Engineering Useful Microbial Species for Pharmaceutical Applications
Abstract
1. Introduction
2. Innovations, Strategies, and Future Directions in Microbial Engineering
2.1. Genetic Engineering
2.2. Synthetic Biology
2.3. Adaptive Evolution
2.4. Artificial Intelligence and Microbial Engineering
2.5. Systems Biology in Microbial Engineering
3. Challenges and Limitations
3.1. Antimicrobial Resistance
3.2. Contamination Risks
3.3. Regulatory Barriers
| Regulatory Barrier | Description | Impact on Development | Mitigation Strategies | Reference |
|---|---|---|---|---|
| Product classification | Ambiguity in categorizing engineered microbes | Divergent review processes and approval delays | Harmonized definitions | [115,116,151] |
| Risk assessment | Extensive preclinical and post-market monitoring required due to unpredictable in vivo behavior and potential off-target effects | Increased development costs and extended timelines | Risk-based approaches; improved in vivo models and rigorous post-market surveillance | [115,116,117] |
| Manufacturing | Challenges ensuring batch-to-batch consistency in live microbial production given inherent biological variability | Variability in product quality and potential for process-related rejections | Advanced GMPs and real-time monitoring systems | [152] |
| Environmental impact | The potential for horizontal gene transfer and unintended environmental release raises significant biosafety concerns | Can lead to increased regulatory scrutiny and the need for extensive environmental risk assessments, potentially delaying product approval | Develop robust biocontainment strategies | [149] |
| International variability | Regulatory requirements vary widely across regions, complicating product development | Complicates the design of global clinical trials and market access strategies, leading to additional time and cost burdens | Promote international harmonization and collaboration among regulatory agencies | [115,116,154] |
International Regulatory Frameworks and Harmonization for GEMs
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ADH6 | Alcohol dehydrogenase gene |
| ADR1 | A transcription factor involved in stress response |
| AI-CRISPR | Artificial intelligence-assisted CRISPR technology |
| ALE | Adaptive laboratory evolution |
| ANTI-SMASH | Antibiotic and Secondary Metabolite Analysis Shell |
| ARTP | Atmospheric and Room Temperature Plasma |
| BGC | Biosynthetic gene cluster |
| CDC | Centers for Disease Control and Prevention |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| Cas | CRISPR-associated protein |
| DL | Deep learning |
| DNA | Deoxyribonucleic Acid |
| DOAJ | Directory of Open Access Journals |
| DSB | Double-strand break |
| EMA | European Medicines Agency |
| FDA | U.S. Food and Drug Administration |
| GAN | Generative adversarial network |
| GEM | Genetically engineered microorganism |
| GMP | Good manufacturing practice |
| GRE2 | A gene involved in stress resistance |
| HEPA | High-efficiency particulate air (filtration) |
| HGT | Horizontal gene transfer |
| LD | Linear Dichroism |
| MDPI | Multidisciplinary Digital Publishing Institute |
| ML | Machine learning |
| NGS | Next-generation sequencing |
| RNA | Ribonucleic Acid |
| TALEN | Transcription Activator-Like Effector Nuclease |
| TLA | Three-Letter Acronym |
| VAE | Variational autoencoder |
| WHO | World Health Organization |
| ZFN | Zinc Finger Nuclease |
| mRNA | Messenger RNA |
References
- Santos-Beneit, F. What is the role of microbial biotechnology and genetic engineering in medicine? Microbiologyopen 2024, 13, e1406. [Google Scholar] [CrossRef] [PubMed]
- Pham, J.V.; Yilma, M.A.; Feliz, A.; Majid, T.T.; Maffetone, N.; Walker, J.R.; Kim, E.; Cho, H.J.; Reynolds, J.M.; Song, M.C.; et al. A review of the microbial production of bioactive natural products and biologics. Front. Microbiol. 2019, 10, 1404. [Google Scholar] [CrossRef] [PubMed]
- Al-Fadhli, A.H.; Jamal, W.Y. Recent advances in gene-editing approaches for tackling antibiotic resistance threats: A review. Front. Cell Infect. Microbiol. 2024, 14, 1410115. [Google Scholar] [CrossRef]
- Yan, X.; Liu, X.; Zhao, C.; Chen, G.Q. Applications of synthetic biology in medical and pharmaceutical fields. Signal Transduct. Target Ther. 2023, 8, 199. [Google Scholar] [CrossRef] [PubMed]
- Barbuto, S.; Cammarota, M.; Schiraldi, C.; Restaino, O.F. Streptomycetes as platform for biotechnological production processes of drugs. Appl. Microbiol. Biotechnol. 2021, 105, 551–568. [Google Scholar] [CrossRef]
- Gust, B.; Chandra, G.; Jakimowicz, D.; Yuqing, T.; Bruton, C.J.; Chater, K.F. Lambda Red-Mediated Genetic Manipulation of Antibiotic-Producing Streptomyces. Adv. Appl. Microbiol. 2004, 54, 107–128. [Google Scholar] [CrossRef]
- Rashid, M.H. Full-length recombinant antibodies from Escherichia coli: Production, characterization, effector function (Fc) engineering, and clinical evaluation. MAbs 2022, 14, 2111748. [Google Scholar] [CrossRef]
- Nielsen, J. Production of biopharmaceutical proteins by yeast: Advances through metabolic engineering. Bioengineered 2013, 4, 207–211. [Google Scholar] [CrossRef]
- Shi, A.; Fan, F.; Broach, J.R. Microbial adaptive evolution. J. Ind. Microbiol. Biotechnol. 2022, 49, kuab076. [Google Scholar] [CrossRef]
- Merzbacher, C.; Oyarzún, D.A. Applications of artificial intelligence and machine learning in dynamic pathway engineering. Biochem. Soc. Trans. 2023, 51, 1871–1879. [Google Scholar] [CrossRef]
- Daboussi, F.; Lindley, N.D. Challenges to ensure a better translation of metabolic engineering for industrial applications. Methods Mol. Biol. 2023, 2553, 1–20. [Google Scholar] [PubMed]
- Crater, J.S.; Lievense, J.C. Scale-up of industrial microbial processes. FEMS Microbiol. Lett. 2018, 365, fny138. [Google Scholar] [CrossRef] [PubMed]
- Yamin, D.; Uskoković, V.; Wakil, A.M.; Goni, M.D.; Shamsuddin, S.H.; Mustafa, F.H.; Alfouzan, W.A.; Alissa, M.; Alshengeti, A.; Almaghrabi, R.H.; et al. Current and future technologies for the detection of antibiotic-resistant bacteria. Diagnostics 2023, 13, 3246. [Google Scholar] [CrossRef]
- Carter, L.; Mankad, A. The promises and realities of integration in synthetic biology: A view from social science. Front. Bioeng. Biotechnol. 2021, 8, 622221. [Google Scholar] [CrossRef]
- Sarsekeyeva, F.K.; Sadvakasova, A.K.; Sandybayeva, S.K.; Kossalbayev, B.D.; Huang, Z.; Zayadan, B.K.; Akmukhanova, N.R.; Leong, Y.K.; Chang, J.-S.; Allakhverdiev, S.I. Microalgae- and Cyanobacteria-Derived Phytostimulants for Mitigation of Salt Stress and Improved Agriculture. Algal Res. 2024, 82, 103686. [Google Scholar] [CrossRef]
- Bansal, A.K. Bioinformatics in microbial biotechnology—A mini review. Microb. Cell Factories 2005, 4, 19. [Google Scholar] [CrossRef]
- Ramírez-Rendon, D.; Passari, A.K.; Ruiz-Villafán, B.; Rodríguez-Sanoja, R.; Sánchez, S.; Demain, A.L. Impact of novel microbial secondary metabolites on the pharma industry. Appl. Microbiol. Biotechnol. 2022, 106, 1855–1878. [Google Scholar] [CrossRef] [PubMed]
- Tyo, K.E.; Kocharin, K.; Nielsen, J. Toward design-based engineering of industrial microbes. Curr. Opin. Microbiol. 2010, 13, 255–262. [Google Scholar] [CrossRef]
- Gelsinger, D.R.; Vo, P.L.H.; Klompe, S.E.; Ronda, C.; Wang, H.H.; Sternberg, S.H. Bacterial genome engineering using CRISPR-associated transposases. Nat. Protoc. 2024, 19, 752–790. [Google Scholar] [CrossRef]
- Fooladi, S.; Rabiee, N.; Iravani, S. Genetically engineered bacteria: A new frontier in targeted drug delivery. J. Mater. Chem. B 2023, 11, 10072–10087. [Google Scholar] [CrossRef]
- Shams, A.; Fischer, A.; Bodnar, A.; Kliegman, M. Perspectives on genetically engineered microorganisms and their regulation in the United States. ACS Synth. Biol. 2024, 13, 1412–1423. [Google Scholar] [CrossRef] [PubMed]
- Tamura, R.; Toda, M. Historic overview of genetic engineering technologies for human gene therapy. Neurol. Med. Chir. 2020, 60, 483–491. [Google Scholar] [CrossRef]
- Carroll, D. Genome engineering with zinc-finger nucleases. Genetics 2011, 188, 773–782. [Google Scholar] [CrossRef]
- Chandrasegaran, S. Recent advances in the use of ZFN-mediated gene editing for human gene therapy. Cell Gene Ther. Insights 2017, 3, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Nemudryi, A.A.; Valetdinova, K.R.; Medvedev, S.P.; Zakian, S.M. TALEN and CRISPR/Cas genome editing systems: Tools of discovery. Acta Naturae 2014, 6, 19–40. [Google Scholar] [CrossRef]
- Joung, J.K.; Sander, J.D. TALENs: A widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013, 14, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Naeem, M.; Majeed, S.; Hoque, M.Z.; Ahmad, I. Latest developed strategies to minimize the off-target effects in CRISPR-Cas-mediated genome editing. Cells 2020, 9, 1608. [Google Scholar] [CrossRef]
- Liu, W.; Li, L.; Jiang, J.; Wu, M.; Lin, P. Applications and challenges of CRISPR-Cas gene-editing to disease treatment in clinics. Precis. Clin. Med. 2021, 4, 179–191. [Google Scholar] [CrossRef]
- Asmamaw, M.; Zawdie, B. Mechanism and applications of CRISPR/Cas-9-mediated genome editing. Biologics 2021, 15, 353–361. [Google Scholar]
- Tavakoli, K.; Pour-Aboughadareh, A.; Kianersi, F.; Poczai, P.; Etminan, A.; Shooshtari, L. Applications of CRISPR-Cas9 as an advanced genome editing system in life sciences. BioTech 2021, 10, 14. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed]
- Munshi, N.V. CRISPR (Clustered Regularly Interspaced Palindromic Repeat)/Cas9 system: A revolutionary disease-modifying technology. Circulation 2016, 11, 777–779. [Google Scholar] [CrossRef]
- Sun, L.; Lutz, B.M.; Tao, Y.X. The CRISPR/Cas9 system for gene editing and its potential application in pain research. Transl. Perioper. Pain Med. 2016, 1, 22–33. [Google Scholar]
- Kim, D.G.; Gu, B.; Cha, Y.; Ha, J.; Lee, Y.; Kim, G.; Cho, B.K.; Oh, M.K. Engineered CRISPR-Cas9 for Streptomyces sp. genome editing to improve specialized metabolite production. Nat. Commun. 2025, 16, 874. [Google Scholar] [CrossRef]
- Lee, D.; Muir, P.; Lundberg, S.; Lundholm, A.; Sandegren, L.; Koskiniemi, S. A CRISPR-Cas9 system protecting E. coli against acquisition of antibiotic resistance genes. Sci. Rep. 2025, 15, 1545. [Google Scholar] [CrossRef]
- Dong, H.; Cui, Y.; Zhang, D. CRISPR/Cas technologies and their applications in Escherichia coli. Front. Bioeng. Biotechnol. 2021, 9, 762676. [Google Scholar] [CrossRef]
- Fellmann, C.; Gowen, B.G.; Lin, P.C.; Doudna, J.A.; Corn, J.E. Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat. Rev. Drug Discov. 2017, 16, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Alberti, F.; Corre, C. Editing Streptomycete genomes in the CRISPR/Cas9 age. Nat. Prod. Rep. 2019, 36, 1237–1248. [Google Scholar] [CrossRef] [PubMed]
- Ameruoso, A.; Claudia, M.; Villegas, K.; Piper, K.; Cohen, J.; Chappell, J. Activating natural product synthesis using CRISPR interference and activation systems in Streptomyces. Nucleic Acids Res. 2022, 50, 7751–7760. [Google Scholar] [CrossRef]
- Azeez, S.S.; Hamad, R.S.; Hamad, B.K.; Shekha, M.S.; Bergsten, P. Advances in CRISPR-Cas technology and its applications: Revolutionising precision medicine. Front. Genome Ed. 2024, 6, 1509924. [Google Scholar] [CrossRef]
- Mahdizade, A.M.; Dadgar, L.; Elahi, Z.; Ghanavati, R.; Taheri, B. Genetically engineered microorganisms and their impact on human health. Int. J. Clin. Pract. 2024, 2024, 6638269. [Google Scholar] [CrossRef]
- Uddin, F.; Rudin, C.M.; Sen, T. CRISPR gene therapy: Applications, limitations, and implications for the future. Front. Oncol. 2020, 10, 1387. [Google Scholar] [CrossRef] [PubMed]
- Chanchal, D.K.; Chaudhary, J.S.; Kumar, P.; Agnihotri, N.; Porwal, P. CRISPR-based therapies: Revolutionizing drug development and precision medicine. Curr. Gene Ther. 2024, 24, 193–207. [Google Scholar] [CrossRef]
- Chancellor, D.; Barrett, D.; Nguyen-Jatkoe, L.; Millington, S.; Eckhardt, F. The state of cell and gene therapy in 2023. Mol. Ther. 2023, 31, 3376–3388. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, C.K.; Khan, M.; Praveen, V.; Khan, S.; Srivastava, A. Enhanced antibiotic production by Streptomyces sindenensis using artificial neural networks coupled with genetic algorithm and Nelder-Mead downhill simplex. J. Microbiol. Biotechnol. 2012, 22, 939–946. [Google Scholar] [CrossRef]
- Ali Agha, A.S.A.; Al-Samydai, A.; Aburjai, T. New Frontiers in CRISPR: Addressing Antimicrobial Resistance with Cas9, Cas12, Cas13, and Cas14. Heliyon 2025, 11, e42013. [Google Scholar] [CrossRef] [PubMed]
- Delpierre, C.; Lefèvre, T. Precision and personalized medicine: Implications for the development of personalized health. Front. Sociol. 2023, 8, 1112159. [Google Scholar] [CrossRef]
- Sorrenti, V.; Burò, I.; Consoli, V.; Vanella, L. Recent advances in health benefits of bioactive compounds from food wastes and by-products: Biochemical aspects. Int. J. Mol. Sci. 2023, 24, 2019. [Google Scholar] [CrossRef]
- Kesik-Brodacka, M. Progress in biopharmaceutical development. Biotechnol. Appl. Biochem. 2018, 65, 306–322. [Google Scholar] [CrossRef]
- Mózsik, L.; Iacovelli, R.; Bovenberg, R.A.L.; Driessen, A.J.M. Transcriptional activation of biosynthetic gene clusters in filamentous fungi. Front. Bioeng. Biotechnol. 2022, 10, 901037. [Google Scholar] [CrossRef]
- Ferrer-Miralles, N.; Saccardo, P.; Corchero, J.L.; Xu, Z.; García-Fruitós, E. General introduction: Recombinant protein production and purification of insoluble proteins. In Insoluble Proteins; Humana Press: New York, NY, USA, 2015; Volume 1258, pp. 1–24. [Google Scholar]
- Landgraf, W.; Sandow, J. Recombinant human insulins—clinical efficacy and safety in diabetes therapy. Eur. Endocrinol. 2016, 12, 12–17. [Google Scholar] [CrossRef]
- Erdoğan, S. Integration of artificial intelligence and genome editing system for determining the treatment of genetic disorders. Balk. Med. J. 2024, 41, 419–420. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Ma, X.; Gao, F.; Guo, Y. Off-Target Effects in CRISPR/Cas9 Gene Editing. Front. Bioeng. Biotechnol. 2023, 11, 1143157. [Google Scholar] [CrossRef] [PubMed]
- Lopes, R.; Prasad, M.K. Beyond the promise: Evaluating and mitigating off-target effects in CRISPR gene editing for safer therapeutics. Front. Bioeng. Biotechnol. 2024, 11, 1339189. [Google Scholar] [CrossRef]
- Yang, P.; Condrich, A.; Lu, L.; Scranton, S.; Hebner, C.; Sheykhhasan, M.; Ali, M.A. Genetic Engineering in Bacteria, Fungi, and Oomycetes, Taking Advantage of CRISPR. DNA 2024, 4, 427–454. [Google Scholar] [CrossRef]
- Inen, J.; Han, C.M.; Farrel, D.M.; Bilousova, G.; Kogut, I. CIRCLE-Seq for Interrogation of Off-Target Gene Editing. J. Vis. Exp. 2024, 213, e67069. [Google Scholar] [CrossRef]
- Nyerges, Á.; Csörgő, B.; Nagy, I.; Bálint, B.; Bihari, P.; Lázár, V.; Apjok, G.; Umenhoffer, K.; Bogos, B.; Pósfai, G.; et al. A Highly Precise and Portable Genome Engineering Method Allows Comparison of Mutational Effects Across Bacterial Species. Proc. Natl. Acad. Sci. USA 2016, 113, 2502–2507. [Google Scholar] [CrossRef]
- Mentani, A.; Maresca, M.; Shiriaeva, A. Prime Editing: Mechanistic Insights and DNA Repair Modulation. Cells 2025, 14, 277. [Google Scholar] [CrossRef]
- Humbert, O.; Samuelson, C.; Kiem, H.P. CRISPR/Cas9 for the Treatment of Haematological Diseases: A Journey from Bacteria to the Bedside. Br. J. Haematol. 2021, 192, 33–49. [Google Scholar] [CrossRef]
- Kaupbayeva, B.; Tsoy, A.; Safarova, Y.; Nurmagambetova, A.; Murata, H.; Matyjaszewski, K.; Askarova, S. Unlocking genome editing: Advances and obstacles in CRISPR/Cas delivery technologies. J. Funct. Biomater. 2024, 15, 324. [Google Scholar] [CrossRef]
- Garner, K.L. Principles of synthetic biology. Essays Biochem. 2021, 65, 791–811. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.V.; Nybo, S.E. Complete sequences of pIJ101-based Streptomyces-Escherichia coli shuttle vectors. Access Microbiol. 2024, 6, 000893.v3. [Google Scholar] [CrossRef] [PubMed]
- Toh, M.; Chengan, K.; Hanson, T.; Freemont, P.S.; Moore, S.J. A high-yield Streptomyces transcription-translation toolkit for synthetic biology and natural product applications. J. Vis. Exp. 2021, 175, e63012. [Google Scholar]
- Liang, J.; Luo, Y.; Zhao, H. Synthetic biology: Putting synthesis into biology. Wiley Interdiscip. Rev. Syst. Biol. Med. 2011, 3, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Horbal, L.; Siegl, T.; Luzhetskyy, A. A set of synthetic versatile genetic control elements for the efficient expression of genes in Actinobacteria. Sci. Rep. 2018, 8, 491. [Google Scholar] [CrossRef]
- Martins, D.P.; Barros, M.T.; O’Sullivan, B.; Seymour, I.; O’Riordan, A.; Coffey, L.; Sweeney, J.; Balasubramaniam, S. Microfluidic-Based Bacterial Molecular Computing on a Chip. IEEE Sens. J. 2022, 22, 16772–16784. [Google Scholar] [CrossRef]
- Breitling, R.; Takano, E. Synthetic biology advances for pharmaceutical production. Curr. Opin. Biotechnol. 2015, 35, 46–51. [Google Scholar] [CrossRef]
- Keasling, J.D. Synthetic biology for synthetic chemistry. ACS Chem. Biol. 2008, 3, 64–76. [Google Scholar] [CrossRef]
- Jones, J.A.; Koffas, M.a.G. Optimizing Metabolic Pathways for the Improved Production of Natural Products. Methods Enzym. 2016, 575, 179–193. [Google Scholar] [CrossRef]
- Perez, R.F.; Pillow, J.J.; Kaur, P. Bioprospecting microbes and enzymes for the production of pterocarpans and coumestans. Front. Bioeng. Biotechnol. 2023, 11, 1154779. [Google Scholar] [CrossRef]
- Bober, J.R.; Beisel, C.L.; Nair, N.U. Synthetic biology approaches to engineer probiotics and members of the human microbiota for biomedical applications. Annu. Rev. Biomed. Eng. 2018, 20, 277–300. [Google Scholar] [CrossRef] [PubMed]
- Abuhena, M.; Zhakypbek, Y.; Yerlan, U.; Aben, A.; Kamarkhan, Z.; Allakhverdiev, S.I.; Assemgul, S.K.; Rashid, J.A.; Karim, M.D.; Kuanysh, T.; et al. An overview of Bacillus species in agriculture for growth promotion, biocontrol and dry tolerance. ES Food Agrofor. 2024, 18, 1321. [Google Scholar] [CrossRef]
- Siddique, A.; Azim, S.; Ali, A.; Adnan, F.; Arif, M.; Imran, M.; Ganda, E.; Rahman, A. Lactobacillus reuteri and Enterococcus faecium from Poultry Gut Reduce Mucin Adhesion and Biofilm Formation of Cephalosporin and Fluoroquinolone-Resistant Salmonella enterica. Animals 2021, 11, 3435. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Kwon, M.; Kim, J.Y. Enhancement of specialized metabolites using CRISPR/Cas gene editing technology in medicinal plants. Front. Plant Sci. 2024, 21, 1279738. [Google Scholar] [CrossRef]
- Ford, T.J.; Silver, P.A. Synthetic biology expands chemical control of microorganisms. Curr. Opin. Chem. Biol. 2015, 28, 20–28. [Google Scholar] [CrossRef]
- Lux, M.W.; Strychalski, E.A.; Vora, G.J. Advancing reproducibility can ease the ‘hard truths’ of synthetic biology. Synth. Biol. 2023, 8, ysad014. [Google Scholar] [CrossRef]
- Schmidt, C.W. Synthetic biology: Environmental health implications of a new field. Environ. Health Perspect. 2010, 118, A118–A123. [Google Scholar] [CrossRef]
- Chandra, S.; Wilson, J.C.; Good, D.; Wei, M.Q. mRNA vaccines: A new era in vaccine development. Oncol. Res. 2024, 32, 1543–1564. [Google Scholar] [CrossRef] [PubMed]
- Carlson, R. Estimating the biotech sector’s contribution to the US economy. Nat. Biotechnol. 2016, 34, 247–255. [Google Scholar] [CrossRef]
- Fernandes, T.; Osório, C.; Sousa, M.J.; Franco-Duarte, R. Contributions of adaptive laboratory evolution towards the enhancement of the biotechnological potential of non-conventional yeast species. J. Fungi 2023, 9, 186. [Google Scholar] [CrossRef]
- Dragosits, M.; Mattanovich, D. Adaptive laboratory evolution—Principles and applications for biotechnology. Microb. Cell Factories 2013, 12, 64. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, P. Current status and applications of adaptive laboratory evolution in industrial microorganisms. J. Microbiol. Biotechnol. 2020, 30, 793–803. [Google Scholar] [CrossRef]
- Zhu, P.; Luo, R.; Li, Y.; Chen, X. Metabolic engineering and adaptive evolution for efficient production of L-lactic acid in Saccharomyces cerevisiae. Microbiol. Spectr. 2022, 10, e0227722. [Google Scholar] [CrossRef] [PubMed]
- Bailey, E.; McGuire, F.E. Genomic analysis of laboratory-evolved, heat-adapted Escherichia coli strains. bioRxiv 2024. 2024-10. [Google Scholar] [CrossRef]
- Marives, T.C.; Ju, E.J.; Lee, J.H. Enhancing microbial resilience: The role of adaptive laboratory evolution in industrial biotechnology. J. Life Sci. 2024, 34, 730–743. [Google Scholar]
- Yao, L.; Jia, Y.; Zhang, Q.; Zheng, X.; Yang, H.; Dai, J.; Chen, X. Adaptive laboratory evolution to obtain furfural tolerant Saccharomyces cerevisiae for bioethanol production and the underlying mechanism. Front. Microbiol. 2024, 14, 1333777. [Google Scholar] [CrossRef]
- Jahn, L.J.; Munck, C.; Ellabaan, M.M.H.; Sommer, M.O.A. Adaptive laboratory evolution of antibiotic resistance using different selection regimes lead to similar phenotypes and genotypes. Front. Microbiol. 2017, 8, 816. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Fang, H.; Cui, F.; Nyabako, B.A.; Tao, T.; Zan, X.; Chen, H.; Sun, W. ARTP mutation and adaptive laboratory evolution improve probiotic performance of Bacillus coagulans. Appl. Microbiol. Biotechnol. 2020, 104, 6363–6373. [Google Scholar] [CrossRef]
- Zhakypbek, Y.; Belkozhayev, A.M.; Kerimkulova, A.; Kossalbayev, B.D.; Murat, T.; Tursbekov, S.; Turysbekova, G.; Tursunova, A.; Tastambek, K.T.; Allakhverdiev, S.I. MicroRNAs in Plant Genetic Regulation of Drought Tolerance and Their Function in Enhancing Stress Adaptation. Plants 2025, 14, 410. [Google Scholar] [CrossRef]
- Vora, L.K.; Gholap, A.D.; Jetha, K.; Thakur, R.R.S.; Solanki, H.K.; Chavda, V.P. Artificial intelligence in pharmaceutical technology and drug delivery design. Pharmaceutics 2023, 15, 1916. [Google Scholar] [CrossRef]
- Shelke, Y.P.; Badge, A.K.; Bankar, N.J. Applications of artificial intelligence in microbial diagnosis. Cureus 2023, 15, e49366. [Google Scholar] [CrossRef] [PubMed]
- Alsulimani, A.; Akhter, N.; Jameela, F.; Ashgar, R.I.; Jawed, A.; Hassani, M.A.; Dar, S.A. The impact of artificial intelligence on microbial diagnosis. Microorganisms 2024, 12, 1051. [Google Scholar] [CrossRef]
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.M.; Macnair, C.R.; French, S.; Carfrae, L.A.; Bloom-Ackermann, Z.; et al. A deep learning approach to antibiotic discovery. Cell 2020, 180, 688–702.e13. [Google Scholar] [CrossRef]
- Jiang, Y.; Luo, J.; Huang, D.; Liu, Y.; Li, D.D. Machine learning advances in microbiology: A review of methods and applications. Front. Microbiol. 2022, 13, 925454. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Zhao, Y.; Qi, Z.; Hou, S.; Chen, J. Machine learning empowering drug discovery: Applications, opportunities and challenges. Molecules 2024, 29, 903. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Ahlgren, N.A.; Lu, Y.Y.; Fuhrman, J.A.; Sun, F. VirFinder: A novel k-mer based tool for identifying viral sequences from assembled metagenomic data. Microbiome 2017, 5, 69. [Google Scholar] [CrossRef]
- Amgarten, D.; Braga, L.P.P.; Da Silva, A.M.; Setubal, J.C. MARVEL, a tool for prediction of bacteriophage sequences in metagenomic bins. Front. Genet. 2018, 9, 304. [Google Scholar] [CrossRef]
- Yu, Y.; Gawlitt, S.; de Andrade e Sousa, L.B. Improved prediction of bacterial CRISPRi guide efficiency from depletion screens through mixed-effect machine learning and data integration. Genome Biol. 2024, 25, 13. [Google Scholar] [CrossRef]
- Wu, Y.; Gadsden, S.A. Machine learning algorithms in microbial classification: A comparative analysis. Front. Artif. Intell. 2023, 6, 1200994. [Google Scholar] [CrossRef]
- Xu, G.; Teng, X.; Gao, X.H.; Zhang, L.; Yan, H.; Qi, R.Q. Advances in machine learning-based bacteria analysis for forensic identification: Identity, ethnicity, and site of occurrence. Front. Microbiol. 2023, 14, 1332857. [Google Scholar] [CrossRef]
- Ahmed, W.; Staley, C.; Kaiser, T.; Sadowsky, M.J.; Kozak, S.; Beale, D. Decay of sewage-associated bacterial communities in fresh and marine environmental waters and sediment. Appl. Microbiol. Biotechnol. 2018, 102, 7159–7170. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cui, X.; Yang, X.; Liu, G.; Zhang, J. Artificial intelligence in predicting pathogenic microorganisms’ antimicrobial resistance: Challenges, progress, and prospects. Front. Cell Infect. Microbiol. 2024, 14, 1482186. [Google Scholar] [CrossRef] [PubMed]
- Alowais, S.A.; Alghamdi, S.S.; Alsuhebany, N. Revolutionizing healthcare: The role of artificial intelligence in clinical practice. BMC Med. Educ. 2023, 23, 689. [Google Scholar] [CrossRef]
- Badwan, B.A.; Liaropoulos, G.; Kyrodimos, E.; Skaltsas, D.; Tsirigos, A.; Gorgoulis, V.G. Machine learning approaches to predict drug efficacy and toxicity in oncology. Cell Rep. Methods 2023, 3, 100413. [Google Scholar] [CrossRef] [PubMed]
- Vilhekar, R.S.; Rawekar, A. Artificial intelligence in genetics. Cureus 2024, 16, e52035. [Google Scholar] [CrossRef]
- Cheng, Y.; Bi, X.; Xu, Y.; Liu, Y.; Li, J.; Du, G.; Lv, X.; Liu, L. Machine learning for metabolic pathway optimization: A review. Comput. Struct. Biotechnol. J. 2023, 21, 2381–2393. [Google Scholar] [CrossRef]
- Sahayasheela, V.J.; Lankadasari, M.B.; Dan, V.M.; Dastager, S.G.; Pandian, G.N.; Sugiyama, H. Artificial intelligence in microbial natural product drug discovery: Current and emerging role. Nat. Prod. Rep. 2022, 39, 2215–2230. [Google Scholar] [CrossRef]
- Quazi, S. Artificial intelligence and machine learning in precision and genomic medicine. Med. Oncol. 2022, 39, 120. [Google Scholar] [CrossRef]
- Al-Shaebi, Z.; Uysal Ciloglu, F.; Nasser, M.; Aydin, O. Highly accurate identification of bacteria’s antibiotic resistance based on Raman spectroscopy and U-Net deep learning algorithms. ACS Omega 2022, 7, 29443–29451. [Google Scholar] [CrossRef]
- Allakhverdiev, E.S.; Kossalbayev, B.D.; Sadvakasova, A.K.; Bauenova, M.O.; Belkozhayev, A.M.; Rodnenkov, O.V.; Martynyuk, T.V.; Maksimov, G.V.; Allakhverdiev, S.I. Spectral Insights: Navigating the Frontiers of Biomedical and Microbiological Exploration with Raman Spectroscopy. J. Photochem. Photobiol. B Biol. 2024, 252, 112870. [Google Scholar] [CrossRef]
- Ho, C.S.; Jean, N.; Hogan, C.A.; Blackmon, L.; Jeffrey, S.S.; Holodniy, M.; Banaei, N.; Saleh, A.A.; Ermon, S.; Dionne, J. Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat. Commun. 2019, 10, 4927. [Google Scholar] [CrossRef] [PubMed]
- Muteeb, G.; Rehman, M.T.; Shahwan, M.; Aatif, M. Origin of antibiotics and antibiotic resistance, and their impacts on drug development: A narrative review. Pharmaceuticals 2023, 16, 1615. [Google Scholar] [CrossRef]
- Woo, H.; Kim, Y.; Kim, D.; Yoon, S.H. Machine learning identifies key metabolic reactions in bacterial growth on different carbon sources. Mol. Syst. Biol. 2024, 20, 170–186. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Petersen, S.D.; Radivojevic, T.; Ramirez, A.; Pérez-Manríquez, A.; Abeliuk, E.; Sánchez, B.J.; Costello, Z.; Chen, Y.; Fero, M.J.; et al. Combining Mechanistic and Machine Learning Models for Predictive Engineering and Optimization of Tryptophan Metabolism. Nat Commun. 2020, 11, 4880. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Rybakova, D.; Fischer, D. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar]
- Liu, J.; Chan, S.H.J.; Chen, J.; Solem, C.; Jensen, P.R. Systems Biology—A Guide for Understanding and Developing Improved Strains of Lactic Acid Bacteria. Front. Microbiol. 2019, 10, 876. [Google Scholar] [CrossRef]
- Ferrer, P. Systems biology and biological systems diversity for the engineering of microbial cell factories. Microb. Cell Fact. 2007, 6, 35. [Google Scholar] [CrossRef]
- Liu, D.; Hoynes-O’Connor, A.; Zhang, F. Bridging the gap between systems biology and synthetic biology. Front. Microbiol. 2013, 4, 211. [Google Scholar] [CrossRef]
- Heinken, A.; Thiele, I. Systems biology of host-microbe metabolomics. Wiley Interdiscip. Rev. Syst. Biol. Med. 2015, 7, 195–219. [Google Scholar] [CrossRef]
- Gray, M.J.; Tavares, N.K.; Escalante-Semerena, J.C. The genome of Rhodobacter sphaeroides strain 2.4.1 encodes functional cobinamide salvaging systems of archaeal and bacterial origins. Mol. Microbiol. 2008, 70, 824–836. [Google Scholar] [CrossRef]
- Imam, S.; Noguera, D.R.; Donohue, T.J. Global insights into energetic and metabolic networks in Rhodobacter sphaeroides. BMC Syst. Biol. 2013, 7, 89. [Google Scholar] [CrossRef] [PubMed]
- Amaning Danquah, C.; Minkah, P.A.B.; Osei Duah Junior, I.; Amankwah, K.B.; Somuah, S.O. Antimicrobial compounds from microorganisms. Antibiotics 2022, 11, 285. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, D.C.; Rocha, V.P.C.; Damasceno, P.K.F.; Barbosa, J.D.V.; Soares, M.B.P. Therapeutic applications of synthetic gene/genetic circuits: A patent review. Front. Bioeng. Biotechnol. 2024, 12, 1425529. [Google Scholar] [CrossRef]
- Sheng, J.; Guo, W.; Ash, C.; Freitas, B.T.; Paoletti, M.; Feng, X. Data-driven prediction of CRISPR-based transcription regulation for programmable control of metabolic flux. bioRxiv 2017. [Google Scholar] [CrossRef]
- Kim, K.; Kang, M.; Cho, B.K. Systems and synthetic biology-driven engineering of live bacterial therapeutics. Front. Bioeng. Biotechnol. 2023, 11, 1267378. [Google Scholar] [CrossRef] [PubMed]
- Shuwen, H.; Yifei, S.; Xinyue, W.; Zhanbo, Q.; Xiang, Y.; Xi, Y. Advances in bacteria-based drug delivery systems for anti-tumor therapy. Clin. Transl. Immunol. 2024, 13, e1518. [Google Scholar] [CrossRef]
- Yan, S.; Gan, Y.; Xu, H.; Piao, H. Bacterial carrier-mediated drug delivery systems: A promising strategy in cancer therapy. Front. Bioeng. Biotechnol. 2025, 12, 1526612. [Google Scholar] [CrossRef]
- Shao, X.; Zhao, X.; Wang, B.; Fan, J.; Wang, J.; An, H. Tumor microenvironment targeted nano-drug delivery systems for multidrug resistant tumor therapy. Theranostics 2025, 15, 1689–1714. [Google Scholar] [CrossRef]
- Deek, R.A.; Ma, S.; Lewis, J.; Li, H. Statistical and computational methods for integrating microbiome, host genomics, and metabolomics data. eLife 2024, 13, e88956. [Google Scholar] [CrossRef]
- Nazir, A.; Hussain, F.H.N.; Raza, A. Advancing microbiota therapeutics: The role of synthetic biology in engineering microbial communities for precision medicine. Front. Bioeng. Biotechnol. 2024, 12, 1511149. [Google Scholar] [CrossRef]
- Leggieri, P.A.; Liu, Y.; Hayes, M.; Connors, B.; Seppälä, S.; O’Malley, M.A.; Venturelli, O.S. Integrating systems and synthetic biology to understand and engineer microbiomes. Annu. Rev. Biomed. Eng. 2021, 23, 169–201. [Google Scholar] [CrossRef] [PubMed]
- Chetty, A.; Blekhman, R. Multi-omic approaches for host-microbiome data integration. Gut Microbes 2024, 16, 2297860. [Google Scholar] [CrossRef]
- Dahal, S.; Yurkovich, J.T.; Xu, H.; Palsson, B.O.; Yang, L. Synthesizing systems biology knowledge from omics using genome-scale models. Proteomics 2020, 20, e1900282. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.; Lorusso, E.; Fosso, B.; Pesole, G. A comprehensive overview of microbiome data in the light of machine learning applications: Categorization, accessibility, and future directions. Front. Microbiol. 2024, 15, 1343572. [Google Scholar] [CrossRef] [PubMed]
- Majzoub, M.E.; Luu, L.D.W.; Haifer, C.; Paramsothy, S.; Borody, T.J.; Leong, R.W.; Thomas, T.; Kaakoush, N.O. Refining microbial community metabolic models derived from metagenomics using reference-based taxonomic profiling. mSystems 2024, 9, e0074624. [Google Scholar] [CrossRef]
- Comparative Analysis of Regulation of GMO Products Worldwide. Available online: https://www.digicomply.com/blog/comparative-analysis-of-regulation-of-gmo-products-worldwide (accessed on 26 February 2025).
- US vs. EU: Getting American Microbial Products into the European Market. Available online: https://www.biosafe.fi/insight/us-vs-eu-getting-american-microbial-products-into-the-european-market (accessed on 26 February 2025).
- World Health Organization (WHO). Available online: https://www.who.int (accessed on 5 February 2025).
- Ryan, M.P.; Adley, C. Ralstonia spp.: Emerging global opportunistic pathogens. J. Med. Microbiol. 2014, 63, 1161–1170. [Google Scholar]
- Driscoll, J.A.; Brody, S.L.; Kollef, M.H. The epidemiology, pathogenesis, and treatment of Pseudomonas aeruginosa infections. Drugs 2007, 67, 351–368. [Google Scholar] [CrossRef]
- LiPuma, J.J. The Burkholderia cepacia complex: A new paradigm of bacterial resistance. Expert Rev. Anti-infect. Ther. 2010, 8, 1445–1459. [Google Scholar]
- Ehling-Schulz, M.; Fricker, M.; Scherer, S. Food- and airborne Bacillus cereus and its toxigenic potential: A review. Int. J. Food Microbiol. 2006, 112, 1–10. [Google Scholar]
- Microbial Monitoring for Biopharma Manufacturing. Available online: http://www.technologynetworks.com (accessed on 5 February 2025).
- Lotfipour, F.; Hallaj-Nezhadi, S. Microbial quality concerns for biopharmaceuticals. In Latest Research into Quality Control; IntechOpen: London, UK, 2012. [Google Scholar]
- Martin, D.; Aebersold, R.; Kovarik, P. The challenge of microbial contamination in biopharmaceutical production. Appl. Microbiol. Biotechnol. 2018, 102, 4023–4034. [Google Scholar]
- Food and Drug Administration. Early Clinical Trials with Live Biotherapeutic Products: Chemistry, Manufacturing, and Control Information. Available online: https://www.fda.gov (accessed on 5 February 2025).
- Quality, Non-Clinical and Clinical Aspects of Medicinal Products Containing Genetically Modified Cells—Scientific Guideline. European Medicines Agency (EMA). Available online: https://www.ema.europa.eu (accessed on 5 February 2025).
- Steidler, L.; Rottiers, P.; Schotte, L.; Remaut, E.; Remon, J.P.; Heremans, K. Treatment of murine colitis by delivery of interleukin-10 via genetically modified Lactococcus lactis. Nat. Biotechnol. 2003, 21, 1022–1028. [Google Scholar] [CrossRef]
- Isabella, V.M.; Ha, B.N.; Castillo, M.J.; Lubkowicz, D.J.; Rowe, S.E.; Millet, Y.A.; Anderson, C.L.; Li, N.; Fisher, A.B.; West, K.A.; et al. Development of a synthetic live bacterial therapeutic for the treatment of phenylketonuria. Sci. Transl. Med. 2018, 10, eaap8602. [Google Scholar]
- Dreher-Lesnick, S.M.; Stibitz, S.; Carlson, P.E.U.S. regulatory considerations for development of live biotherapeutic products as drugs. Microbiol. Spectr. 2017, 5, 409–416. [Google Scholar] [CrossRef]
- Keasling, J.D. Manufacturing molecules through metabolic engineering. Science 2010, 330, 1355–1358. [Google Scholar] [CrossRef] [PubMed]
- Dueber, J.E.; Wu, G.C.; Malmirchegini, G.R.; Moon, T.S.; Petzold, C.J.; Ullal, A.V.; Prather, K.L.J.; Keasling, J.D. Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 2009, 27, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Cameron, D.E.; Bashor, C.J.; Collins, J.J. A brief history of synthetic biology. Nat. Rev. Microbiol. 2014, 12, 381–390. [Google Scholar] [CrossRef]
- Liu, Y.; Feng, J.; Pan, H.; Zhang, X.; Zhang, Y. Genetically engineered bacterium: Principles, practices, and prospects. Front. Microbiol. 2022, 13, 997587. [Google Scholar] [CrossRef]
- Lange, L.; Berg, G.; Cernava, T.; Champomier-Vergès, M.C.; Charles, T.; Cocolin, L.; Cotter, P.; D’hondt, K.; Kostic, T.; Maguin, E.; et al. Microbiome ethics, guiding principles for microbiome research, use and knowledge management. Environ. Microbiome 2022, 17, 50. [Google Scholar] [CrossRef]
- Jagger, K.S.; Furlong, J. Infusing bioethics into biology and microbiology courses and curricula: A vertical approach. J. Microbiol. Biol. Educ. 2014, 15, 213–217. [Google Scholar] [CrossRef][Green Version]
- Anderson, J.; Strelkowa, N.; Stan, G.B.; Douglas, T.; Savulescu, J.; Barahona, M.; Papachristodoulou, A. Engineering and ethical perspectives in synthetic biology. EMBO Rep. 2012, 13, 584–590. [Google Scholar] [CrossRef]
- Mandrioli, M. Genome editing among bioethics and regulatory practices. Biomolecules 2021, 12, 13. [Google Scholar] [CrossRef]
- Lerner, A.; Benzvi, C.; Vojdani, A. The potential harmful effects of genetically engineered microorganisms (GEMs) on the intestinal microbiome and public health. Microorganisms 2024, 12, 238. [Google Scholar] [CrossRef] [PubMed]
- Rebello, S.; Nathan, V.K.; Sindhu, R.; Binod, P.; Awasthi, M.K.; Pandey, A. Bioengineered microbes for soil health restoration: Present status and future. Bioengineered 2021, 12, 12839–12853. [Google Scholar] [CrossRef]
- Sudheer, S.; Bai, R.G.; Usmani, Z.; Sharma, M. Insights on engineered microbes in sustainable agriculture: Biotechnological developments and future prospects. Curr. Genom. 2020, 21, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, G.R.; Toranzos, G.A.; Bhatti, A.R. Novel method for monitoring genetically engineered microorganisms in the environment. Appl. Environ. Microbiol. 1989, 55, 1301–1304. [Google Scholar] [CrossRef] [PubMed]
- Buckner, M.M.C.; Ciusa, M.L.; Piddock, L.J.V. Strategies to combat antimicrobial resistance: Anti-plasmid and plasmid curing. FEMS Microbiol. Rev. 2018, 42, 781–804. [Google Scholar] [CrossRef]
- Omer, R.; Mohsin, M.Z.; Mohsin, A.; Mushtaq, B.S.; Huang, X.; Guo, M.; Zhuang, Y.; Huang, J. Engineered bacteria-based living materials for biotherapeutic applications. Front. Bioeng. Biotechnol. 2022, 10, 870675. [Google Scholar] [CrossRef]
- Jin, K.; Huang, Y.; Che, H.; Wu, Y. Engineered bacteria for disease diagnosis and treatment using synthetic biology. Microb. Biotechnol. 2025, 18, e70080. [Google Scholar] [CrossRef]
- Jeong, S.H.; Lee, H.J.; Lee, S.J. Recent advances in CRISPR-Cas technologies for synthetic biology. J. Microbiol. 2023, 61, 13–36. [Google Scholar] [CrossRef]
- Loureiro, A.; da Silva, G.J. CRISPR-Cas: Converting a bacterial defense mechanism into a state-of-the-art genetic manipulation tool. Antibiotics 2019, 8, 18. [Google Scholar] [CrossRef]
- Dey, A. CRISPR/Cas genome editing to optimize pharmacologically active plant natural products. Pharmacol. Res. 2021, 164, 105359. [Google Scholar] [CrossRef] [PubMed]
- Smanski, M.J.; Bhatia, S.; Zhao, D.; Park, Y.; Woodruff, L.B.A.; Giannoukos, G.; Chandran, S. Synthetic biology to access and expand nature’s chemical diversity. Nat. Rev. Microbiol. 2014, 12, 709–723. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, A.A.K.; Der, B.S.; Shin, J.; Vaidyanathan, P.; Paralanov, V.; Strychalski, E.A.; Voigt, C.A. Genetic circuit design automation. Science 2016, 352, aac7341. [Google Scholar] [CrossRef] [PubMed]
- Sewell, F.; Alexander-White, C.; Brescia, S.; Currie, R.A.; Roberts, R.; Roper, C.; Vickers, C.; Westmoreland, C.; Kimber, I. New approach methodologies (NAMs): Identifying and overcoming hurdles to accelerated adoption. Toxicol. Res. 2024, 13, tfae044. [Google Scholar] [CrossRef]
- Corporate Europe Observatory. Industry Edited EFSA’s Glyphosate Evaluation Ahead of Publication. Available online: https://corporateeurope.org/en/efsa/2017/07/industry-edited-efsa-glyphosate-evaluation-ahead-publication (accessed on 26 February 2025).
- Gupta, A.; Falkner, R. The Influence of the Cartagena Protocol on Biosafety: Comparing Mexico, China and South Africa. Glob. Environ. Politics 2006, 6, 23–55. [Google Scholar] [CrossRef]
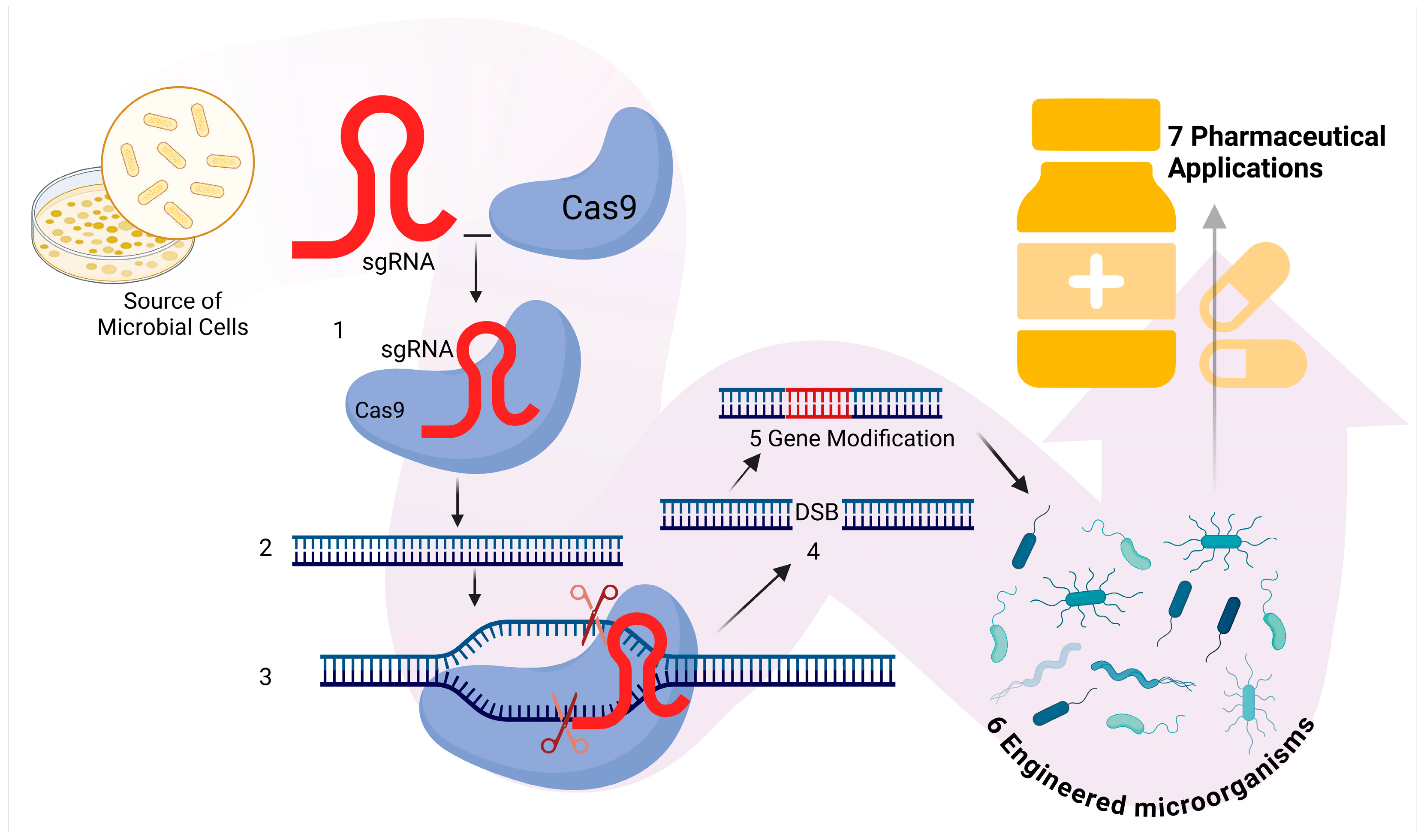
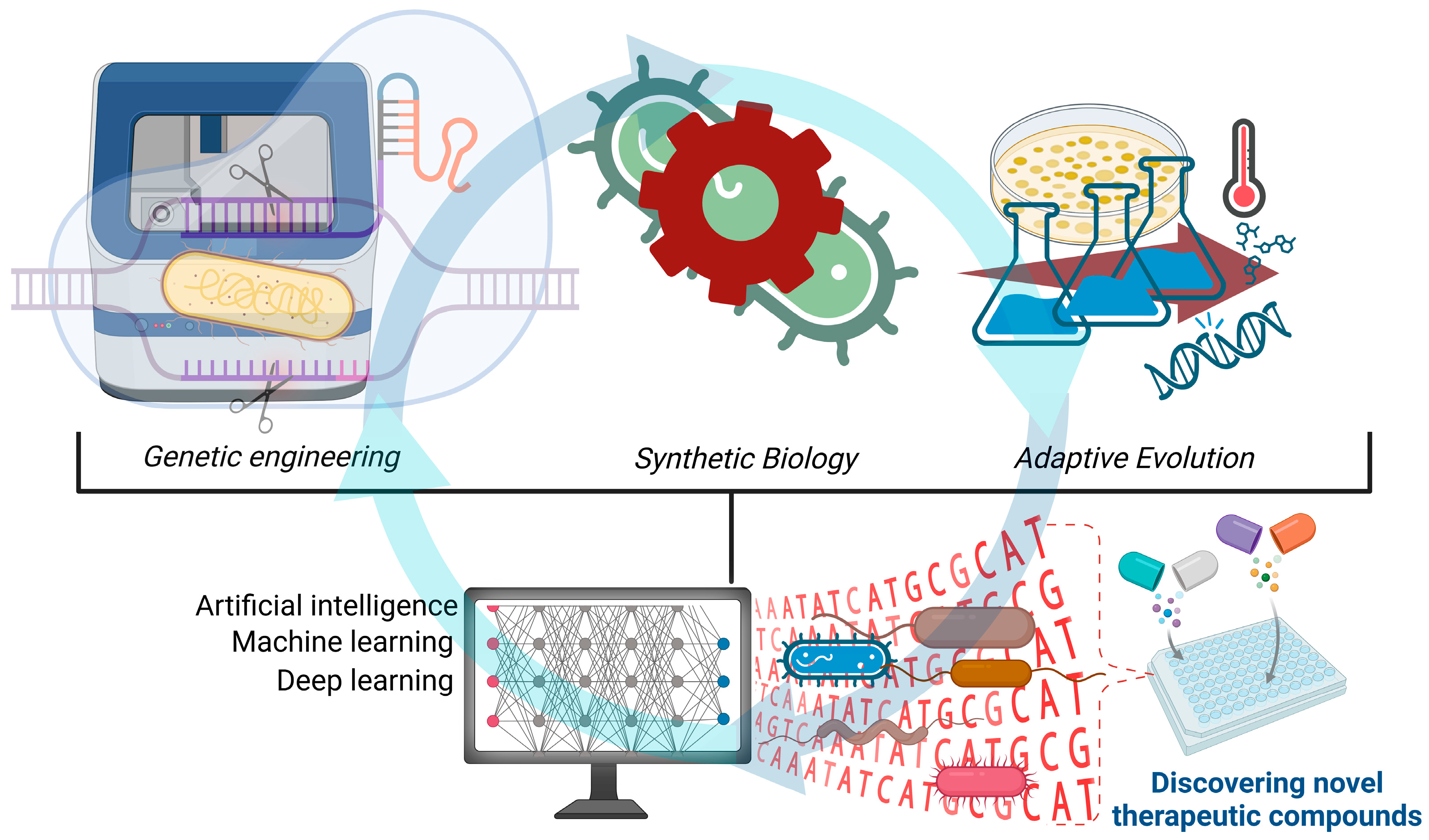

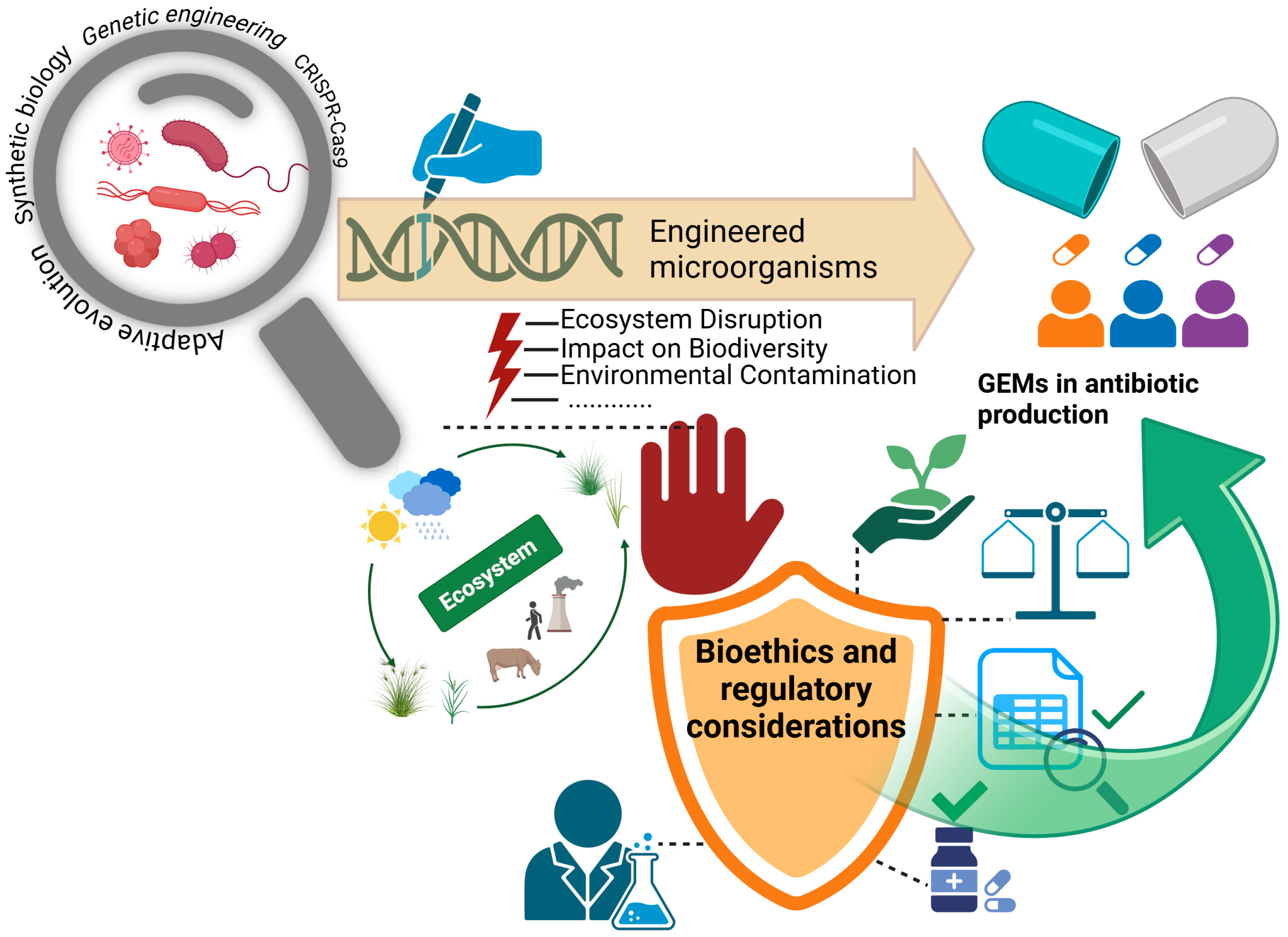
| Future Direction | Objective and Application Areas | Reference |
|---|---|---|
| CRISPR-based drug development | Precise treatment of genetic diseases and gene therapy, targeting rare genetic disorders | [43] |
| Cell and gene therapies | Correction of mutant genes, cell regeneration for cancer and hereditary diseases | [44] |
| Enhanced antibiotic production | Combating antibiotic-resistant bacteria, development of new antibiotics | [45,46] |
| Personalized medicine | Tailoring treatments based on patient genetics, precision pharmacotherapy | [47] |
| Biopharmaceutical active compounds | Increasing production of biologically derived drugs, vaccines, insulin, hormones | [48,49] |
| Activation of biosynthetic gene clusters | Discovery of novel natural compounds, antibiotics, antiviral drugs | [50] |
| Recombinant protein production | Artificial synthesis of essential human proteins, insulin, therapeutic enzymes | [51,52] |
| AI and CRISPR integration | Automating genome editing to accelerate new drug development | [53] |
| Microorganism | Contamination Source | Product | Strategies | Reference |
|---|---|---|---|---|
| Ralstonia pickettii | Contaminated water systems used in bioreactors and manufacturing equipment | Can lead to product spoilage and endotoxin contamination; may cause batch failures | Enhanced water system sterilization; use of ultrafiltration and validated decontamination protocols; routine environmental monitoring | [140] |
| Pseudomonas aeruginosa | Environmental sources (air, surfaces, equipment) within manufacturing facilities | Biofilm formation on equipment surfaces; production downtime; potential endotoxin release affecting product safety | Strict facility hygiene practices; routine disinfection; installation of high-efficiency particulate air (HEPA) filtration and regular environmental monitoring | [141] |
| Burkholderia cepacia complex | Contaminated raw materials or water used in production processes | Leads to production delays, product recalls, and poses risks of patient infection due to its intrinsic resistance mechanisms | Rigorous quality control for raw materials and water; validated cleaning procedures; regular microbial testing of production environments | [142] |
| Bacillus cereus | Airborne spores entering sterile manufacturing areas during production | Potential for pyrogenic reactions; product contamination may lead to recalls and safety concerns | Implementation of HEPA filtration; strict environmental monitoring; effective sanitization protocols and controlled air-handling systems | [143] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sadanov, A.K.; Baimakhanova, B.B.; Orasymbet, S.E.; Ratnikova, I.A.; Turlybaeva, Z.Z.; Baimakhanova, G.B.; Amitova, A.A.; Omirbekova, A.A.; Aitkaliyeva, G.S.; Kossalbayev, B.D.; et al. Engineering Useful Microbial Species for Pharmaceutical Applications. Microorganisms 2025, 13, 599. https://doi.org/10.3390/microorganisms13030599
Sadanov AK, Baimakhanova BB, Orasymbet SE, Ratnikova IA, Turlybaeva ZZ, Baimakhanova GB, Amitova AA, Omirbekova AA, Aitkaliyeva GS, Kossalbayev BD, et al. Engineering Useful Microbial Species for Pharmaceutical Applications. Microorganisms. 2025; 13(3):599. https://doi.org/10.3390/microorganisms13030599
Chicago/Turabian StyleSadanov, Amankeldi K., Baiken B. Baimakhanova, Saltanat E. Orasymbet, Irina A. Ratnikova, Zere Z. Turlybaeva, Gul B. Baimakhanova, Aigul A. Amitova, Anel A. Omirbekova, Gulzat S. Aitkaliyeva, Bekzhan D. Kossalbayev, and et al. 2025. "Engineering Useful Microbial Species for Pharmaceutical Applications" Microorganisms 13, no. 3: 599. https://doi.org/10.3390/microorganisms13030599
APA StyleSadanov, A. K., Baimakhanova, B. B., Orasymbet, S. E., Ratnikova, I. A., Turlybaeva, Z. Z., Baimakhanova, G. B., Amitova, A. A., Omirbekova, A. A., Aitkaliyeva, G. S., Kossalbayev, B. D., & Belkozhayev, A. M. (2025). Engineering Useful Microbial Species for Pharmaceutical Applications. Microorganisms, 13(3), 599. https://doi.org/10.3390/microorganisms13030599






