In Infants with Neuroblastoma Standard Therapy Only Partially Reverts the Fecal Microbiome Dysbiosis Present at Diagnosis
Abstract
1. Introduction
2. Methods
2.1. Study Design
2.2. Alpha and Beta Diversity Analysis
2.3. Bioinformatics and Statistics
3. Results
3.1. Alpha and Beta Diversity at Onset and During Treatment
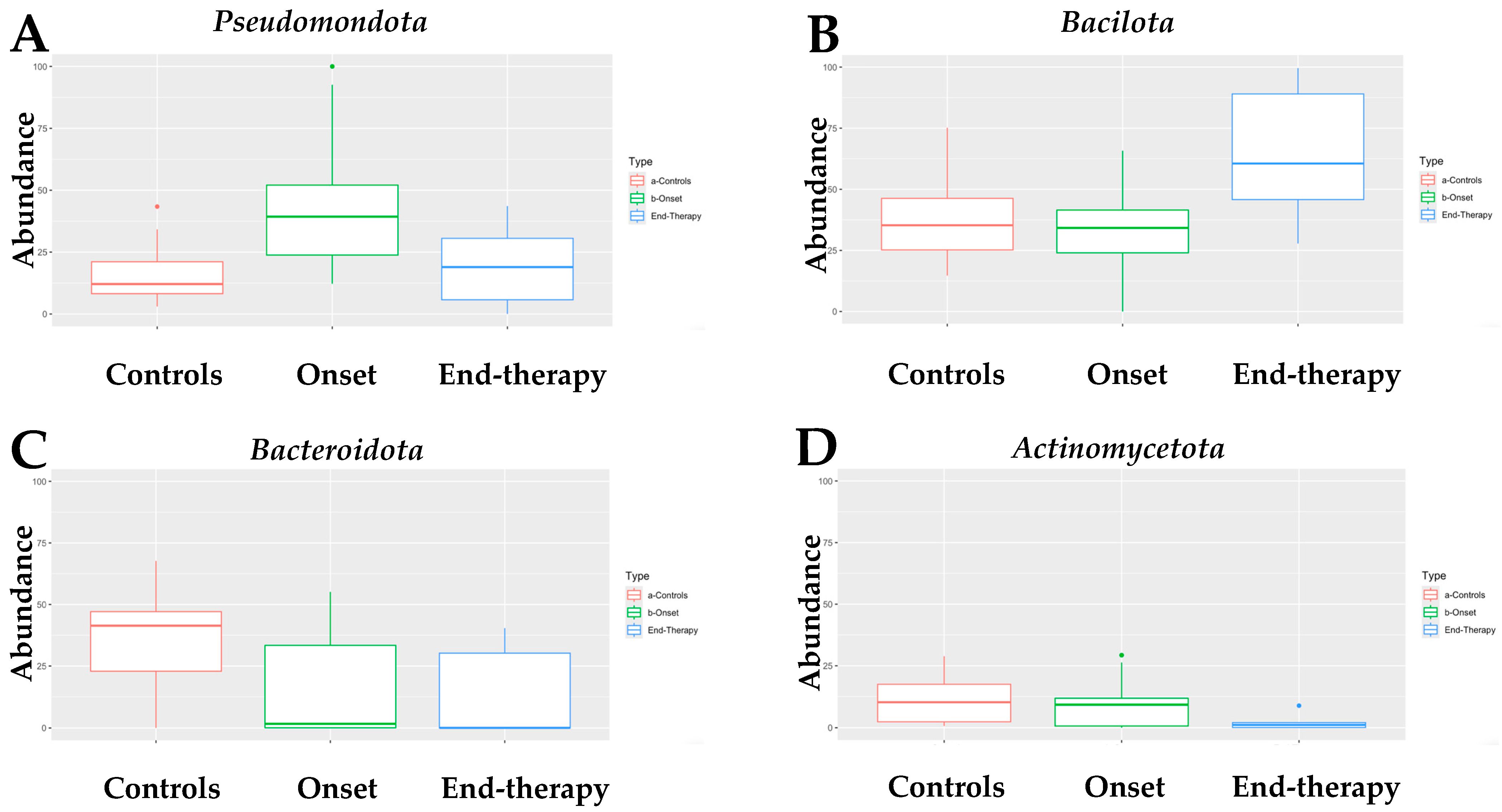
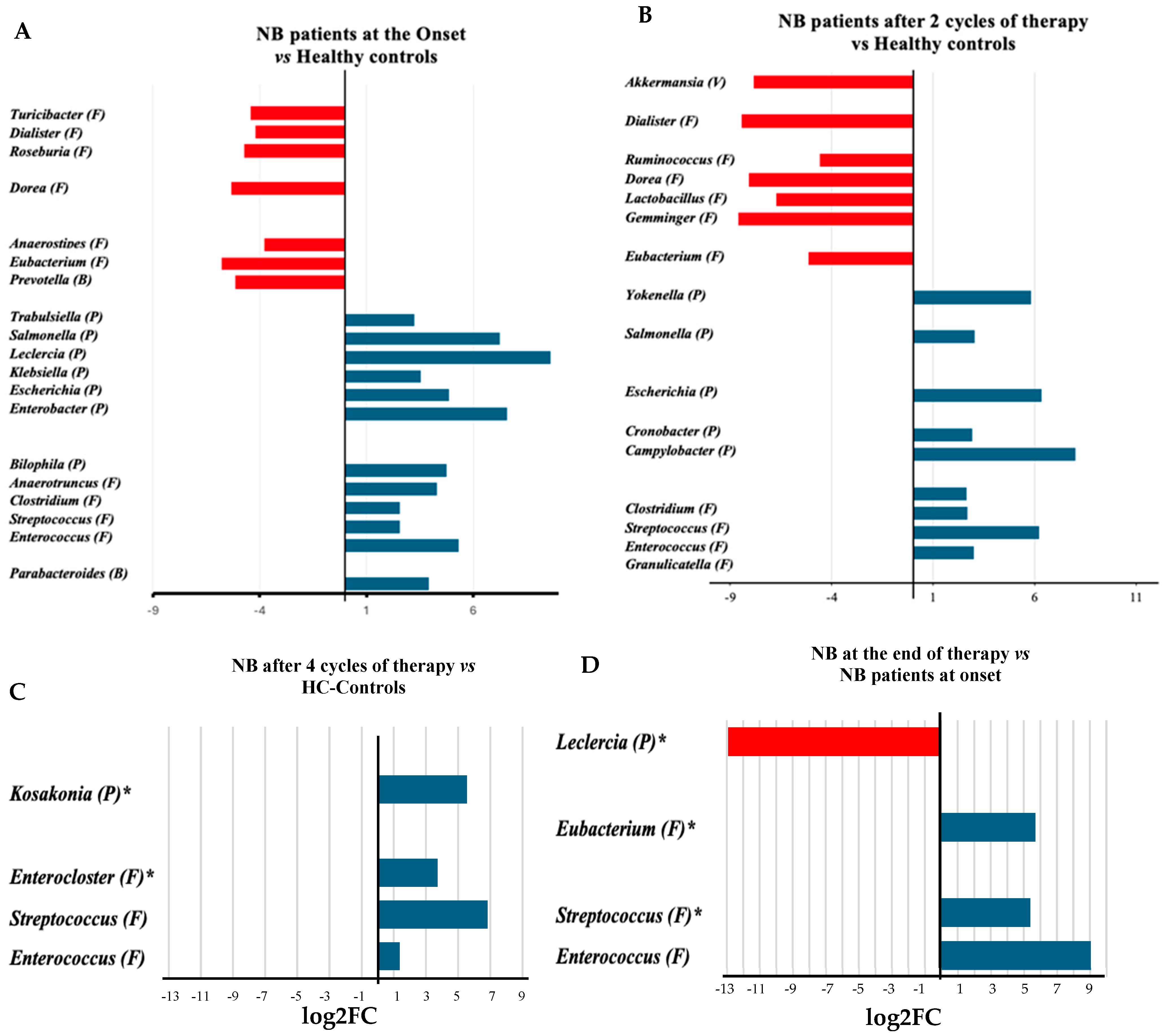
3.2. Microbiome Compositional Analysis of Healthy Infants and Infants with NB
3.3. Supervised Analysis with Random Forest Learning Algorithm
3.4. Heat Tree Analysis
3.5. Sparse Correlations for Compositional Data (SparCC) Analysis

3.6. Microbial Dysbiosis Index
3.7. Weighted Correlation Network Analysis
3.8. Differentially Expressed Inferred Pathways by the Fecal Microbiome of Infants with NB
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Park, J.R.; Eggert, A.; Caron, H. Neuroblastoma: Biology, prognosis, and treatment. Hematol. Oncol. Clin. N. Am. 2010, 24, 65–86. [Google Scholar] [CrossRef] [PubMed]
- Cheung, N.K.; Dyer, M.A. Neuroblastoma: Developmental biology, cancer genomics and immunotherapy. Nat. Rev. Cancer 2013, 13, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Matthay, K.K.; Maris, J.M.; Schleiermacher, G.; Nakagawara, A.; Mackall, C.L.; Diller, L.; Weiss, W.A. Neuroblastoma. Nat. Rev. Dis. Primers 2016, 2, 16078. [Google Scholar] [CrossRef]
- Yan, P.; Qi, F.; Bian, L.; Xu, Y.; Zhou, J.; Hu, J.; Ren, L.; Li, M.; Tang, W. Comparison of Incidence and Outcomes of Neuroblastoma in Children, Adolescents, and Adults in the United States: A Surveillance, Epidemiology, and End Results (SEER) Program Population Study. Med. Sci. Monit. 2020, 26, e927218. [Google Scholar] [CrossRef]
- Helmink, B.A.; Khan, M.A.W.; Hermann, A.; Gopalakrishnan, V.; Wargo, J.A. The microbiome, cancer, and cancer therapy. Nat. Med. 2019, 25, 77–88. [Google Scholar] [CrossRef]
- Valles-Colomer, M.; Manghi, P.; Cumbo, F.; Masetti, G.; Armanini, F.; Asnicar, F.; Blanco-Miguez, A.; Pinto, F.; Punčochář, M.; Garaventa, A.; et al. Neuroblastoma is associated with alterations in gut microbiome composition subsequent to maternal microbial seeding. EBioMedicine 2024, 99, 104917. [Google Scholar] [CrossRef]
- Stewart CJAjami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; Muzny, D.; et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef]
- Parte, A.C.; Sardà Carbasse, J.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef]
- Chao, A.; Lee, S.-M. Estimating the Number of Classes via Sample Coverage. J. Am. Stat. Assoc. 1992, 87, 210–217. [Google Scholar] [CrossRef]
- Shannon, C.E. A Mathematical Theory of Communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Simpson, E.H. Measurement of Diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Bray, J.R.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst—A web-based tool for comprehensive statistical, visual, and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef]
- Lu, Y.; Zhou, G.; Ewald, J.; Pang, Z.; Shiri, T.; Xia, J. MicrobiomeAnalyst 2.0: Comprehensive statistical, functional, and integrative analysis of microbiome data. Nucleic Acids Res. 2023, 51, 310–318. [Google Scholar] [CrossRef]
- Paulson, J.N.; Stine, O.C.; Bravo, H.C.; Pop, M. Differential abundance analysis for microbial marker-gene surveys. Nat. Meth. 2013, 10, 1200–1202. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Foster, Z.S.; Sharpton, T.J.; Grünwald, N.J. Metacoder: An R package for visualization and manipulation of community taxonomic diversity data. PLoS Comput. Biol. 2017, 13, e1005404. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.; Alm, E.J. Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 2012, 8, e1002687. [Google Scholar] [CrossRef] [PubMed]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Tanaka, M.; Nakayama, J. Development of the gut microbiota in infancy and its impact on health in later life. Allergol. Int. 2017, 66, 515–522. [Google Scholar] [CrossRef]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- Shin, N.R.; Whon, T.W.; Bae, J.W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Rizzatti, G.; Lopetuso, L.R.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A Common Factor in Human Diseases. BioMed Res. Int. 2017, 9351507. [Google Scholar] [CrossRef]
- García-Solache, M.; Rice, L.B. The Enterococcus: A Model of Adaptability to its Environment. Clin. Microbiol. Rev. 2019, 32, e00058-18. [Google Scholar] [CrossRef] [PubMed]
- Barco, S.; Lavarello, C.; Cangelosi, D.; Morini, M.; Eva, A.; Oneto, L.; Uva, P.; Tripodi, G.; Garaventa, A.; Conte, M.; et al. Untargeted LC-HRMS Based-Plasma Metabolomics Reveals 3-O-Methyldopa as a New Biomarker of Poor Prognosis in High-Risk Neuroblastoma. Front. Oncol. 2022, 12, 845936. [Google Scholar] [CrossRef] [PubMed]
- Castellani, C.; Singer, G.; Eibisberger, M.; Obermüller, B.; Warncke, G.; Miekisch, W.; Kolb-Lenz, D.; Summer, G.; Pauer, T.M.; ElHaddad, A.; et al. The effects of neuroblastoma and chemotherapy on metabolism, fecal microbiome, volatile organic compounds, and gut barrier function in a murine model. Pediatr. Res. 2019, 85, 546–555. [Google Scholar] [CrossRef]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibility for its control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef]
- Sagar, N.A.; Tarafdar, S.; Agarwal, S.; Tarafdar, A.; Sharma, S. Polyamines: Functions, Metabolism, and Role in Human Disease Management. Med. Sci. 2021, 9, 44. [Google Scholar] [CrossRef]
- Holbert, C.E.; Cullen, M.T.; Casero, R.A., Jr.; Stewart, T.M. Polyamines in cancer: Integrating organismal metabolism and antitumour immunity. Nat. Rev. Cancer 2022, 22, 467–480. [Google Scholar] [CrossRef]
- Altves, S.; Guclu, E.; Cinar Ayan, I.; Bilecen, K.; Vural, H. Lysates from the probiotic bacterium Streptococcus thermophilus enhances the survival of T cells and triggers programmed cell death in neuroblastoma cells. Med. Oncol. 2023, 40, 315. [Google Scholar] [CrossRef]
- Bock, H.J.; Lee, H.W.; Lee, N.K.; Paik, H.D. Probiotic Lactiplantibacillus plantarum KU210152 and its fermented soy milk attenuates oxidative stress in neuroblastoma cells. Food Res. Int. 2024, 177, 113868. [Google Scholar] [CrossRef]

| (A) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample | Sex | Phase | Stage | Age (Months) | Mode of Delivery | Mode of Feeding | Bristol Score | READS | OTUs |
| P1 | F | onset | MS | 6.6 | eutocic | breast | 7 | 240,098 | 1879 |
| P2 | M | onset | L1 | 10.9 | eutocic | formula | 4 | 232,017 | 1491 |
| P3 | M | onset | L1 | 10.9 | c-section | formula | 2 | 224,975 | 1578 |
| P5 | F | onset | L2 | 8.4 | eutocic | formula | 1 | 349,389 | 2567 |
| P7 | F | onset | L2 | 22.0 | eutocic | formula | 2 | 184,108 | 1435 |
| P8 | F | onset | M | 19.8 | c-section | breast | 6 | 228,071 | 1623 |
| P10 | M | onset | MS | 4.8 | eutocic | breast | 6 | 197,682 | 1311 |
| P11 | M | onset | L1 | 2.6 | c-section | breast | 3 | 313,173 | 2306 |
| P12 | F | onset | MS | 4.9 | c-section | formula | 7 | 186,662 | 1445 |
| P15 | M | onset | M | 22.1 | eutocic | formula | 3 | 228,290 | 1488 |
| P19 | F | onset | M | 5.8 | c-section | breast | 6 | 130,251 | 842 |
| P21 | F | onset | L2 | 6.0 | eutocic | breast | 6 | 240,067 | 1891 |
| P22 | M | onset | M | 8.6 | eutocic | breast | 6 | 182,989 | 1364 |
| P24 | M | onset | L1 | 5.0 | eutocic | breast | 6 | 128,618 | 971 |
| P25 | M | onset | L2 | 6.5 | eutocic | formula | 6 | 208,146 | 1419 |
| (B) | |||||||||
| Sample | Sex | Phase | Stage | Age (Months) | Drug Therapy | Bristol Score | READS | OTUs | |
| P1 B | F | CP/VP16 | MS | 9.6 | 2 cycles—half treatment | 7 | 251,064 | 1660 | |
| P7 A | F | CP/VP16 | L2 | 24 | 2 cycles—half treatment | 6 | 229,871 | 1722 | |
| P21 A | F | CP/VP16 | L2 | 8.4 | 2 cycles—half treatment | 6 | 197,013 | 1714 | |
| P22 A | M | CP/VP16 | M | 9.6 | 2 cycles—half treatment | 6 | 182,989 | 1683 | |
| P24 A | M | CP/VP16 | L1 | 8.4 | 2 cycles—half treatment | 6 | 130,474 | 1355 | |
| P1 C | F | CP/VP16 | MS | 12 | 4 cycles—end of treatment | 6 | 145,892 | 970 | |
| P5 A | F | CP/VP16 | L2 | 21.6 | 4 cycles—end of treatment | 4 | 109,008 | 944 | |
| P7 B | F | CP/VP16 | L | 31.2 | 4 cycles—end of treatment | 5 | 217,098 | 1438 | |
| P12 A | F | CP/VP16 | MS | 8.4 | 4 cycles—end of treatment | 6 | 239,966 | 1654 | |
| P22 B | M | CP/VP16 | M | 13.2 | 4 cycles—end of treatment | 6 | 221,960 | 1619 | |
| P24 B | M | CP/VP16 | M | 20.4 | 4 cycles—end of treatment | 7 | 152,753 | 1253 | |
| Age | Bristol Score | ||||
|---|---|---|---|---|---|
| Family | Pearson Corr. Coeff. | p-Value | Pearson Corr. Coeff. | p-Value | |
| Cluster 7 | Christensenellaceae, Desulfohalobiaceae, Desulfovibrionaceae, Erysipelotrichaceae, Eubacteriaceae, Oscillospiraceae, Paenibacillaceae, Peptococcaceae, Porphyromonadaceae, Rikenellaceae, Verrucomicrobiaceae | 0.59 | 0.020 | ||
| Cluster 8 | Bacteroidaceae, Campylobacteraceae, Dethiosulfovibrionaceae, Hyphomicrobiaceae, Lachnospiraceae, Micrococcaceae, Oscillospiraceae, Prevotellaceae, Synergistaceae | 0.54 | 0.04 | ||
| genus | |||||
| Cluster 1 | Akkermansia, Alistipes, Anaerofustis, Anaerotruncus, Barnesiella, Bilophila, Desulfovibrio, Flavonifractor, Hydrogenoanaerobacterium, Moryella, Oscillibacter, Parabacteroides, Subdoligranulum | 0.58 | 0.02 | ||
| Cluster 7 | Abiotrophia, Butyricicoccus, Eggerthella, Enterococcus, Howardella, Megamonas, Parasutterella, Pseudomonas | 0.54 | 0.04 | ||
| species | |||||
| Cluster 6 | Clostridioides_difficile, Kluyvera_georgiana | −0.53 | 0.04 | ||
| Cluster 12 | Abiotrophia_defectiva, Bacteroides_ovatus, Bifidobacterium_bifidum, Bifidobacterium_breve, Butyricicoccus_pullicaecorum, Clostridium_innocuum, Eggerthella_lenta, Enterocloster_citroniae, Enterococcus_avium, Enterococcus_faecium, Enterococcus_lemanii, Megamonas_funiformis, Megamonas_rupellensis, Pseudoflavonifractor_capillosus, Pseudomonas_aeruginosa, Pseudomonas_indica | 0.52 | 0.04 | ||
| Cluster 17 | Akkermansia_muciniphila, Anaerofustis_stercorihominis, Anaerotruncus_colihominis, Barnesiella_intestinihominis, Blautia_hansenii, Eggerthella_sinensis, Eubacterium_callanderi, Flavonifractor_plautii, Moryella_indoligenes, Subdoligranulum_variabile | 0.55 | 0.04 | ||
| ko | DESeq2 | EdgeR | Pathways | Orthology | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| log2FC | lfcSE | p-Values | FDR | log2FC | logCPM | p-Values | FDR | |||
| K03190 | 32.498 | 0.62533 | 2.03 × 10−3 | 0.00079121 | 3.2919 | 6.1374 | 3.3685 × 10−11 | 2.0242 × 10−9 | ureD, ureH; urease accessory protein | |
| K01428 | 31.685 | 0.62601 | 4.16 × 10−3 | 0.0008125 | 3.2364 | 6.1378 | 6.5176 × 10−11 | 3.1912 × 10−9 | ko:K01428 ureC; urease subunit alpha [EC:3.5.1.5] | |
| K01071 | 40.126 | 0.8589 | 2.98 × 10−2 | 0.0038861 | 5.4371 | 8.745 | 1.2689 × 10−12 | 2.9721 × 10−10 | ko:K01071 MCH; medium-chain acyl-[acyl-carrier-protein] hydrolase [EC:3.1.2.21] | |
| K07146 | 26.275 | 0.5798 | 5.85 × 10−2 | 0.0057137 | 4.0782 | 8.8766 | 5.3469 × 10−12 | 6.5049 × 10−10 | K07146; UPF0176 protein | |
| K00563 | 19.082 | 0.42737 | 8.00 × 10−2 | 0.0061473 | 3.4302 | 9.0049 | 2.6382 × 10−12 | 4.9071 × 10−10 | rlmA1; 23S rRNA (guanine745-N1)-methyltransferase [EC:2.1.1.187] | |
| K01478 | 21.936 | 0.49523 | 9.44 × 10−2 | 0.0061473 | 4.6804 | 8.3424 | 4.0374 × 10−15 | 3.9425 × 10−12 | ko:K01478 arcA; arginine deiminase [EC:3.5.3.6] | |
| K07586 | 34.572 | 0.78697 | 1.12 × 10−1 | 0.0062367 | 4.9499 | 8.7905 | 7.5016 × 10−12 | 8.1392 × 10−10 | ygaC; uncharacterized protein | |
| K00383 | 23.032 | 0.52916 | 1.35 × 10−1 | 0.0065712 | 4.246 | 9.607 | 3.161 × 10−13 | 1.5434 × 10−10 | ko:K00383 GSR; glutathione reductase (NADPH) [EC:1.8.1.7] | mprF, fmtC; phosphatidylglycerol lysyltransferase [EC:2.3.2.3] |
| K02052 | −16.149 | 0.37679 | 1.82 × 10−1 | 0.0078962 | −2.3178 | 8.0724 | 0.00012877 | 0.001267 | ko:K02052 ABC.SP.A; putative spermidine/putrescine transport system ATP-binding protein | |
| K01674 | 30.408 | 0.7332 | 3.36 × 10−1 | 0.013141 | 5.3147 | 8.3658 | 1.6729 × 10−14 | 1.089 × 10−11 | ko:K01674 cah; carbonic anhydrase [EC:4.2.1.1] | |
| K04784 | −44.021 | 10.957 | 5.88 × 10−1 | 0.019938 | −5.266 | 8.7619 | 0.00014225 | 0.0013821 | ko:K04784 irp2; yersinia b-actin nonribosomal peptide synthetase | |
| K00244 | 14.086 | 0.35146 | 6.13 × 10−1 | 0.019938 | 1.7709 | 9.5778 | 0.00092 | 0.0052846 | ko:K00244 frdA; succinate dehydrogenase flavoprotein subunit [EC:1.3.5.1] | |
| K15051 | 27.683 | 0.6988 | 7.45 × 10−1 | 0.022379 | 3.159 | 9.3922 | 1.0309 × 10−5 | 0.00015025 | endA; DNA-entry nuclease | |
| K14260 | 1.606 | 0.40857 | 8.46 × 10−1 | 0.023616 | 2.9388 | 9.2072 | 9.3617 × 10−10 | 3.4826 × 10−8 | ko:K14260 alaA; alanine-synthesizing transaminase [EC:2.6.1.66 2.6.1.2] | |
| K02798 | 1.963 | 0.51119 | 0.00012294 | 0.029987 | 4.6477 | 8.9458 | 5.5942 × 10−15 | 4.3702 × 10−12 | ko:K02798 cmtB; mannitol PTS system EIIA component [EC:2.7.1.197] | |
| K03483 | 31.123 | 0.81399 | 0.00013157 | 0.029987 | 5.6633 | 8.1866 | 3.3345 × 10−15 | 3.9425 × 10−12 | mtlR; mannitol operon transcriptional activator | |
| K02810 | 14.768 | 0.38651 | 0.00013305 | 0.029987 | 2.0846 | 10.2 | 0.00036687 | 0.0029185 | ko:K02810 scrA; sucrose PTS system EIIBCA or EIIBC component [EC:2.7.1.211] | |
| K02053 | −15.036 | 0.39573 | 0.00014499 | 0.029987 | −2.0887 | 7.779 | 0.00025198 | 0.0021727 | ko:K02053 ABC.SP.P; putative spermidine/putrescine transport system permease protein | |
| K08234 | 16.875 | 0.44431 | 0.00014587 | 0.029987 | 2.9769 | 9.1374 | 4.4347 × 10−9 | 1.4933 × 10−7 | yaeR; glyoxylase I family protein | |
| K01963 | 12.884 | 0.3427 | 0.00017017 | 0.032387 | 2.0293 | 9.7298 | 1.3397 × 10−5 | 0.0001886 | ko:K01963 accD; acetyl-CoA carboxylase carboxyl transferase subunit beta [EC:6.4.1.2 2.1.3.15] | |
| K03785 | 16.993 | 0.45269 | 0.00017412 | 0.032387 | 3.0777 | 9.2258 | 2.5298 × 10−10 | 1.0625 × 10−8 | ko:K03785 aroD; 3-dehydroquinate dehydratase I [EC:4.2.1.10] | |
| K02443 | −22.726 | 0.61306 | 0.0002097 | 0.036299 | −2.1083 | 7.6313 | 0.0047712 | 0.020616 | glpP; glycerol uptake operon antiterminator | |
| K02054 | −13.832 | 0.37362 | 0.00021374 | 0.036299 | −1.7256 | 7.3454 | 0.0005016 | 0.0037598 | ko:K02054 ABC.SP.P1; putative spermidine/putrescine transport system permease protein | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corrias, M.V.; Di Marco, E.; Bonaretti, C.; Squillario, M.; Amoroso, L.; Conte, M.; Ponzoni, M.; Biassoni, R. In Infants with Neuroblastoma Standard Therapy Only Partially Reverts the Fecal Microbiome Dysbiosis Present at Diagnosis. Microorganisms 2025, 13, 691. https://doi.org/10.3390/microorganisms13030691
Corrias MV, Di Marco E, Bonaretti C, Squillario M, Amoroso L, Conte M, Ponzoni M, Biassoni R. In Infants with Neuroblastoma Standard Therapy Only Partially Reverts the Fecal Microbiome Dysbiosis Present at Diagnosis. Microorganisms. 2025; 13(3):691. https://doi.org/10.3390/microorganisms13030691
Chicago/Turabian StyleCorrias, Maria Valeria, Eddi Di Marco, Carola Bonaretti, Margherita Squillario, Loredana Amoroso, Massimo Conte, Mirco Ponzoni, and Roberto Biassoni. 2025. "In Infants with Neuroblastoma Standard Therapy Only Partially Reverts the Fecal Microbiome Dysbiosis Present at Diagnosis" Microorganisms 13, no. 3: 691. https://doi.org/10.3390/microorganisms13030691
APA StyleCorrias, M. V., Di Marco, E., Bonaretti, C., Squillario, M., Amoroso, L., Conte, M., Ponzoni, M., & Biassoni, R. (2025). In Infants with Neuroblastoma Standard Therapy Only Partially Reverts the Fecal Microbiome Dysbiosis Present at Diagnosis. Microorganisms, 13(3), 691. https://doi.org/10.3390/microorganisms13030691






