fuh, N.J.; Christiana, O.M.; Attah, O.G.; Uteh, U.P.; Dantani, O.D.; Kolawole, F.V.; Ogechi, U.M. Risk Analysis and Antibiogram Spectrum of Escherichia coli O157: H7 Serotype from Children Stool and Raw Bovine Meat in Households Across Cross River State, Nigeria. Eur. J. Clin. Res. 2018, 4, 39–45. [Google Scholar] [CrossRef]
- Abong’o, B.; Momba, M. Prevalence and potential link between E. coli O157: H7 isolated from drinking water, meat and vegetables and stools of diarrhoeic confirmed and non-confirmed HIV/AIDS patients in the Amathole District–South Africa. J. Appl. Microbiol. 2008, 105, 424–431. [Google Scholar] [CrossRef]
- Ateba, C.N.; Mbewe, M. Detection of Escherichia coli O157: H7 virulence genes in isolates from beef, pork, water, human and animal species in the northwest province, South Africa: Public health implications. Res. Microbiol. 2011, 162, 240–248. [Google Scholar] [CrossRef]
- Ateba, C.N.; Mbewe, M. Determination of the genetic similarities of fingerprints from Escherichia coli O157: H7 isolated from different sources in the North West Province, South Africa using ISR, BOXAIR and REP-PCR analysis. Microbiol. Res. 2013, 168, 438–446. [Google Scholar] [CrossRef]
- Ateba, C.N.; Mbewe, M. Genotypic characterization of Escherichia coli O157: H7 isolates from different sources in the north-west province, South Africa, using enterobacterial repetitive intergenic consensus PCR analysis. Int. J. Mol. Sci. 2014, 15, 9735–9747. [Google Scholar] [CrossRef] [PubMed]
- Duvenage, S.; Korsten, L. Assessment of foodborne pathogen presence in the peach supply chain and its potential risk to the end consumer. Food Control 2017, 78, 374–382. [Google Scholar] [CrossRef]
- Msolo, L.; Igbinosa, E.O.; Okoh, A. Prevalence and antibiogram profiles of Escherichia coli O157: H7 isolates recovered from three selected dairy farms in the Eastern Cape Province, South Africa. Asian Pac. Trop. Dis. 2016, 6, 990–995. [Google Scholar] [CrossRef]
- Müller, E.; Taylor, M.; Grabow, W.; Ehlers, M.J.W.S.; Supply, T.W. Isolation and characterization of Escherichia coli O157: H7 and shiga toxin-converting bacteriophages from strains of human, bovine and porcine origin. Water 2002, 2, 29–38. [Google Scholar] [CrossRef]
- Ogundare, S.T.; Fasina, F.O.; Makumbi, J.-P.; van der Zel, G.A.; Geertsma, P.F.; Kock, M.M.; Smith, A.M.; Ehlers, M.M. Epidemiology and antimicrobial resistance profiles of pathogenic Escherichia coli from commercial swine and poultry abattoirs and farms in South Africa: A One Health approach. Sci. Total Environ. 2024, 951, 175705. [Google Scholar] [CrossRef] [PubMed]
- Ateba, C.; Mbewe, M.; Bezuidenhout, C. Prevalence of Escherichia coli O157 strains in cattle, pigs and humans in North West province, South Africa: Research in action. S. Afr. J. Sci. 2008, 104, 7–8. [Google Scholar]
- Momba, M.; Abong’o, B.; Mwambakana, J.N. Prevalence of enterohaemorrhagic Escherichia coli O157: H7 in drinking water and its predicted impact on diarrhoeic HIV/AIDS patients in the Amathole District, Eastern Cape Province, South Africa. Water SA 2008, 34, 365–372. [Google Scholar] [CrossRef]
- Myataza, A.; Igbinosa, E.O.; Igumbor, E.U.; Nontongana, N.; Okoh, A. Incidence and antimicrobial susceptibility of Escherichia coli O157: H7 isolates recovered from dairy farms in amathole district municipality, Eastern Cape, South Africa. Asian Pac. Trop. Dis. 2017, 7, 765–770. [Google Scholar] [CrossRef]
- Obi, C.; Green, E.; Bessong, P.; De Villiers, B.; Hoosen, A.; Igumbor, E.; Potgieter, N.J. Gene encoding virulence markers among Escherichia coli isolates from diarrhoeic stool samples and river sources in rural Venda communities of South Africa. Water SA 2004, 30, 37–42. [Google Scholar] [CrossRef]
- Abdissa, R.; Haile, W.; Fite, A.T.; Beyi, A.F.; Agga, G.E.; Edao, B.M.; Tadesse, F.; Korsa, M.G.; Beyene, T.; Beyene, T. Prevalence of Escherichia coli O157: H7 in beef cattle at slaughter and beef carcasses at retail shops in Ethiopia. BMC Infect. Dis. 2017, 17, 277. [Google Scholar] [CrossRef]
- Abunna, F.; Yimana, M.; Waketole, H.; Beyene, T.; Megersa, B. Detection and Antimicrobial Resistance Profile of E. coli O157: H7 from slaughterhouses and Butcher shops in Bishoftu Town, Central Oromia, Ethiopia. J. Food Microbiol. Saf. Hyg. 2023, 8, 269–280. [Google Scholar] [CrossRef]
- Assefa, A. Prevalence of Escherichia coli O157: H7 in foods of animal origin in Ethiopia: A meta-analysis. Cogent Food Agric. 2019, 5, 1642981. [Google Scholar] [CrossRef]
- Biruhtesfa Atnafie, B.A.; Degmawi Paulos, D.P.; Mesele Abera, M.A.; Genene Tefera, G.T.; Dereje Hailu, D.H.; Surafel Kasaye, S.K.; Kebede Amenu, K.A. Occurrence of Escherichia coli O157: H7 in cattle feces and contamination of carcass and various contact surfaces in abattoir and butcher shops of Hawassa, Ethiopia. BMC Microbiol. 2017, 17, 24. [Google Scholar] [CrossRef]
- Beyi, A.F.; Fite, A.T.; Tora, E.; Tafese, A.; Genu, T.; Kaba, T.; Beyene, T.J.; Beyene, T.; Korsa, M.G.; Tadesse, F. Prevalence and antimicrobial susceptibility of Escherichia coli O157 in beef at butcher shops and restaurants in central Ethiopia. BMC Microbiol. 2017, 17, 49. [Google Scholar] [CrossRef] [PubMed]
- Dejene, H.; Abunna, F.; Tuffa, A.C.; Gebresenbet, G. Epidemiology and antimicrobial susceptibility pattern of E. coli O157: H7 along dairy milk supply chain in Central Ethiopia. Vet. Med. Res. Rep. 2022, 13, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Fikadu, Y.; Kabeta, T.; Diba, D.; Waktole, H. Antimicrobial Profiles and Conventional PCR Assay of Shiga Toxigenic Escherichia coli O157: H7 (STEC) Isolated from Cattle Slaughtered at Bedele Municipal Abattoir, South West Ethiopia. Infect. Drug Resist. 2023, 16, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Mengistu, S.; Abayneh, E.; Shiferaw, D. E. coli O157: H7 and Salmonella species: Public health importance and microbial safety in beef at selected slaughter houses and retail shops in eastern Ethiopia. J. Vet. Sci. Technol. 2017, 8, 2. [Google Scholar] [CrossRef]
- Sebsibe, M.A.; Asfaw, E.T. Occurrence of multi-drug resistant Escherichia coli and Escherichia coli O157: H7 in meat and swab samples of various contact surfaces at abattoir and butcher shops in Jimma town, Southwest district of Ethiopia. Infect. Drug Resist. 2020, 13, 3853–3862. [Google Scholar] [CrossRef]
- Tarekegn, A.A.; Mitiku, B.A.; Alemu, Y. Escherichia coli O157: H7 beef carcass contamination and its antibiotic resistance in Awi Zone, Northwest Ethiopia. Food Sci. Nutr. 2023, 11, 6140–6150. [Google Scholar] [CrossRef] [PubMed]
- Edget, A.; Shiferaw, D.; Mengistu, S. Microbial safety and its public health concern of E. coli O157: H7 and Salmonella spp. in beef at Dire Dawa administrative city and Haramaya University, Ethiopia. J. Vet. Med. Anim. Health 2017, 9, 213–227. [Google Scholar] [CrossRef]
- Mesele, F.; Leta, S.; Amenu, K.; Abunna, F. Occurrence of Escherichia coli O157: H7 in lactating cows and dairy farm environment and the antimicrobial susceptibility pattern at Adami Tulu Jido Kombolcha District, Ethiopia. BMC Vet. Res. 2023, 19, 6. [Google Scholar] [CrossRef]
- Gutema, F.D.; De Zutter, L.; Piérard, D.; Hinckel, B.; Imamura, H.; Rasschaert, G.; Abdi, R.D.; Agga, G.E.; Crombé, F. Core Genome Sequencing Analysis of E. coli O157: H7 Unravelling Genetic Relatedness among Strains from Cattle, Beef, and Humans in Bishoftu, Ethiopia. Microbiol. Res. 2023, 14, 148–160. [Google Scholar] [CrossRef]
- Gutema, F.D.; Rasschaert, G.; Agga, G.E.; Jufare, A.; Duguma, A.B.; Abdi, R.D.; Duchateau, L.; Crombe, F.; Gabriël, S.; De Zutter, L. Occurrence, molecular characteristics, and antimicrobial resistance of Escherichia coli O157 in cattle, beef, and humans in Bishoftu Town, Central Ethiopia. Foodborne Pathog. Dis. 2021, 18, 1–7. [Google Scholar] [CrossRef]
- Gemeda, B.A.; Wieland, B.; Alemayehu, G.; Knight-Jones, T.J.; Wodajo, H.D.; Tefera, M.; Kumbe, A.; Olani, A.; Abera, S.; Amenu, K. Antimicrobial resistance of Escherichia coli isolates from Livestock and the environment in extensive smallholder Livestock production systems in Ethiopia. Antibiotics 2023, 12, 941. [Google Scholar] [CrossRef] [PubMed]
- Engda, T.; Tessema, B.; Mesifin, N.; Nuru, A.; Belachew, T.; Moges, F. Shiga toxin-producing Escherichia coli O157: H7 among diarrheic patients and their cattle in Amhara National Regional State, Ethiopia. PLoS ONE 2023, 18, e0295266. [Google Scholar] [CrossRef] [PubMed]
- Kunadu, A.P.-H.; Otwey, R.Y.; Mosi, L. Microbiological quality and Salmonella prevalence, serovar distribution and antimicrobial resistance associated with informal raw chicken processing in Accra, Ghana. Food Control 2020, 118, 107440. [Google Scholar] [CrossRef]
- Donkor, E.; Lanyo, R.; Akyeh, M.; Kayang, B.; Boniface, J.; Quaye, J. Monitoring enterohaemorrhagic Escherichia coli 0157: H7 in the vegetable food chain in Ghana. Res. J. Microbiol. 2008, 3, 423–428. [Google Scholar] [CrossRef]
- Roughyatou, K. Recherche d’Escherichia coli O157 dans les selles et les produits carnés. Rev. Afr. Malgache Rech. Sci. Sci. Santé 2015, 2. ISSN 2630-1113. [Google Scholar]
- Bankole, H.S.; Dougnon, V.T.; Johnson, R.C.; Dougnon, T.; Yehouenou, B.; Kougblenou, S.; Agonsa, M.; Legonou, M.; Dadie, T.; Baba-Moussa, L. Assessment of the contamination of some foodstuffs by Escherichia coli O157 in Benin, West Africa. Int. J. Microbiol. 2014, 2014, 417848. [Google Scholar] [CrossRef]
- Uthman, O.A.; Uthman, M.B. Geography of Africa biomedical publications: An analysis of 1996–2005 PubMed papers. Afr. J. Food Agri. Nutr. Dev. 2008, 8, 252–263. [Google Scholar] [CrossRef][Green Version]
- Somda, N.; Tankoano, A.; Métuor-Dabiré, A.; Kaboré, D.; Bonkoungou, J.; Kpoda, D.; Sambe-Ba, B.; Dabiré, Y.; Saba, C.; Ouoba, I. A systematic review and meta-analysis of antibiotic resistance of foodborne pathogenic bacteria in West Africa between 2010 and 2020. J. Food Prot. 2023, 86, 100061. [Google Scholar] [CrossRef]
- Adzitey, F.; Huda, N.; Ali, G.R. Molecular techniques for detecting and typing of bacteria, advantages and application to foodborne pathogens isolated from ducks. 3 Biotech 2013, 3, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Dossouvi, K.M.; Bakpatina-Batako, K.D. Carbapenem resistance in West Africa: A systematic review. Microbiol. Indep. Res. J. 2024, 11, 25–56. [Google Scholar] [CrossRef]
- Abdel-Aziz, M.A.; Eid, R.A. Detection of Escherichia coli O157: H7 from patients with Gastroenteritis. Egypt. J. Med. Microbiol. 2024, 33, 145–152. [Google Scholar] [CrossRef]
- Alua, A.J.; Omeizaa, G.K.; Amehb, J.A.; Enem, S.I. Prevalence and antibiotic resistance profile of Shiga-toxigenic Escherichia coli O157 (STEC) from retailed miscellaneous meat and fish types in Abuja, Nigeria. Vet. World 2021, 2, 37–43. [Google Scholar] [CrossRef]
- Diab, M.S.; Tarabees, R.; Elnaker, Y.F.; Hadad, G.A.; Saad, M.A.; Galbat, S.A.; Albogami, S.; Hassan, A.M.; Dawood, M.A.; Shaaban, S.I. Molecular detection, serotyping, and antibiotic resistance of shiga toxigenic Escherichia coli isolated from she-camels and in-contact humans in Egypt. Antibiotics 2021, 10, 1021. [Google Scholar] [CrossRef]
- Iweriebor, B.C.; Iwu, C.J.; Obi, L.C.; Nwodo, U.U.; Okoh, A.I. Multiple antibiotic resistances among Shiga toxin producing Escherichia coli O157 in feces of dairy cattle farms in Eastern Cape of South Africa. BMC Microbiol. 2015, 15, 213. [Google Scholar] [CrossRef]
- Iwu, C.D.; du Plessis, E.; Korsten, L.; Okoh, A. Prevalence of E. coli O157:H7 strains in irrigation water and agricultural soil in two district municipalities in South Africa. Int. J. Environ. Studies 2021, 78, 474–483. [Google Scholar] [CrossRef]
- de Assis, D.C.S.; da Silva, T.M.L.; Brito, R.F.; da Silva, L.C.G.; Lima, W.G.; Brito, J.C. Shiga toxin-producing Escherichia coli (STEC) in bovine meat and meat products over the last 15 years in Brazil: A systematic review and meta-analysis. Meat Sci. 2021, 173, 108394. [Google Scholar] [CrossRef] [PubMed]
- Ayalew Assefa, A.A.; Amare Bihon, A.B. A systematic review and meta-analysis of prevalence of Escherichia coli in foods of animal origin in Ethiopia. Heliyon 2018, 4, e00716. [Google Scholar] [CrossRef] [PubMed]
- Tayh, G.; Boubaker, S.M.; Khedher, R.B.; Jbeli, M.; Chehida, F.B.; Mamlouk, A.; Dâaloul-Jedidi, M.; Messadi, L. Prevalence, virulence genes, and antimicrobial profiles of Escherichia coli O157: H7 isolated from healthy cattle in Tunisia. J. Infect. Dev. Ctries. 2022, 16, 1308–1316. [Google Scholar] [CrossRef]
- Abreham, S.; Teklu, A.; Cox, E.; Sisay Tessema, T. Escherichia coli O157:H7: Distribution, molecular characterization, antimicrobial resistance patterns and source of contamination of sheep and goat carcasses at an export abattoir, Mojdo, Ethiopia. BMC Microbiol. 2019, 19, 215. [Google Scholar] [CrossRef]
- Saxena, T.; Kaushik, P.; Mohan, M.K. Prevalence of E. coli O157:H7 in water sources: An overview on associated diseases, outbreaks and detection methods. Diagn. Microbiol. Infect. Dis. 2015, 82, 249–264. [Google Scholar] [CrossRef]
- Somda, N.S.; Bonkoungou, I.J.O.; Sambe-Ba, B.; Drabo, M.S.; Wane, A.A.; Sawadogo-Lingani, H.; Savadogo, A. Diversity and antimicrobial drug resistance of non-typhoid Salmonella serotypes isolated in lettuce, irrigation water and clinical samples in Burkina Faso. J. Agric. Res. 2021, 5, 100167. [Google Scholar] [CrossRef]
- Dallman, T.J.; Ashton, P.M.; Byrne, L.; Perry, N.T.; Petrovska, L.; Ellis, R.; Allison, L.; Hanson, M.; Holmes, A.; Gunn, G. Applying phylogenomics to understand the emergence of Shiga-toxin-producing Escherichia coli O157: H7 strains causing severe human disease in the UK. Microb. Genom. 2015, 1, e000029. [Google Scholar] [CrossRef]
- Phu, D.H.; Wongtawan, T.; Truong, D.B.; Van Cuong, N.; Carrique-Mas, J.; Thomrongsuwannakij, T. A systematic review and meta-analysis of integrated studies on antimicrobial resistance in Vietnam, with a focus on Enterobacteriaceae, from a One Health perspective. One Health 2022, 15, 100465. [Google Scholar] [CrossRef]
- Fujita, A.W.; Werner, K.; Jacob, J.T.; Tschopp, R.; Mamo, G.; Mihret, A.; Abdissa, A.; Kempker, R.; Rebolledo, P.A. Antimicrobial resistance through the lens of one health in Ethiopia: A review of the literature among humans, animals, and the environment. Int. J. Infect. Dis. 2022, 119, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Hounkpe, C.E.; Sessou, P.; Farougou, S.; Daube, G.; Delcenserie, V.; Azokpota, P.; Korsak, N. Prevalence, antibiotic resistance, and virulence gene profile of Escherichia coli strains shared between food and other sources in Africa: A systematic review. Vet. World 2023, 16, 2016–2028. [Google Scholar] [CrossRef]
- Pormohammad, A.; Nasiri, M.J.; Azimi, T. Prevalence of antibiotic resistance in Escherichia coli strains simultaneously isolated from humans, animals, food, and the environment: A systematic review and meta-analysis. Infect. Drug Resist. 2019, 12, 1181–1197. [Google Scholar] [CrossRef] [PubMed]
- ANSES; National Agency for Sanitary Safety of Food, the Environment and Work. Review of Monitoring of Antibiotic Resistance in Animal Health and the Sale of Antibiotics for Veterinary Use; ANSES: Maisons-Alfort, France, 2023; Available online: https://www.anses.fr/fr/system/files/Press2023DPA01.pdf (accessed on 26 October 2024).
- Tozluyurt, A. Fosfomycin in the treatment of New Delhi Metallo-β-Lactamase-5 (blaNDM-5)-producing Escherichia coli infection. Ger. J. Microbiol. 2024, 4, 1–5. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).