The Impact of DNA Methylation in Trichoderma reesei on Cellulase Production and Strain Degeneration
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Strains
2.2. Growth Conditions
2.3. Construction of Deletion Cassettes for dim2 and rid1
2.4. Fungal Transformation
2.5. Genotyping of Deletion Strains
2.6. Induced Strain Degeneration (ISD) Protocol
2.7. Cellulase Activity Assay
2.8. Global DNA Methylation Assay
2.9. Transcript Analysis
2.10. Statistical Analysis
3. Results
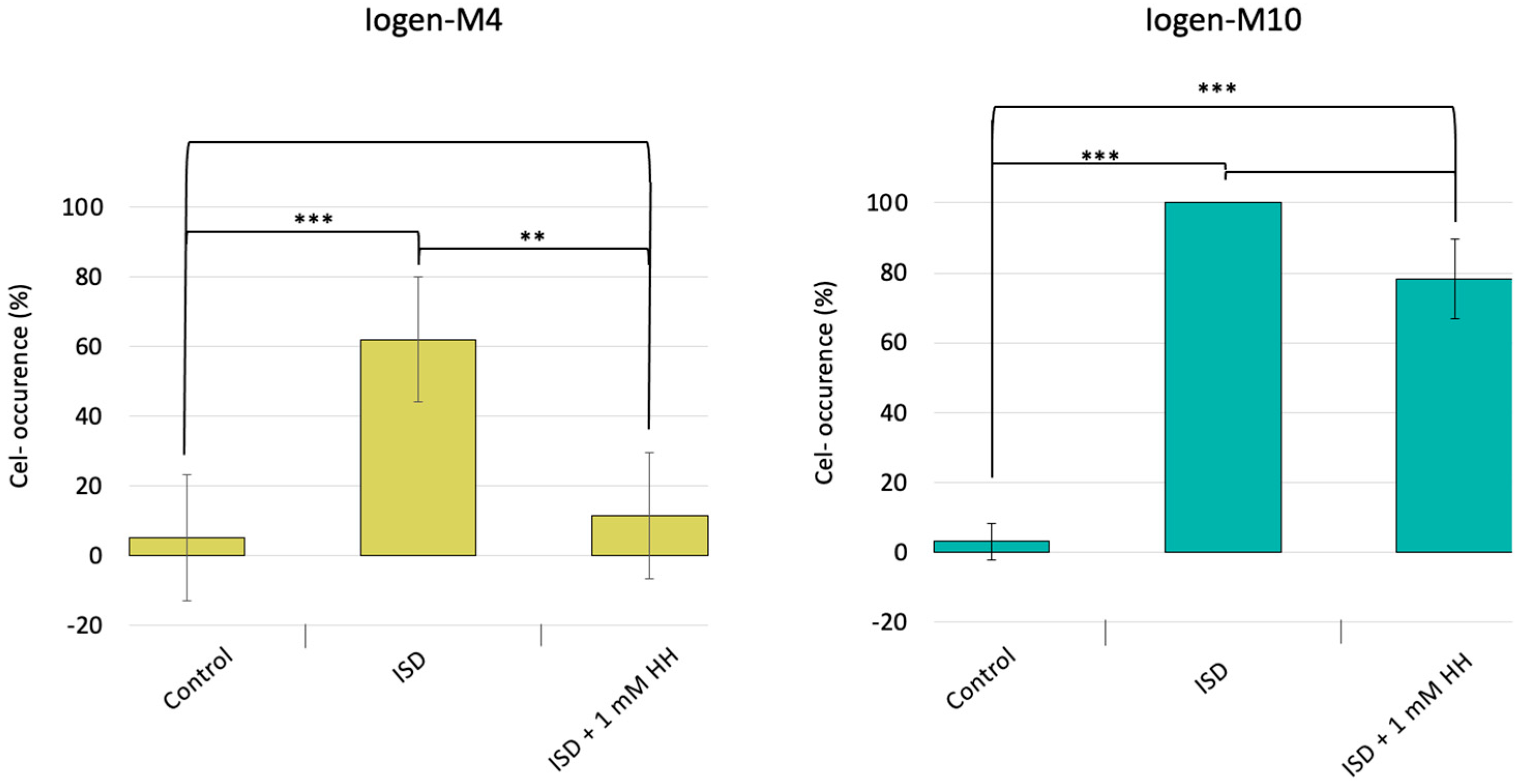
3.1. DNA Hypomethylation Partly Prevents the Degeneration Phenomenon
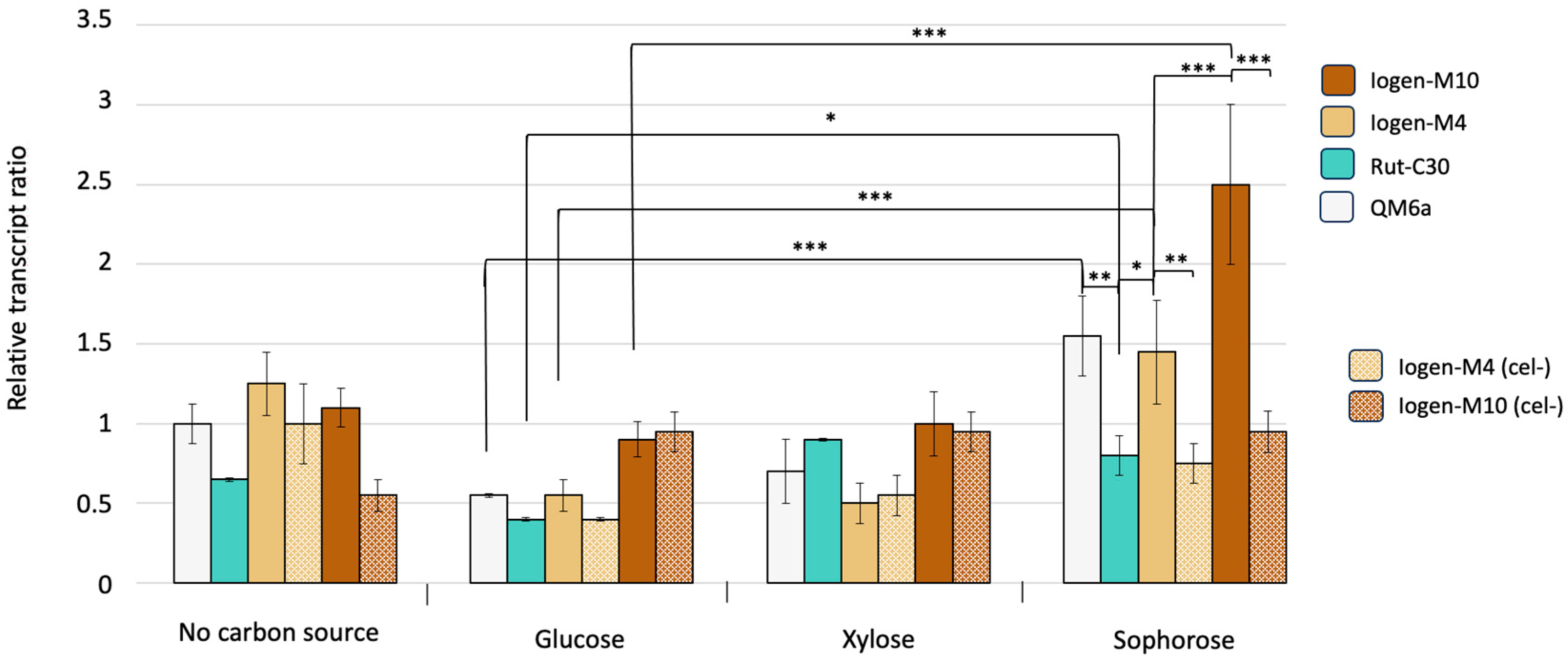
3.2. Global Methylation Pattern of Differently Productive T. reesei Strains
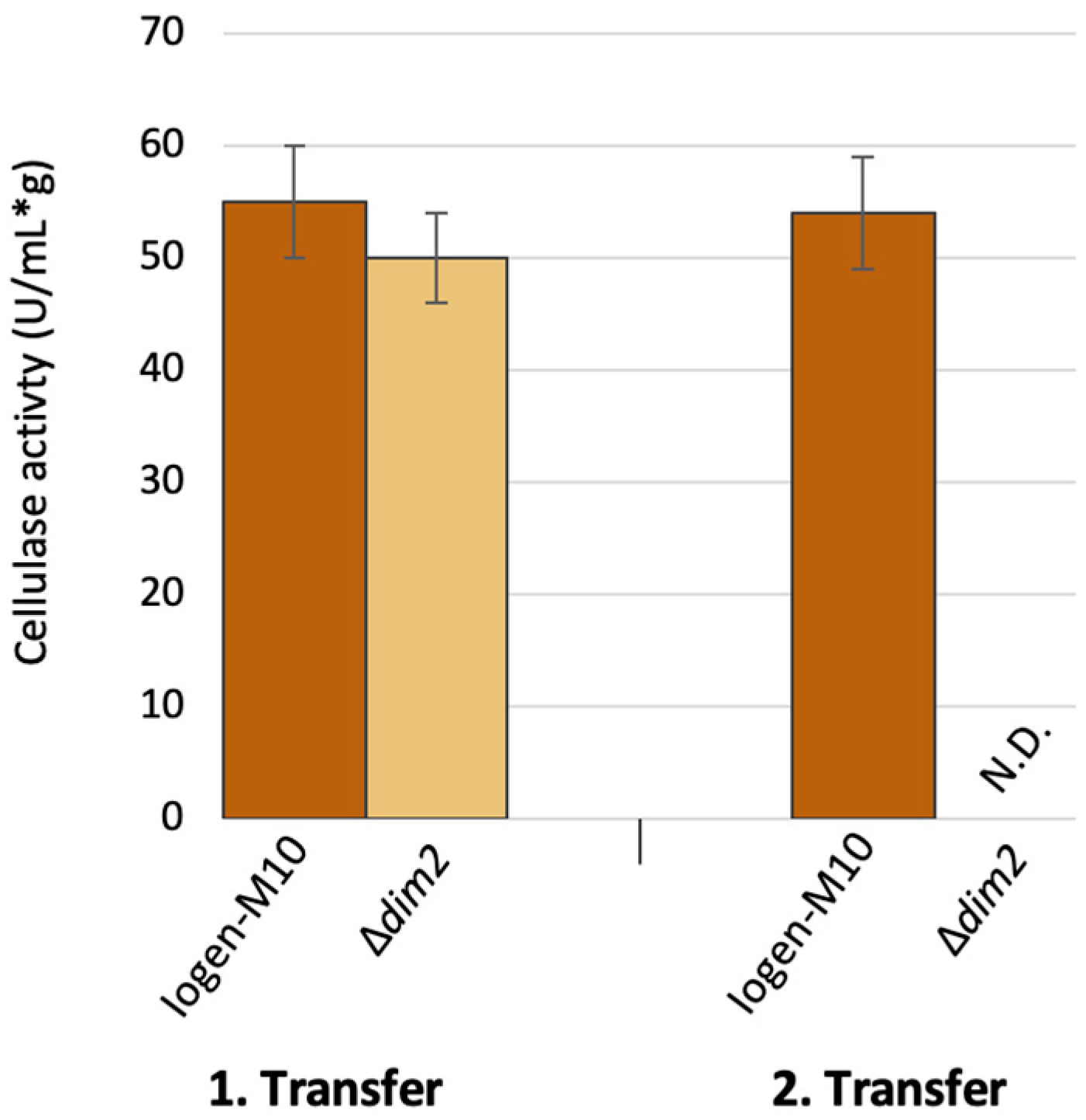
3.3. The Role of the Putative DNA Methyltransferases Dim2 and Rid1 in Strain Degeneration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhardwaj, N.; Kumar, B.; Agrawal, K.; Verma, P. Current Perspective on Production and Applications of Microbial Cellulases: A Review. Bioresour. Bioprocess. 2021, 8, 95. [Google Scholar] [CrossRef] [PubMed]
- Margolles-Clark, E.; Ilmén, M.; Penttilä, M. Expression Patterns of Ten Hemicellulase Genes of the Filamentous Fungus Trichoderma reesei on Various Carbon Sources. J. Biotechnol. 1997, 57, 167–179. [Google Scholar] [CrossRef]
- Yan, S.; Xu, Y.; Yu, X.W. From Induction to Secretion: A Complicated Route for Cellulase Production in Trichoderma reesei. Bioresour. Bioprocess. 2021, 8, 107. [Google Scholar] [CrossRef] [PubMed]
- Strauss, J.; Mach, R.L.; Zeilinger, S.; Hartler, G.; Stöffler, G.; Wolschek, M.; Kubicek, C.P. Crel, the Carbon Catabolite Repressor Protein from Trichoderma reesei. FEBS Lett. 1995, 376, 103–107. [Google Scholar] [CrossRef]
- Derntl, C.; Gudynaite-Savitch, L.; Calixte, S.; White, T.; Mach, R.L.; Mach-Aigner, A.R. Mutation of the Xylanase Regulator 1 Causes a Glucose Blind Hydrolase Expressing Phenotype in Industrially Used Trichoderma Strains. Biotechnol. Biofuels 2013, 6, 62. [Google Scholar] [CrossRef]
- Cherry, J.R.; Fidantsef, A.L. Directed Evolution of Industrial Enzymes: An Update. Curr. Opin. Biotechnol. 2003, 14, 438–443. [Google Scholar] [CrossRef]
- Hui, J.P.M.; Lanthier, P.; White, T.C.; McHugh, S.G.; Yaguchi, M.; Roy, R.; Thibault, P. Characterization of Cellobiohydrolase I (Cel7A) Glycoforms from Extracts of Trichoderma reesei Using Capillary Isoelectric Focusing and Electrospray Mass Spectrometry. J. Chromatogr. B Biomed. Sci. Appl. 2001, 752, 349–368. [Google Scholar] [CrossRef] [PubMed]
- Martzy, R.; Mello-de-Sousa, T.M.; Mach, R.L.; Yaver, D.; Mach-Aigner, A.R. The Phenomenon of Degeneration of Industrial Trichoderma reesei Strains. Biotechnol. Biofuels 2021, 14, 193. [Google Scholar] [CrossRef]
- Peterson, R.; Nevalainen, H. Trichoderma reesei RUT-C30—Thirty Years of Strain Improvement. Microbiology 2012, 158, 58–68. [Google Scholar] [CrossRef]
- Ilmen, M.; Thrane, C.; Penttilla, M. The Glucose Repressor Gene Crel of Trichoderma: Isolation and Expression of a Full-Length and a Truncated Mutant Form. Mol. Gen. Genet. MGG 1996, 251, 451–460. [Google Scholar] [CrossRef]
- Danner, C.; Mach, R.L.; Mach-Aigner, A.R. The Phenomenon of Strain Degeneration in Biotechnologically Relevant Fungi. Appl. Microbiol. Biotechnol. 2023, 107, 4745–4758. [Google Scholar] [CrossRef]
- Xie, H.; Ma, Q.; Wei, D.Z.; Wang, F.Q. Transcriptomic Analysis of Aspergillus Niger Strains Reveals the Mechanism Underlying High Citric Acid Productivity. Bioresour. Bioprocess. 2018, 5, 21. [Google Scholar] [CrossRef]
- Hui, X.; Fengqing, W.; Dongzhi, W. Morphological Phenotypes of an Aspergillus Niger Strain with High Citric Acid Production Capacity. Res. J. Biotechnol. 2017, 12, 1–7. [Google Scholar]
- Douma, R.D.; Batista, J.M.; Touw, K.M.; Kiel, J.A.K.W.; Krikken, A.M.; Zhao, Z.; Veiga, T.; Klaassen, P.; Bovenberg, R.A.L.; Daran, J.M.; et al. Degeneration of Penicillin Production in Ethanol-Limited Chemostat Cultivations of Penicillium Chrysogenum: A Systems Biology Approach. BMC Syst. Biol. 2011, 5, 132. [Google Scholar] [CrossRef]
- Christensen, L.H.; Henriksen, C.M.; Nielsen, J.; Villadsen, J.; Egel-Mitani, M. Continuous Cultivation of Penicillium Chrysogenum. Growth on Glucose and Penicillin Production. J. Biotechnol. 1995, 42, 5–107. [Google Scholar] [CrossRef]
- Gupta, V.K.; Steindorff, A.S.; de Paula, R.G.; Silva-Rocha, R.; Mach-Aigner, A.R.; Mach, R.L.; Silva, R.N. The Post-Genomic Era of Trichoderma reesei: What’s Next? Trends Biotechnol. 2016, 34, 970–982. [Google Scholar] [CrossRef] [PubMed]
- Mello-De-Sousa, T.M.; Rassinger, A.; Derntl, C.; Poças-Fonseca, M.J.; Mach, R.L.; Mach-Aigner, A.R. The Relation Between Promoter Chromatin Status, Xyr1 and Cellulase Expression in Trichoderma reesei. Curr. Genom. 2016, 17, 142–152. [Google Scholar] [CrossRef]
- Ries, L.; Belshaw, N.J.; Ilmén, M.; Penttilä, M.E.; Alapuranen, M.; Archer, D.B. The Role of CRE1 in Nucleosome Positioning within the Cbh1 Promoter and Coding Regions of Trichoderma reesei. Appl. Microbiol. Biotechnol. 2014, 98, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Zheng, F.; Zhang, W.; Meng, X.; Liu, W. Trichoderma reesei XYR1 Recruits SWI/SNF to Facilitate Cellulase Gene Expression. Mol. Microbiol. 2019, 112, 1145–1162. [Google Scholar] [CrossRef]
- Zeilinger, S.; Schmoll, M.; Pail, M.; Mach, R.L.; Kubicek, C.P. Nucleosome Transactions on the Hypocrea Jecorina (Trichoderma reesei) Cellulase Promoter Cbh2 Associated with Cellulase Induction. Mol. Genet. Genom. 2003, 270, 46–55. [Google Scholar] [CrossRef]
- Mello-de-Sousa, T.M.; Rassinger, A.; Pucher, M.E.; dos Santos Castro, L.; Persinoti, G.F.; Silva-Rocha, R.; Poças-Fonseca, M.J.; Mach, R.L.; Nascimento Silva, R.; Mach-Aigner, A.R. The Impact of Chromatin Remodelling on Cellulase Expression in Trichoderma reesei. BMC Genom. 2015, 16, 1–11. [Google Scholar] [CrossRef]
- Mello-de-Sousa, T.M.; Gorsche, R.; Rassinger, A.; Poças-Fonseca, M.J.; Mach, R.L.; Mach-Aigner, A.R. A Truncated Form of the Carbon Catabolite Repressor 1 Increases Cellulase Production in Trichoderma reesei. Biotechnol. Biofuels 2014, 7, 129. [Google Scholar] [CrossRef] [PubMed]
- Portnoy, T.; Margeot, A.; Linke, R.; Atanasova, L.; Fekete, E.; Sándor, E.; Hartl, L.; Karaffa, L.; Druzhinina, I.S.; Seiboth, B.; et al. The CRE1 Carbon Catabolite Repressor of the Fungus Trichoderma reesei: A Master Regulator of Carbon Assimilation. BMC Genom. 2011, 12, 269. [Google Scholar] [CrossRef] [PubMed]
- Bird, A. The Essentials of DNA Methylation Minireview. Cell 1992, 70, 5–8. [Google Scholar] [CrossRef]
- Bewick, A.J.; Hofmeister, B.T.; Powers, R.A.; Mondo, S.J.; Grigoriev, I.V.; James, T.Y.; Stajich, J.E.; Schmitz, R.J. Diversity of Cytosine Methylation across the Fungal Tree of Life. Nat. Ecol. Evol. 2019, 3, 479–490. [Google Scholar] [CrossRef]
- Galagan, J.E.; Henn, M.R.; Ma, L.J.; Cuomo, C.A.; Birren, B. Genomics of the Fungal Kingdom: Insights into Eukaryotic Biology. Genome Res. 2005, 15, 1620–1631. [Google Scholar] [CrossRef]
- Bonasio, R.; Tu, S.; Reinberg, D. Molecular Signals of Epigenetic States. Science 2010, 330, 612–616. [Google Scholar] [CrossRef]
- Moller, M.; Habig, M.; Lorrain, C.; Feurtey, A.; Haueisen, J.; Fagundes, W.C.; Alizadeh, A.; Freitag, M.; Stukenbrock, E.H. Recent Loss of the Dim2 DNA Methyltransferase Decreases Mutation Rate in Repeats and Changes Evolutionary Trajectory in a Fungal Pathogen. PLoS Genet. 2021, 17, e1009448. [Google Scholar] [CrossRef] [PubMed]
- Li, W.C.; Chen, C.L.; Wang, T.F. Repeat-Induced Point (RIP) Mutation in the Industrial Workhorse Fungus Trichoderma reesei. Appl. Microbiol. Biotechnol. 2017, 102, 1567–1574. [Google Scholar] [CrossRef]
- Khalil, M.I. In Silico Detection of DNA Methylation in Fungi Neurospora Crassa Genes, Rid-1 and Dim-2. AIP Conf. Proc. 2020, 2213, 020072. [Google Scholar]
- Montenecourt, B.S.; Eveleigh, D.E. Selective Screening Methods for the Isolation of High Yielding Cellulase Mutants of Trichoderma reesei. Adv. Chem. 1979, 181, 289–301. [Google Scholar]
- Mandels, M.; Reese, E.T. Induction of Cellulase in Trichoderma Viride as Influenced by Carbon Sources and Metals. J. Bacteriol. 1957, 73, 269–278. [Google Scholar] [CrossRef]
- Schamhart, S.; Mandels, M. Applications of Cellulases; Plenum Press: New York, NY, USA, 1985; Volume 70. [Google Scholar]
- Derntl, C.; Kiesenhofer, D.P.; Mach, R.L.; Mach-Aigner, A.R. Novel Strategies for Genomic Manipulation of Trichoderma reesei with the Purpose of Strain Engineering. Appl. Environ. Microbiol. 2015, 81, 6314–6323. [Google Scholar] [CrossRef] [PubMed]
- Gruber, E.; Kubicek, C.; de Graaff, L. The Development of a Heterologous Transformation System for the Cellulolytic Fungus Trichoderma reesei Based on a PyrG-Negative Mutant Strain. Curr. Genet. 1990, 18, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Steiger, M.G.; Mach, R.L.; Mach-Aigner, A.R. An Accurate Normalization Strategy for RT-QPCR in Hypocrea Jecorina (Trichoderma reesei). J. Biotechnol. 2010, 145, 30–37. [Google Scholar] [CrossRef]
- Segura-Pacheco, B.; Trejo-Becerril, C.; Perez-Cardenas, E.; Taja-Chayeb, L.; Mariscal, I.; Chavez, A.; Acuñ, C.; Salazar, A.M.; Lizano, M.; Dueñ As-Gonzalez, A. Reactivation of Tumor Suppressor Genes by the Cardiovascular Drugs Hydralazine and Procainamide and Their Potential Use in Cancer Therapy. Clin. Cancer Res. 2003, 9, 1596–1603. [Google Scholar]
- Singh, N.; Dueñas-González, A.; Lyko, F.; Medina-Franco, J.L. Molecular Modeling and Molecular Dynamics Studies of Hydralazine with Human DNA Methyltransferase 1. ChemMedChem 2009, 4, 792–799. [Google Scholar] [CrossRef]
- Deng, C.; Lu, Q.; Zhang, Z.; Rao, T.; Attwood, J.; Yung, R.; Richardson, B. Hydralazine May Induce Autoimmunity by Inhibiting Extracellular Signal-Regulated Kinase Pathway Signaling. Arthritis Rheum. 2003, 48, 746–756. [Google Scholar] [CrossRef]
- Arce, C.; Segura-Pacheco, B.; Perez-Cardenas, E.; Taja-Chayeb, L.; Candelaria, M.; Dueñnas-Gonzalez, A. Hydralazine Target: From Blood Vessels to the Epigenome. J. Transl. Med. 2006, 4, 10. [Google Scholar] [CrossRef]
- Ruiz-Magaña, M.J.; Martínez-Aguilar, R.; Lucendo, E.; Campillo-Davo, D.; Schulze-Osthoff, K.; Ruiz-Ruiz, C. The Antihypertensive Drug Hydralazine Activates the Intrinsic Pathway of Apoptosis and Causes DNA Damage in Leukemic T Cells. Oncotarget 2016, 7, 21875–21886. [Google Scholar] [CrossRef]
- Seiboth, B.; Karimi, R.A.; Phatale, P.A.; Linke, R.; Hartl, L.; Sauer, D.G.; Smith, K.M.; Baker, S.E.; Freitag, M.; Kubicek, C.P. The Putative Protein Methyltransferase LAE1 Controls Cellulase Gene Expression in Trichoderma reesei. Mol. Microbiol. 2012, 84, 1150–1164. [Google Scholar] [CrossRef] [PubMed]
- Fekete, E.; Karaffa, L.; Karimi Aghcheh, R.; Németh, Z.; Fekete, É.; Orosz, A.; Paholcsek, M.; Stágel, A.; Kubicek, C.P. The Transcriptome of Lae1 Mutants of Trichoderma reesei Cultivated at Constant Growth Rates Reveals New Targets of LAE1 Function. BMC Genom. 2014, 15, 447. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Chen, Y.; Zhang, L.; Fang, X.; Wang, W. A Three-Gene Cluster in Trichoderma reesei Reveals a Potential Role of Dmm2 in DNA Repair and Cellulase Production. Biotechnol. Biofuels Bioprod. 2022, 15, 34. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, K.; Zhang, X.; Song, X.; Qin, Y. Different Putative Methyltransferases Have Different Effects on the Expression Patterns of Cellulolytic Genes. J. Fungi 2023, 9, 1118. [Google Scholar] [CrossRef] [PubMed]
- Lewis, Z.A.; Honda, S.; Khlafallah, T.K.; Jeffress, J.K.; Freitag, M.; Mohn, F.; Schübeler, D.; Selker, E.U. Relics of Repeat-Induced Point Mutation Direct Heterochromatin Formation in Neurospora Crassa. Genome Res. 2009, 19, 427–437. [Google Scholar] [CrossRef]
- Ikeda, K.I.; Van Vu, B.; Kadotani, N.; Tanaka, M.; Murata, T.; Shiina, K.; Chuma, I.; Tosa, Y.; Nakayashiki, H. Is the Fungus Magnaporthe Losing DNA Methylation? Genetics 2013, 195, 845–855. [Google Scholar] [CrossRef]
- Lewis, Z.A.; Adhvaryu, K.K.; Honda, S.; Shiver, A.L.; Knip, M.; Sack, R.; Selker, E.U. DNA Methylation and Normal Chromosome Behavior in Neurospora Depend on Five Components of a Histone Methyltransferase Complex, DCDC. PLoS Genet. 2010, 6, e1001196. [Google Scholar] [CrossRef]
- Aramayo, R.; Selker, E.U. Neurospora Crassa, a Model System for Epigenetics Research. Cold Spring Harb. Perspect. Biol. 2013, 5, a017921. [Google Scholar] [CrossRef]



| Strain Name | Origin | Public Availability | Cellulase Expression Capacity | Protein Titer (g/g Biomass) 1 | Sensitivity to Carbon Sources | Reference |
|---|---|---|---|---|---|---|
| QM6a | Wild-type isolate | Yes | Low | 1 | Sensitive to CCR 3 | [32] |
| Rut-C30 | 3 rounds of RM 2 from QM6a | Yes | Moderate | 12 | Partial release of CCR due to cre1 truncation | [31] |
| Iogen-M4 | 1 round of RM from Rut-C30 | No | High | 25 | Strong release of CCR, full length cre1 is missing | [7] |
| Iogen-M10 | 3 rounds of RM from Rut-C30 | No | Very high | 40 | Strong release of CCR, full length cre1 is missing | [8] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Danner, C.; Mello de Sousa, T.M.; Mach, R.L.; Mach-Aigner, A.R. The Impact of DNA Methylation in Trichoderma reesei on Cellulase Production and Strain Degeneration. Microorganisms 2025, 13, 584. https://doi.org/10.3390/microorganisms13030584
Danner C, Mello de Sousa TM, Mach RL, Mach-Aigner AR. The Impact of DNA Methylation in Trichoderma reesei on Cellulase Production and Strain Degeneration. Microorganisms. 2025; 13(3):584. https://doi.org/10.3390/microorganisms13030584
Chicago/Turabian StyleDanner, Caroline, Thiago M. Mello de Sousa, Robert L. Mach, and Astrid R. Mach-Aigner. 2025. "The Impact of DNA Methylation in Trichoderma reesei on Cellulase Production and Strain Degeneration" Microorganisms 13, no. 3: 584. https://doi.org/10.3390/microorganisms13030584
APA StyleDanner, C., Mello de Sousa, T. M., Mach, R. L., & Mach-Aigner, A. R. (2025). The Impact of DNA Methylation in Trichoderma reesei on Cellulase Production and Strain Degeneration. Microorganisms, 13(3), 584. https://doi.org/10.3390/microorganisms13030584






