Safety Assessment of Lactiplantibacillus plantarum GUANKE Based on Whole-Genome Sequencing, Phenotypic, and Anti-Inflammatory Capacity Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatic Analysis of the Genome of L. plantarum GUANKE
2.2. Functional Genome Annotation
2.3. Bacteria and Cell Culture
2.4. Phenotypic Characterization
2.5. Hemolytic Activity Assessment
2.6. Antibiotic Sensitivity Analysis
2.7. Bacterial Adhesion Capacity Test
2.8. In Vitro Cytotoxicity Assays
2.9. Isolation and Culture of Mice Bone Marrow-Derived Macrophage (mBMDMs) Cells
2.10. In Vitro Anti-Inflammatory Capacity Testing
2.11. Inflammatory Cytokine Assay
2.12. Animals
2.13. RNA Sequence
2.14. Statistical Analysis
3. Results and Discussion
3.1. Genomic Characterization of L. plantarum GUANKE
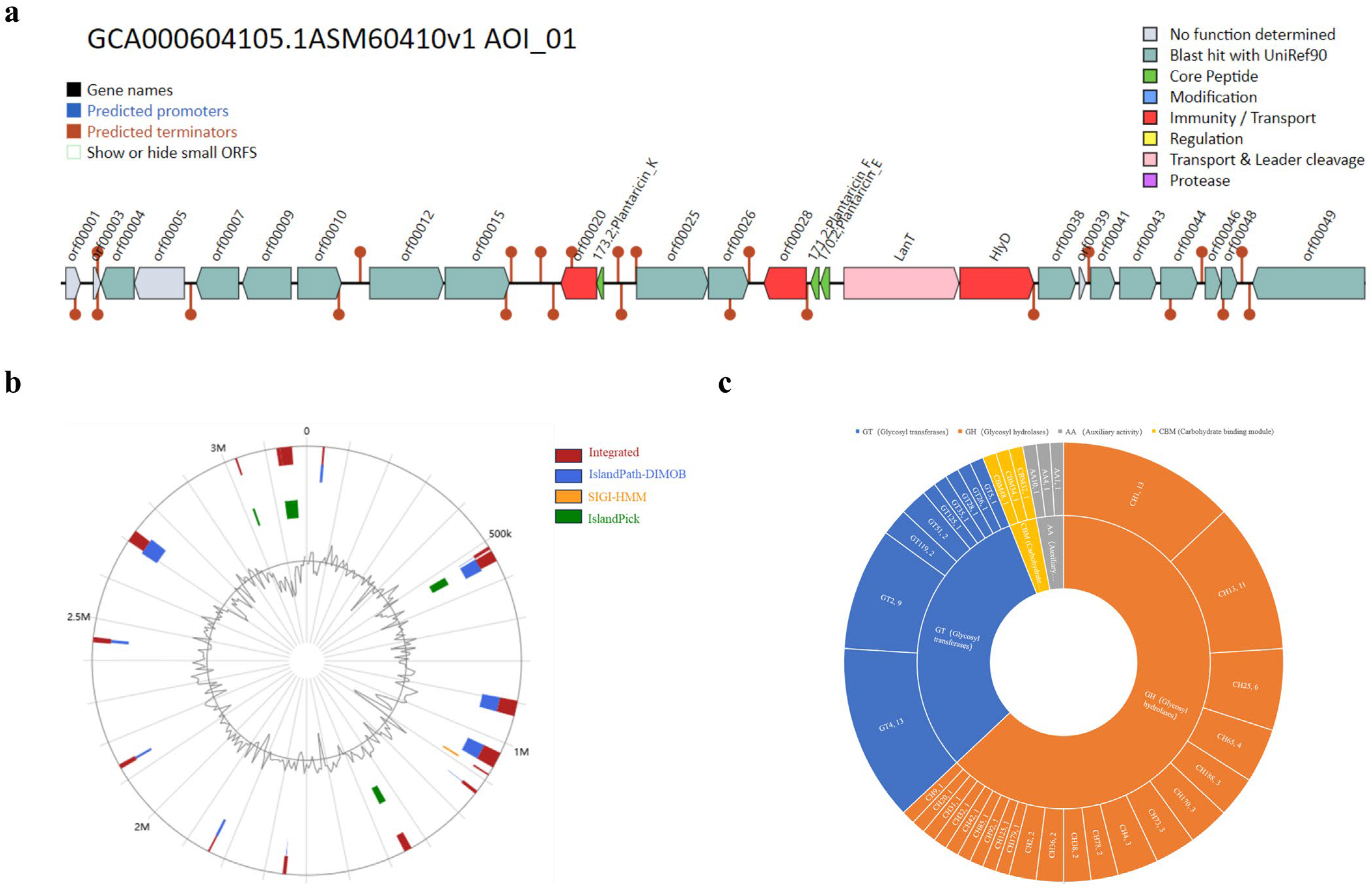
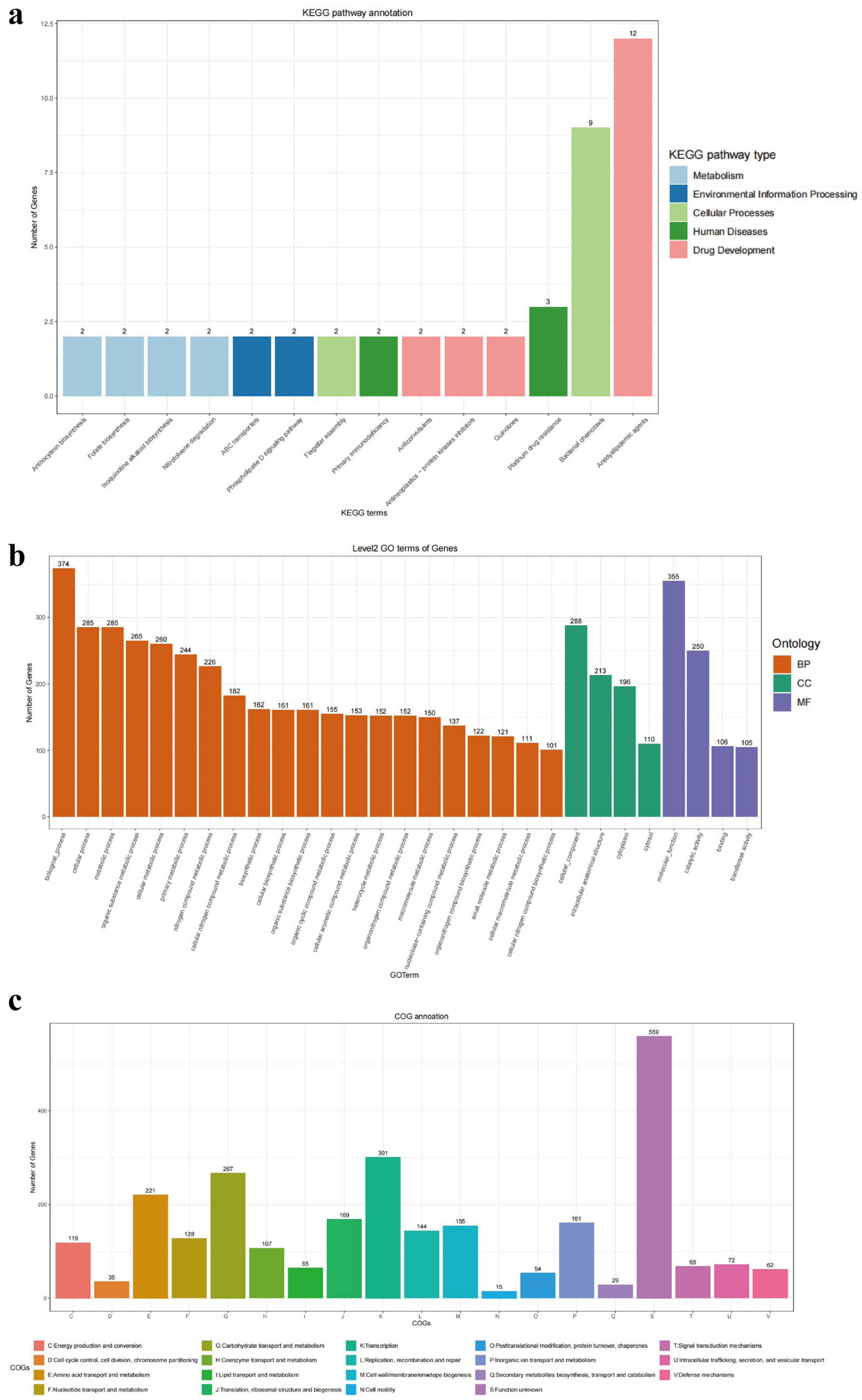
3.2. Genomic Function in L. plantarum GUANKE
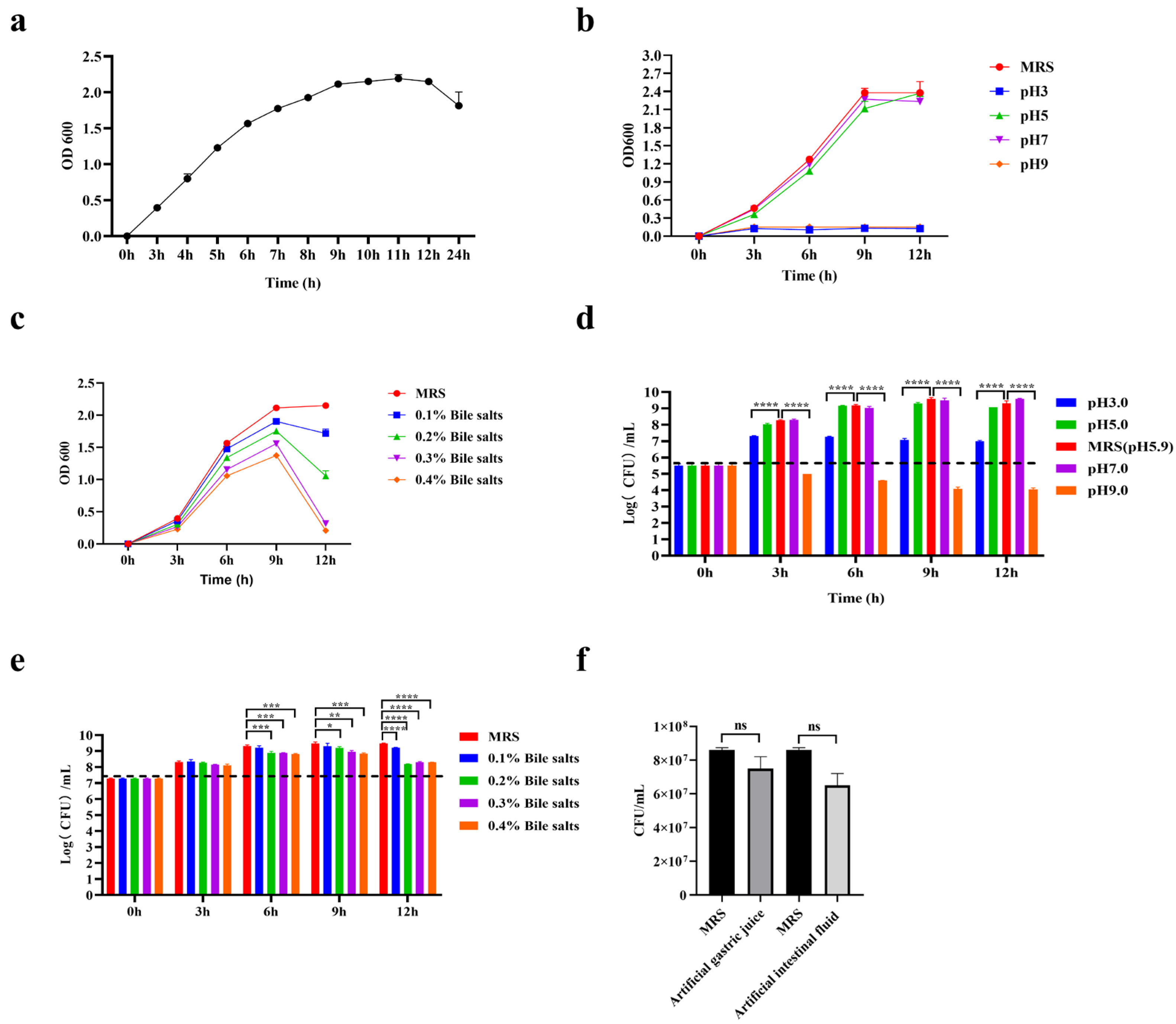
3.3. General Characteristics of L. plantarum GUANKE
3.4. Hemolysis Analysis, Antibiotic Susceptibility Analysis, and Adhesion Capacity of L. plantarum GUANKE
3.5. Cytotoxicity of L. plantarum GUANKE
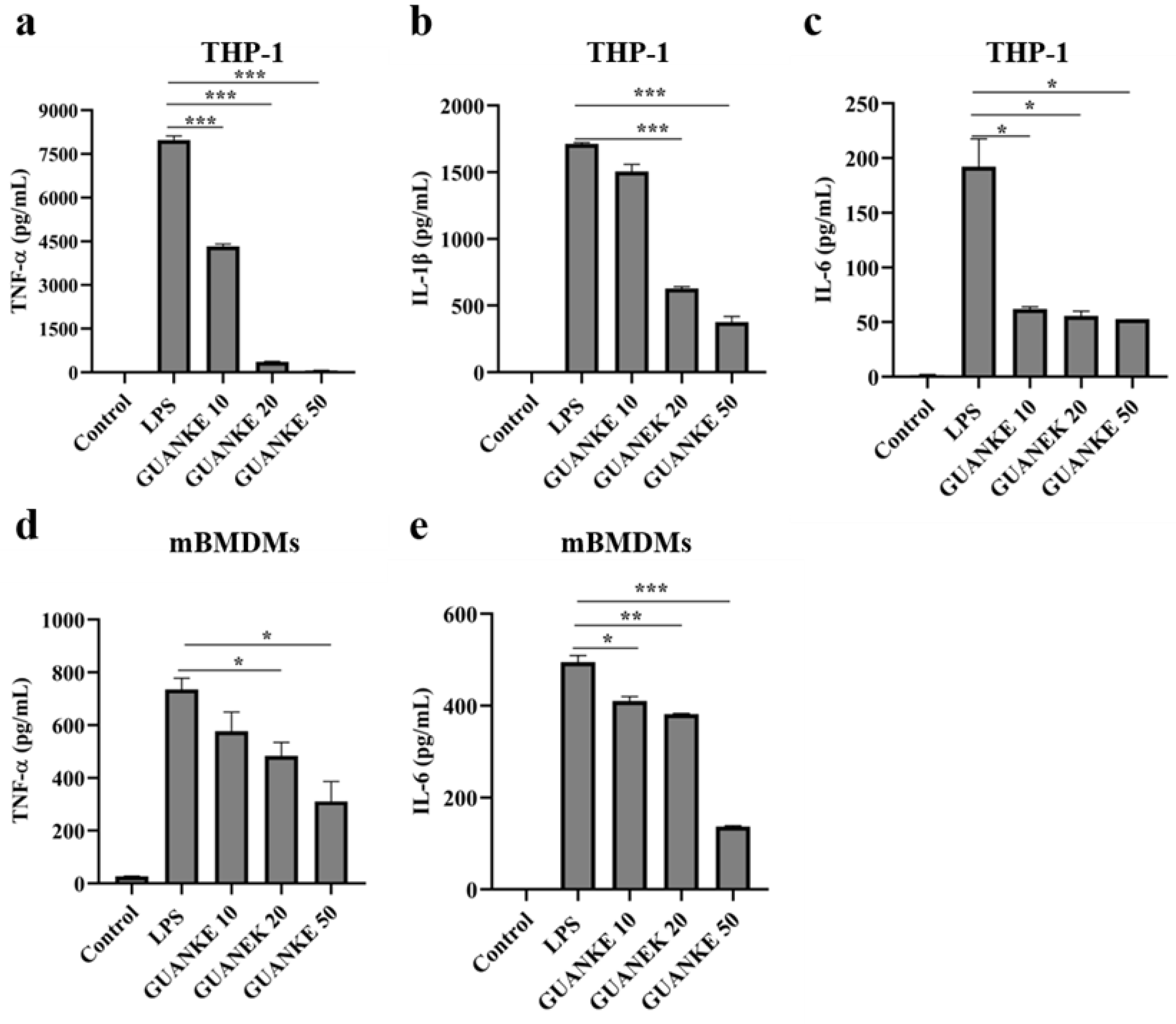
3.6. In Vitro Anti-Inflammatory Ability of L. plantarum GUANKE
3.7. L. plantarum GUANKE Enhances Apical Junction Pathway and Interferon Response Pathway in Mouse Colon Tissue
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| Cas | CRISPR-associated |
| LPS | Lipopolysaccharide |
| CAZymes | Carbohydrate-active enzymes |
| FAO | Food and Agriculture Organization of the United Nations (FAO) |
| WHO | World Health Organization |
| WGS | Whole-genome sequencing (WGS) |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| CARD | Comprehensive Antibiotic Resistance Database |
| PHASTER | Phage Search Tool-Enhanced Release |
| FBS | Fetal bovine serum |
| PMA | Phorbol 12-myristate 13-acetate |
| OD600 | Optical density 600 |
| MIC | Minimum inhibitory concentration |
| EFSA | European Union Commission recommendations for probiotic safety |
| LDH | Lactate dehydrogenase |
| ELISA | Enzyme-linked immunosorbent assay |
| M-CSF | Macrophage-stimulating factor |
| GSEA | Gene Set Enrichment Analysis |
References
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert Consensus Document. The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Marteau, P. Safety Aspects of Probiotic Products. Näringsforskning 2001, 45, 22–24. [Google Scholar] [CrossRef]
- Vahabnezhad, E.; Mochon, A.B.; Wozniak, L.J.; Ziring, D.A. Lactobacillus Bacteremia Associated with Probiotic Use in a Pediatric Patient with Ulcerative Colitis. J. Clin. Gastroenterol. 2013, 47, 437–439. [Google Scholar] [CrossRef] [PubMed]
- Merenstein, D.; Pot, B.; Leyer, G.; Ouwehand, A.C.; Preidis, G.A.; Elkins, C.A.; Hill, C.; Lewis, Z.T.; Shane, A.L.; Zmora, N.; et al. Emerging Issues in Probiotic Safety: 2023 Perspectives. Gut Microbes 2023, 15, 2185034. [Google Scholar] [CrossRef]
- Liu, B.; Zhong, X.; Liu, Z.; Guan, X.; Wang, Q.; Qi, R.; Zhou, X.; Huang, J. Probiotic Potential and Safety Assessment of Lactiplantibacillus plantarum Cqf-43 and Whole-Genome Sequence Analysis. Int. J. Mol. Sci. 2023, 24, 17570. [Google Scholar] [CrossRef]
- Mileriene, J.; Aksomaitiene, J.; Kondrotiene, K.; Asledottir, T.; Vegarud, G.E.; Serniene, L.; Malakauskas, M. Whole-Genome Sequence of Lactococcus lactis subsp. lactis LL16 Confirms Safety, Probiotic Potential, and Reveals Functional Traits. Microorganisms 2023, 11, 1034. [Google Scholar] [CrossRef]
- Stergiou, O.S.; Tegopoulos, K.; Kiousi, D.E.; Tsifintaris, M.; Papageorgiou, A.C.; Tassou, C.C.; Chorianopoulos, N.; Kolovos, P.; Galanis, A. Whole-Genome Sequencing, Phylogenetic and Genomic Analysis of Lactiplantibacillus pentosus L33, a Potential Probiotic Strain Isolated From Fermented Sausages. Front. Microbiol. 2021, 12, 746659. [Google Scholar] [CrossRef]
- Echegaray, N.; Yilmaz, B.; Sharma, H.; Kumar, M.; Pateiro, M.; Ozogul, F.; Lorenzo, J.M. A Novel Approach to Lactiplantibacillus plantarum: From Probiotic Properties to the Omics Insights. Microbiol. Res. 2023, 268, 127289. [Google Scholar] [CrossRef]
- Prete, R.; Long, S.L.; Gallardo, A.L.; Gahan, C.G.; Corsetti, A.; Joyce, S.A. Beneficial Bile Acid Metabolism from Lactobacillus plantarum of Food Origin. Sci. Rep. 2020, 10, 1165. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, H.; Rehman, M.U.; Mehmood, K.; Jiang, X.; Iqbal, M.; Tong, X.; Gao, X.; Li, J. Antibacterial Activity of Lactobacillus plantarum Isolated from Tibetan Yaks. Microb. Pathog. 2018, 115, 293–298. [Google Scholar] [CrossRef]
- Zhang, N.; Li, C.; Niu, Z.; Kang, H.; Wang, M.; Zhang, B.; Tian, H. Colonization and Immunoregulation of Lactobacillus plantarum BF_15, a Novel Probiotic Strain from the Feces of Breast-Fed Infants. Food Funct. 2020, 11, 3156–3166. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhao, Q.; Li, T.; Lu, L.; Wang, F.; Zhang, H.; Liu, Z.; Ma, H.; Zhu, Q.; Wang, J.; et al. Lactobacillus plantarum-Derived Indole-3-Lactic Acid Ameliorates Colorectal Tumorigenesis via Epigenetic Regulation of CD8+ T Cell Immunity. Cell Metab. 2023, 35, 943–960.e9. [Google Scholar] [CrossRef] [PubMed]
- Padro, T.; Santisteban, V.; Huedo, P.; Puntes, M.; Aguiló, M.; Espadaler-Mazo, J.; Badimon, L. Lactiplantibacillus plantarum Strains KABP011, KABP012, and KABP013 Modulate Bile Acids and Cholesterol Metabolism in Humans. Cardiovasc. Res. 2024, 120, 708–722. [Google Scholar] [CrossRef] [PubMed]
- Kamil, R.Z.; Murdiati, A.; Juffrie, M.; Rahayu, E.S. Gut Microbiota Modulation of Moderate Undernutrition in Infants Through Gummy Lactobacillus plantarum Dad-13 Consumption: A Randomized Double-Blind Controlled Trial. Nutrients 2022, 14, 1049. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A General Classification Scheme for Bacterial Virulence Factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2019, 48, gkz935. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an Update of CRISRFinder, Includes a Portable Version, Enhanced Performance and Integrates Search for Cas Proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A Better, Faster Version of the PHAST Phage Search Tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A User-Friendly Web Server to Thoroughly Mine RiPPs and Bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Dhillon, B.K.; Chiu, T.A.; Laird, M.R.; Langille, M.G.I.; Brinkman, F.S.L. IslandViewer Update: Improved Genomic Island Discovery and Visualization. Nucleic Acids Res. 2013, 41, W129–W132. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-Mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The Carbohydrate-Active Enzymes Database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Pascal Andreu, V.; Augustijn, H.E.; Chen, L.; Zhernakova, A.; Fu, J.; Fischbach, M.A.; Dodd, D.; Medema, M.H. gutSMASH Predicts Specialized Primary Metabolic Pathways from the Human Gut Microbiota. Nat. Biotechnol. 2023, 41, 1416–1423. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Ren, Z.; Cao, K.; Li, X.; Yang, J.; Luo, X.; Zhu, L.; Wang, X.; Ding, L.; Liang, J.; et al. Boosting Vaccine-Elicited Respiratory Mucosal and Systemic COVID-19 Immunity in Mice with the Oral Lactobacillus plantarum. Front. Nutr. 2021, 8, 789242. [Google Scholar] [CrossRef]
- Guo, M.; Liu, H.; Yu, Y.; Zhu, X.; Xie, H.; Wei, C.; Mei, C.; Shi, Y.; Zhou, N.; Qin, K.; et al. Lactobacillus rhamnosus GG Ameliorates Osteoporosis in Ovariectomized Rats by Regulating the Th17/Treg Balance and Gut Microbiota Structure. Gut Microbes 2023, 15, 2190304. [Google Scholar] [CrossRef]
- Chen, T.; Wang, L.; Li, Q.; Long, Y.; Lin, Y.; Yin, J.; Zeng, Y.; Huang, L.; Yao, T.; Abbasi, M.N.; et al. Functional Probiotics of Lactic Acid Bacteria from Hu Sheep Milk. BMC Microbiol. 2020, 20, 228. [Google Scholar] [CrossRef]
- He, S.; Wang, C.; Huang, Y.; Lu, S.; Li, W.; Ding, N.; Chen, C.; Wu, Y. Chlamydia psittaci Plasmid-Encoded CPSIT_P7 Induces Macrophage Polarization to Enhance the Antibacterial Response Through TLR4-Mediated MAPK and NF-κB Pathways. Biochim. Biophys. Acta Mol. Cell Res. 2022, 1869, 119324. [Google Scholar] [CrossRef]
- Lu, S.; He, S.; Yue, K.; Mi, J.; Huang, Y.; Song, L.; Yang, T.; Ren, Z.; Ren, L.; Xu, J. Lactobacillus plantarum GUANKE Modulate Anti-Viral Function of Dendritic Cells in Mice. Int. Immunopharmacol. 2024, 134, 112169. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A Reference Database for Bacterial Virulence Factors. Nucleic Acids Res. 2004, 33, D325–D328. [Google Scholar] [CrossRef]
- Abbas, A.; Barkhouse, A.; Hackenberger, D.; Wright, G.D. Antibiotic Resistance: A Key Microbial Survival Mechanism That Threatens Public Health. Cell Host Microbe 2024, 32, 837–851. [Google Scholar] [CrossRef]
- Tóth, A.G.; Judge, M.F.; Nagy, S.Á.; Papp, M.; Solymosi, N. A Survey on Antimicrobial Resistance Genes of Frequently Used Probiotic Bacteria, 1901 to 2022. Eurosurveillance 2023, 28, 2200272. [Google Scholar] [CrossRef] [PubMed]
- Deghorain, M.; Goffin, P.; Fontaine, L.; Mainardi, J.-L.; Daniel, R.; Errington, J.; Hallet, B.; Hols, P. Selectivity for D-Lactate Incorporation into the Peptidoglycan Precursors of Lactobacillus plantarum: Role of Aad, a VanX-like D-Alanyl-D-Alanine Dipeptidase. J. Bacteriol. 2007, 189, 4332–4337. [Google Scholar] [CrossRef] [PubMed]
- Kahne, D.; Leimkuhler, C.; Lu, W.; Walsh, C. Glycopeptide and Lipoglycopeptide Antibiotics. Chem. Rev. 2005, 105, 425–448. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Li, F.; Zhang, M.; Xia, Y.; Ai, L.; Wang, G. Effect of D-Ala-Ended Peptidoglycan Precursors on the Immune Regulation of Lactobacillus plantarum Strains. Front. Immunol. 2022, 12, 825825. [Google Scholar] [CrossRef]
- Cui, Y.; Qu, X. CRISPR-Cas Systems of Lactic Acid Bacteria and Applications in Food Science. Biotechnol. Adv. 2024, 71, 108323. [Google Scholar] [CrossRef]
- Roberts, A.; Barrangou, R. Applications of CRISPR-Cas Systems in Lactic Acid Bacteria. FEMS Microbiol. Rev. 2020, 44, 523–537. [Google Scholar] [CrossRef]
- Crawley, A.B.; Henriksen, E.D.; Stout, E.; Brandt, K.; Barrangou, R. Characterizing the Activity of Abundant, Diverse and Active CRISPR-Cas Systems in Lactobacilli. Sci. Rep. 2018, 8, 11544. [Google Scholar] [CrossRef]
- Zhong, Z.; Liu, G.; Tang, Z.; Xiang, S.; Yang, L.; Huang, L.; He, Y.; Fan, T.; Liu, S.; Zheng, X.; et al. Efficient Plant Genome Engineering Using a Probiotic Sourced CRISPR-Cas9 System. Nat. Commun. 2023, 14, 6102. [Google Scholar] [CrossRef]
- Chen, X.; Wei, Y.; Ji, X. Research Progress of Prophages. Yi Chuan 2021, 43, 240–248. [Google Scholar] [CrossRef]
- Pei, Z.; Sadiq, F.A.; Han, X.; Zhao, J.; Zhang, H.; Ross, R.P.; Lu, W.; Chen, W. Comprehensive Scanning of Prophages in Lactobacillus: Distribution, Diversity, Antibiotic Resistance Genes, and Linkages with CRISPR-Cas Systems. mSystems 2021, 6, e01211-20. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Hsiao, W.W.L.; Brinkman, F.S.L. Detecting Genomic Islands Using Bioinformatics Approaches. Nat. Rev. Microbiol. 2010, 8, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Tarrah, A.; Da Silva Duarte, V.; Pakroo, S.; Corich, V.; Giacomini, A. Genomic and Phenotypic Assessments of Safety and Probiotic Properties of Streptococcus macedonicus Strains of Dairy Origin. Food Res. Int. 2020, 130, 108931. [Google Scholar] [CrossRef] [PubMed]
- Garbacz, K. Anticancer Activity of Lactic Acid Bacteria. Semin. Cancer Biol. 2022, 86, 356–366. [Google Scholar] [CrossRef] [PubMed]
- Heeney, D.D.; Zhai, Z.; Bendiks, Z.; Barouei, J.; Martinic, A.; Slupsky, C.; Marco, M.L. Lactobacillus plantarum Bacteriocin Is Associated with Intestinal and Systemic Improvements in Diet-Induced Obese Mice and Maintains Epithelial Barrier Integrity in Vitro. Gut Microbes 2019, 10, 382–397. [Google Scholar] [CrossRef]
- Martinez, F.A.C.; Balciunas, E.M.; Converti, A.; Cotter, P.D.; De Souza Oliveira, R.P. Bacteriocin Production by Bifidobacterium spp. A Review. Biotechnol. Adv. 2013, 31, 482–488. [Google Scholar] [CrossRef]
- Kos, B.; Šušković, J.; Vuković, S.; Šimpraga, M.; Frece, J.; Matošić, S. Adhesion and Aggregation Ability of Probiotic Strain Lactobacillus acidophilus M92. J. Appl. Microbiol. 2003, 94, 981–987. [Google Scholar] [CrossRef]
- Xu, H.; Ma, C.; Zhao, F.; Chen, P.; Liu, Y.; Sun, Z.; Cui, L.; Kwok, L.-Y.; Zhang, H. Adjunctive Treatment with Probiotics Partially Alleviates Symptoms and Reduces Inflammation in Patients with Irritable Bowel Syndrome. Eur. J. Nutr. 2021, 60, 2553–2565. [Google Scholar] [CrossRef]
- Casaro, M.B.; Thomas, A.M.; Mendes, E.; Fukumori, C.; Ribeiro, W.R.; Oliveira, F.A.; Crisma, A.R.; Murata, G.M.; Bizzarro, B.; Sá-Nunes, A.; et al. A Probiotic Has Differential Effects on Allergic Airway Inflammation in A/J and C57BL/6 Mice and Is Correlated with the Gut Microbiome. Microbiome 2021, 9, 134. [Google Scholar] [CrossRef]
- Salaris, C.; Scarpa, M.; Elli, M.; Bertolini, A.; Guglielmetti, S.; Pregliasco, F.; Brun, P.; Castagliuolo, I. Lacticaseibacillus paracasei DG Enhances the Lactoferrin Anti-SARS-CoV-2 Response in Caco-2 Cells. Gut Microbes 2021, 13, 1961970. [Google Scholar] [CrossRef]
- Baum, B.; Georgiou, M. Dynamics of Adherens Junctions in Epithelial Establishment, Maintenance, and Remodeling. J. Cell Biol. 2011, 192, 907–917. [Google Scholar] [CrossRef]
- Denker, B.M.; Nigam, S.K. Molecular Structure and Assembly of the Tight Junction. Am. J. Physiol.-Ren. Physiol. 1998, 274, F1–F9. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.J.; Kim, W.-K.; Han, D.H.; Lee, K.; Ko, G. Lactobacillus fermentum Species Ameliorate Dextran Sulfate Sodium-Induced Colitis by Regulating the Immune Response and Altering Gut Microbiota. Gut Microbes 2019, 10, 696–711. [Google Scholar] [CrossRef] [PubMed]
- Orlando, A.; Linsalata, M.; Bianco, G.; Notarnicola, M.; D’Attoma, B.; Scavo, M.P.; Tafaro, A.; Russo, F. Lactobacillus rhamnosus GG Protects the Epithelial Barrier of Wistar Rats from the Pepsin-Trypsin-Digested Gliadin (PTG)-Induced Enteropathy. Nutrients 2018, 10, 1698. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W. Interferon-Stimulated Genes: What Do They All Do? Annu. Rev. Virol. 2019, 6, 567–584. [Google Scholar] [CrossRef]
- Kim, S.; Lee, S.; Kim, T.-Y.; Lee, S.-H.; Seo, S.-U.; Kweon, M.-N. Newly Isolated Lactobacillus paracasei Strain Modulates Lung Immunity and Improves the Capacity to Cope with Influenza Virus Infection. Microbiome 2023, 11, 260. [Google Scholar] [CrossRef]




| Gene | Identity | Mode Type | Drug Class | Mechanism | Gene Family |
|---|---|---|---|---|---|
| vanY | 27.35% | protein homolog model | glycopeptide antibiotic | antibiotic target alteration | glycopeptide resistance gene cluster |
| vanH | 35.02% | protein homolog model | glycopeptide antibiotic | antibiotic target alteration | glycopeptide resistance gene cluster |
| Region | Length | Completeness | Total Proteins | Start | End | GC% |
|---|---|---|---|---|---|---|
| 1 | 18.1 kb | questionable | 23 | 37,804 | 55,910 | 42.44% |
| 2 | 38.9 kb | Intact | 52 | 513,739 | 552,681 | 41.47% |
| 3 | 42.7 kb | Intact | 53 | 1,021,684 | 1,064,390 | 41.50% |
| Type | Class | From | To | Core Biosynthetic Gene | Similarity |
|---|---|---|---|---|---|
| Nitrate reductase | E-MGC | 1,339,351 | 1,365,822 | ctg1_1263, ctg1_1264, ctg1_1265, ctg1_1266 | 40% |
| Pyruvate to acetate-formate | SCFA | 2,855,852 | 2,878,968 | ctg1_2687, ctg1_2688 | 100% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, S.; Yue, K.; He, S.; Huang, Y.; Ren, Z.; Xu, J. Safety Assessment of Lactiplantibacillus plantarum GUANKE Based on Whole-Genome Sequencing, Phenotypic, and Anti-Inflammatory Capacity Analysis. Microorganisms 2025, 13, 873. https://doi.org/10.3390/microorganisms13040873
Lu S, Yue K, He S, Huang Y, Ren Z, Xu J. Safety Assessment of Lactiplantibacillus plantarum GUANKE Based on Whole-Genome Sequencing, Phenotypic, and Anti-Inflammatory Capacity Analysis. Microorganisms. 2025; 13(4):873. https://doi.org/10.3390/microorganisms13040873
Chicago/Turabian StyleLu, Simin, Kun Yue, Siqin He, Yuanming Huang, Zhihong Ren, and Jianguo Xu. 2025. "Safety Assessment of Lactiplantibacillus plantarum GUANKE Based on Whole-Genome Sequencing, Phenotypic, and Anti-Inflammatory Capacity Analysis" Microorganisms 13, no. 4: 873. https://doi.org/10.3390/microorganisms13040873
APA StyleLu, S., Yue, K., He, S., Huang, Y., Ren, Z., & Xu, J. (2025). Safety Assessment of Lactiplantibacillus plantarum GUANKE Based on Whole-Genome Sequencing, Phenotypic, and Anti-Inflammatory Capacity Analysis. Microorganisms, 13(4), 873. https://doi.org/10.3390/microorganisms13040873




