Relevancy Prediction of the Emerging Pathogens with Porcine Diarrhea by Logistic Regression Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and Multiplex PCR Assay
2.2. Statistical Analysis
3. Results
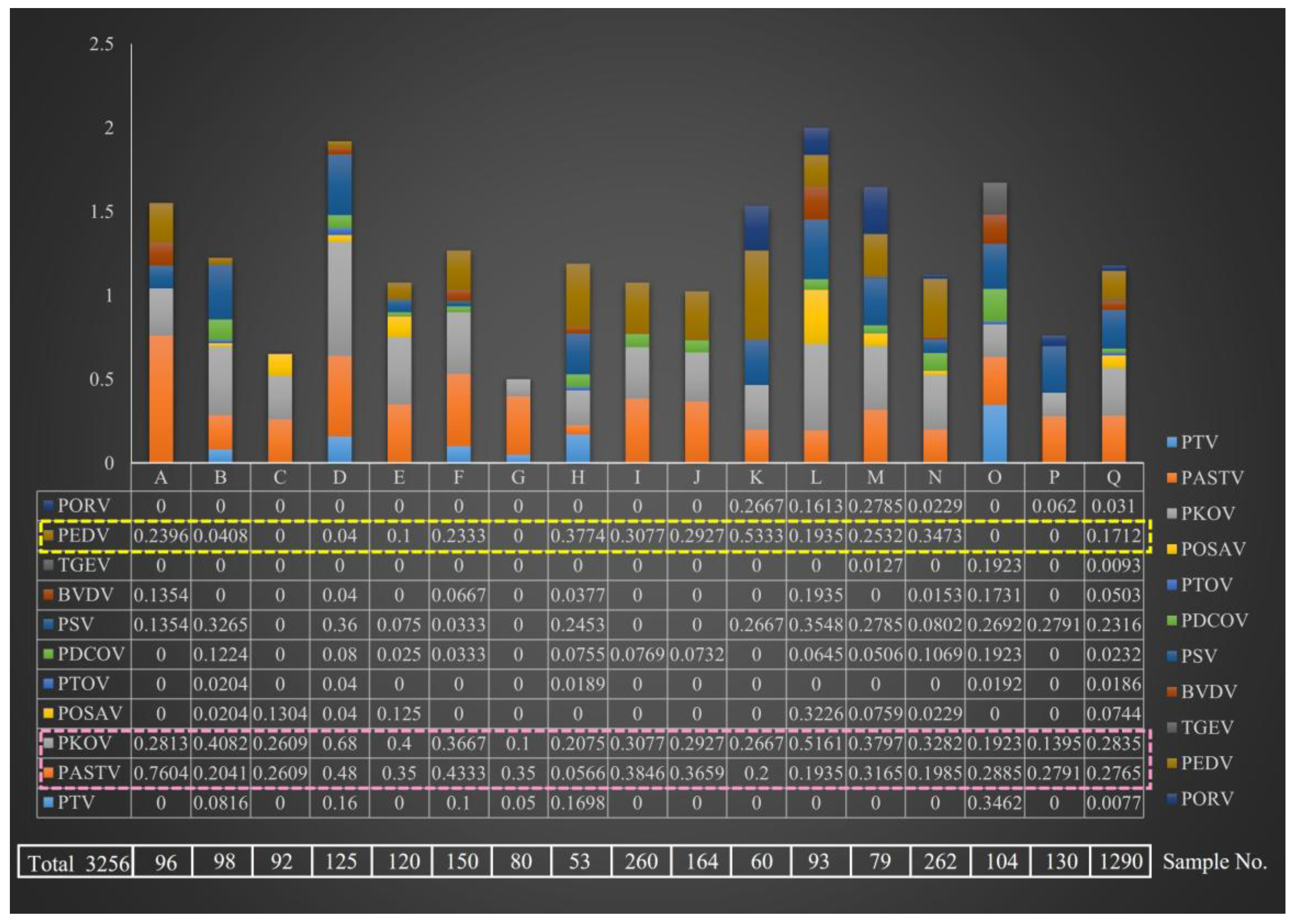
3.1. Comparative Analysis of Diarrheal Pathogen Prevalence in Different Pig Farms
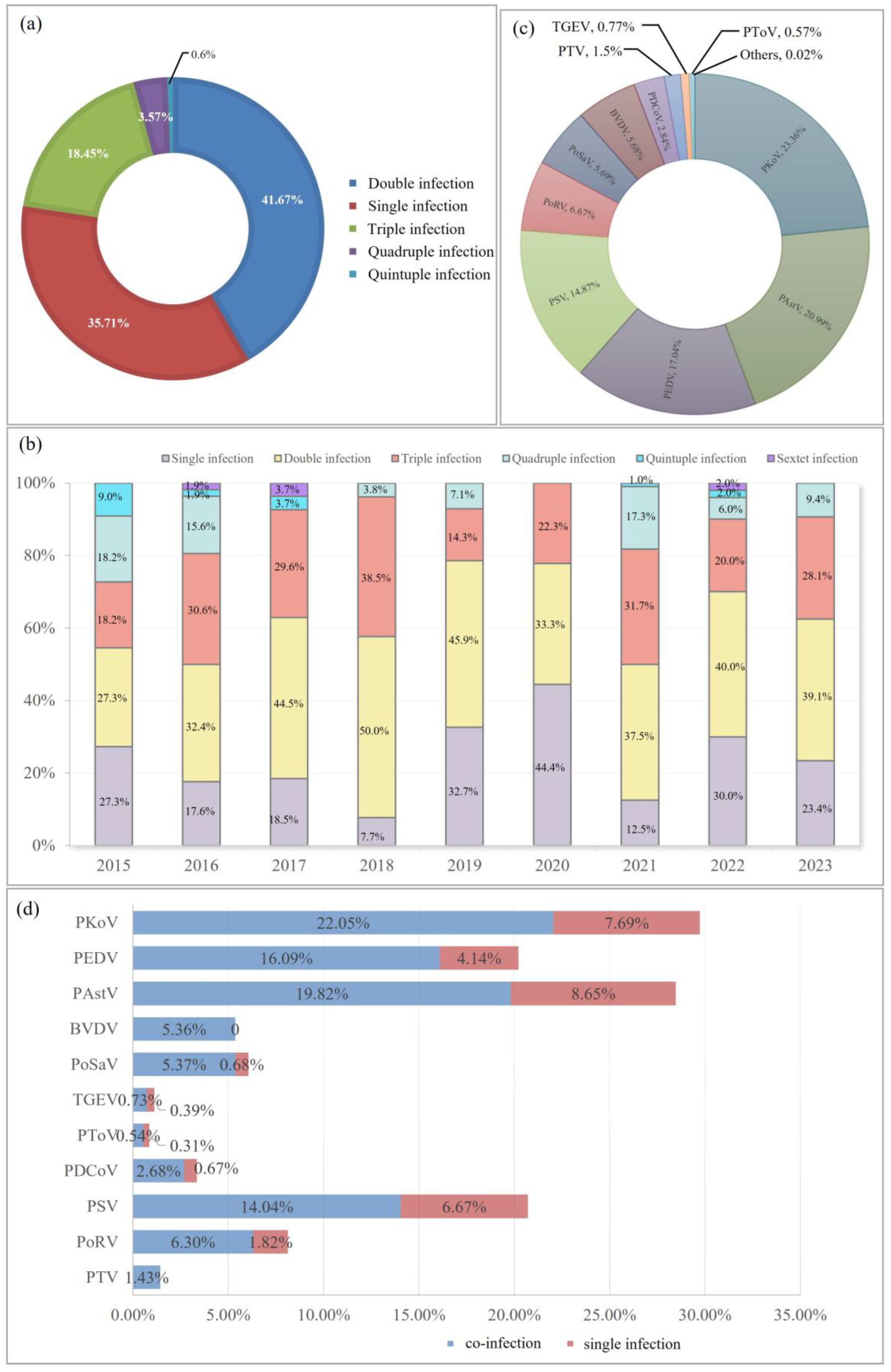
3.2. Overall Epidemic Trend of Porcine Diarrheal Pathogens in Shanghai from 2015 to 2023
3.3. Analysis of the Mixed Infection in Porcine Diarrhea Samples
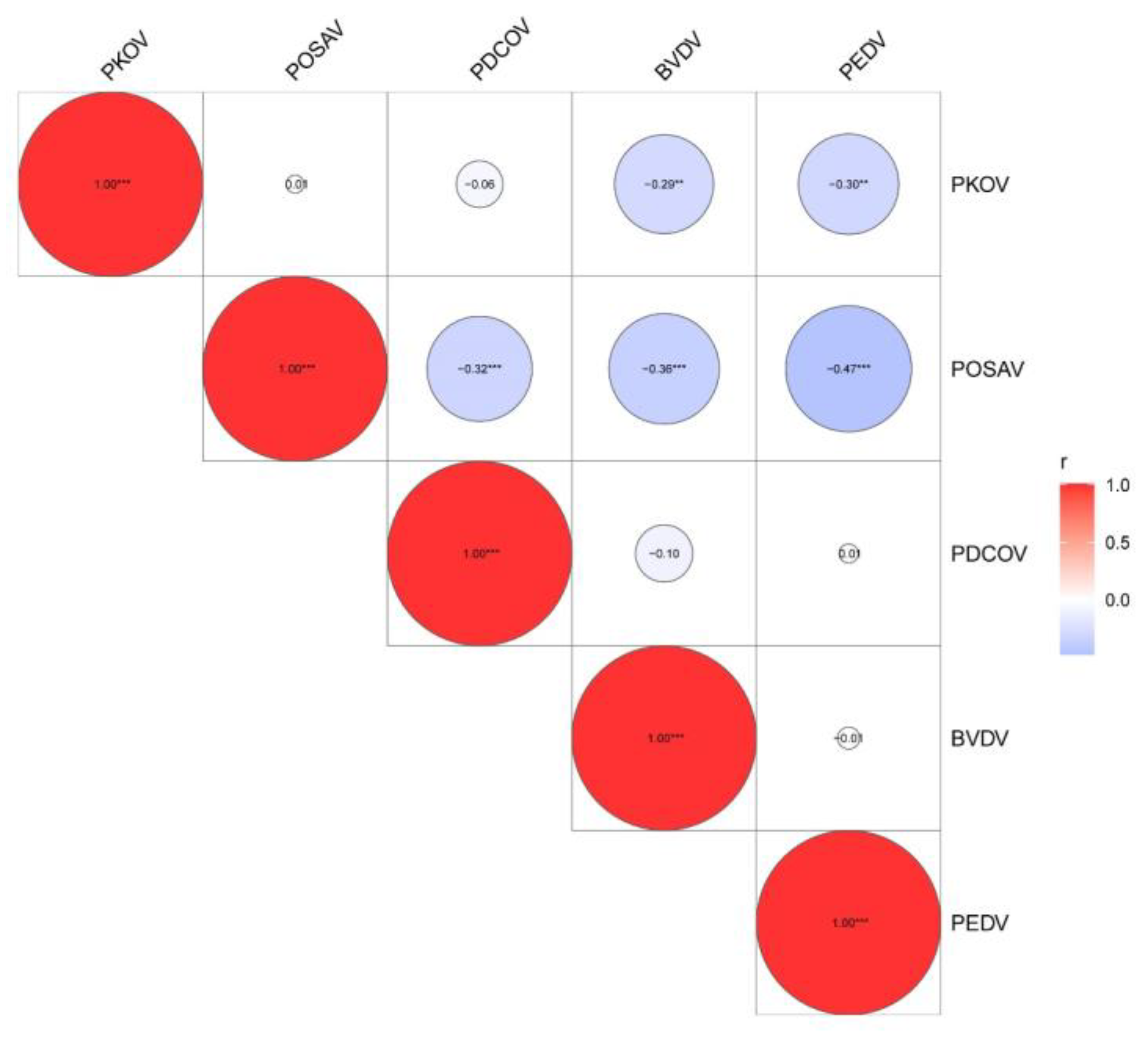
3.4. Identification of Mixed-Infection Models of PEDV and Emerging Diarrhea Pathogens
3.5. Logistic Regression Analysis of the Porcine Diarrhea Pathogens
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, Q.; Hu, R.; Tang, X.; Wu, C.; He, Q.; Zhao, Z.; Chen, H.; Wu, B. Occurrence and investigation of enteric viral infections in pigs with diarrhea in China. Arch. Virol. 2013, 158, 1631–1636. [Google Scholar] [CrossRef]
- Wang, M.; Zheng, H.; Wang, S.W.; Luo, H.Q.; Li, Z.W.; Song, X.Z.; Xu, H.X.; Li, P.D.; Sun, S.Y.; Wang, Y.; et al. Comparative analysis of changes in diarrhea and gut microbiota in Beigang pigs. Microb. Pathog. 2023, 185, 106441. [Google Scholar] [CrossRef]
- Cortey, M.; Díaz, I.; Vidal, A.; Martín-Valls, G.; Franzo, G.; Gómez de Nova, P.J.; Darwich, L.; Puente, H.; Carvajal, A.; Martín, M.; et al. High levels of unreported intraspecific diversity among RNA viruses in faeces of neonatal piglets with diarrhoea. BMC Vet. Res. 2019, 15, 441. [Google Scholar] [CrossRef]
- Li, M.; Pan, Y.; Xi, Y.; Wang, M.; Zeng, Q. Insights and progress on epidemic characteristics, genotyping, and preventive measures of PEDV in China: A review. Microb. Pathog. 2023, 181, 106185. [Google Scholar] [CrossRef]
- Dortmans, J.; Li, W.; van der Wolf, P.J.; Buter, G.J.; Franssen, P.J.M.; van Schaik, G.; Houben, M.; Bosch, B.J. Porcine epidemic diarrhea virus (PEDV) introduction into a naive Dutch pig population in 2014. Vet. Microbiol. 2018, 221, 13–18. [Google Scholar] [CrossRef]
- Zhang, B.; Tang, C.; Yue, H.; Ren, Y.; Song, Z. Viral metagenomics analysis demonstrates the diversity of viral flora in piglet diarrhoeic faeces in China. J. Gen. Virol. 2014, 95, 1603–1611. [Google Scholar] [CrossRef]
- Qian, L.; Tang, C.; Yue, H.; Ren, Y.; Song, Z. Metagenomic survey of viral diversity obtained from feces of piglets with diarrhea. Heliyon 2024, 10, e25616. [Google Scholar] [CrossRef]
- Tao, J.; Li, B.Q.; Cheng, J.H.; Shi, Y.; Liu, P.H.; He, G.X.; Xu, W.J.; Liu, H.L. Viral metagenome analysis of the viral community composition of the porcine diarrhea feaces. Biodivers. Sci. 2023, 31, 23170. [Google Scholar] [CrossRef]
- Bouyer, J. Logistics regression in epidemiology. II. Rev. Epidemiol. Sante Publique 1991, 39, 183–196. [Google Scholar]
- Rattenborg, E.; Chriel, M.; Dietz, H.H. Influence of farm, feed-producer and season on incidence of gastrointestinal disorders in Danish farm mink. Prev. Vet. Med. 1999, 38, 231–237. [Google Scholar] [CrossRef]
- Birkegard, A.C.; Halasa, T.; Toft, N. Sampling pig farms at the abattoir in a cross-sectional study—Evaluation of a sampling method. Prev. Vet. Med. 2017, 145, 83–90. [Google Scholar] [CrossRef]
- Herve-Claude, L.P.; Lwanga-Iga, I.; Kroll-Lwanga-Iga, S.; Nyangiwe, N.; Ruddat, I.; Kreienbrock, L. Village livestock population and sampling strategies in communal areas in the Eastern Cape Province, South Africa. Trop. Anim. Health Prod. 2011, 43, 573–580. [Google Scholar] [CrossRef]
- Shi, Y.; Li, B.Q.; Tao, J.; Cheng, J.H. The Complex Co-infections of Multiple Porcine Diarrhea Viruses in Local Area Based on the Luminex xTAG Multiplex Detection Method. Front. Vet. Sci. 2021, 8, 602866. [Google Scholar] [CrossRef]
- Homwong, N.; Diaz, A.; Rossow, S.; Ciarlet, M.; Marthaler, D. Three-Level Mixed-Effects Logistic Regression Analysis Reveals Complex Epidemiology of Swine Rotaviruses in Diagnostic Samples from North America. PLoS ONE 2016, 11, e0154734. [Google Scholar] [CrossRef]
- Zhang, F.; Luo, S.; Gu, J.; Li, Z.; Li, K.; Yuan, W.; Ye, Y.; Li, H.; Ding, Z.; Song, D.; et al. Prevalence and phylogenetic analysis of porcine diarrhea associated viruses in southern China from 2012 to 2018. BMC Vet. Res. 2019, 15, 470. [Google Scholar] [CrossRef]
- Su, M.; Qi, S.; Yang, D.; Guo, D.; Yin, B.; Sun, D. Coinfection and Genetic Characterization of Porcine Astrovirus in Diarrheic Piglets in China From 2015 to 2018. Front. Vet. Sci. 2020, 7, 462. [Google Scholar] [CrossRef]
- Li, Y.; Liang, J.; Wu, S.; Yan, Z.; Zhang, W. Complete genomic sequence analysis and intestinal tissue localization of a porcine Kobuvirus variant in China. Infect. Genet. Evol. 2022, 104, 105362. [Google Scholar] [CrossRef]
- Staubli, T.; Rickli, C.I.; Torgerson, P.R.; Fraefel, C.; Lechmann, J. Porcine teschovirus, sapelovirus, and enterovirus in Swiss pigs: Multiplex RT-PCR investigation of viral frequencies and disease association. J. Vet. Diagn. Investig. 2021, 33, 864–874. [Google Scholar] [CrossRef]
- Kumthip, K.; Khamrin, P.; Saikruang, W.; Kongkaew, A.; Vachirachewin, R.; Ushijima, H.; Maneekarn, N. Detection and genetic characterization of porcine astroviruses in piglets with and without diarrhea in Thailand. Arch. Virol. 2018, 163, 1823–1829. [Google Scholar] [CrossRef]
- Nantel-Fortier, N.; Gauthier, M.; L’Homme, Y.; Lachapelle, V.; Fravalo, P.; Brassard, J. The swine enteric virome in a com mercial production system and its association with neonatal diarrhea. Vet. Microbiol. 2022, 266, 109366. [Google Scholar] [CrossRef]
- Qiu, M.; Li, S.; Xiao, Y.; Li, J.; Zhang, Y.; Li, X.; Feng, B.; Li, C.; Lin, H.; Zhu, J.; et al. Pathogenic and metagenomic evaluations reveal the correlations of porcine epidemic diarrhea virus, porcine kobuvirus and porcine astroviruses with neonatal piglet diarrhea. Microb. Pathog. 2022, 170, 105703. [Google Scholar] [CrossRef]
- Chen, J.; Suo, X.; Cao, L.; Yuan, C.; Shi, L.; Duan, Y.; Zheng, H.; Wang, Q. Virome Analysis for Identification of a Novel Porcine Sapelovirus Isolated in Western China. Microbiol. Spectr. 2022, 10, e0180122. [Google Scholar] [CrossRef]
- Wu, S.; Gou, F.; Meng, J.; Jin, X.; Liu, W.; Ding, W.; Xu, W.; Gu, C.; Hu, X.; Cheng, G.; et al. Porcine kobuvirus enhances porcine epidemic diarrhea virus pathogenicity and alters the number of intestinal lymphocytes in piglets. Vet. Microbiol. 2024, 293, 110100. [Google Scholar] [CrossRef]
- An, D.J.; Jeoung, H.Y.; Jeong, W.; Lee, H.S.; Park, J.Y.; Kim, B. Porcine kobuvirus from pig stool in Korea. Virus Genes 2011, 42, 208–211. [Google Scholar] [CrossRef]
- Park, J.S.; Jeong, C.G.; Chae, S.B.; Yang, M.S.; Oh, B.; Lee, S.Y.; Oem, J.K. Porcine Astrovirus Infection in Brains of Pigs in Korea. Viruses 2024, 16, 1372. [Google Scholar] [CrossRef]
- Chuchaona, W.; Khamrin, P.; Yodmeeklin, A.; Kongkaew, A.; Vachirachewin, R.; Kumthip, K.; Ushijima, H.; Maneekarn, N. Detection and molecular characterization of porcine kobuvirus in piglets in 2009–2013 in northern Thailand. Trop. Anim. Health Prod. 2017, 49, 1077–1080. [Google Scholar] [CrossRef]
- Capai, L.; Piorkowski, G.; Maestrini, O.; Casabianca, F.; Masse, S.; de Lamballerie, X.; Charrel, R.N.; Falchi, A. Detection of porcine enteric viruses (Kobuvirus, Mamastrovirus and Sapelovirus) in domestic pigs in Corsica, France. PLoS ONE 2022, 17, e0260161. [Google Scholar] [CrossRef]
- Okitsu, S.; Khamrin, P.; Thongprachum, A.; Hidaka, S.; Kongkaew, S.; Kongkaew, A.; Maneekarn, N.; Mizuguchi, M.; Hayakawa, S.; Ushijima, H. Sequence analysis of porcine kobuvirus VP1 region detected in pigs in Japan and Thailand. Virus Genes 2012, 44, 253–257. [Google Scholar] [CrossRef]
- Verma, H.; Mor, S.K.; Abdel-Glil, M.Y.; Goyal, S.M. Identification and molecular characterization of porcine kobuvirus in U. S. swine. Virus Genes 2013, 46, 551–553. [Google Scholar] [CrossRef]
- Matias Ferreyra, F.; Harmon, K.; Bradner, L.; Burrough, E.; Derscheid, R.; Magstadt, D.R.; Michael, A.; de Almeida, M.N.; Schumacher, L.; Siepker, C.; et al. Comparative Analysis of Novel Strains of Porcine Astrovirus Type 3 in the USA. Viruses 2021, 13, 1859. [Google Scholar] [CrossRef]
- Mor, S.K.; Chander, Y.; Marthaler, D.; Patnayak, D.P.; Goyal, S.M. Detection and molecular characterization of Porcine astrovirus strains associated with swine diarrhea. J. Vet. Diagn. Investig. 2012, 24, 1064–1067. [Google Scholar] [CrossRef] [PubMed]
- Chelli, E.; De Sabato, L.; Vaccari, G.; Ostanello, F.; Di Bartolo, I. Detection and Characterization of Porcine Sapelovirus in Italian Pig Farms. Animals 2020, 10, 966. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Saikumar, G.; Desingu, P.A.; Das, T.; Singh, R. Immunohistochemical detection of naturally occurring porcine Sapelovirus infection in Indian pigs. J. Immunoass. Immunochem. 2019, 40, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Vaishali; Gupta, R.; Kumar, M.; Bansal, N.; Vivek; Kumar, P.; Kumar, P.; Jindal, N. Coinfection of porcine astrovirus and other porcine viruses in diarrheic pigs in Haryana, India. Arch. Virol. 2023, 168, 246. [Google Scholar] [CrossRef]
- Brnic, D.; Prpić, J.; Keros, T.; Roić, B.; Starešina, V.; Jemeršić, L. Porcine astrovirus viremia and high genetic variability in pigs on large holdings in Croatia. Infect. Genet. Evol. 2013, 14, 258–264. [Google Scholar] [CrossRef]


| Names | Gene | Primer Sequences | Length (bp) |
|---|---|---|---|
| PEDV-F | M | GATACTTTGGCCTCTTGTGT | 332 |
| PEDV-R | ATTGACTTACCTGTACGC | ||
| TGEV-F | N | ACCAGATAGAAGTCACGTT | 226 |
| TGEV-R | TCAACCTGTGTGTCATCAAA | ||
| PoRV-F | VP6 | ATTTATATTYCATGCTAC | 167 |
| PoRV-R | CTGTCCAATTCATYCCT | ||
| BVDV-F | Npro | TTACGACATCAACGGAT | 372 |
| BVDV-R | TGGTCCCTAGTCGCTCT | ||
| PTV-F | 3D | ATGGGACTCTAGATCTCGT | 176 |
| PTV-R | ACGCCTCTGTAGTTCTCTCTT | ||
| PAstV-F | ORF2 | GGATTTACAGTTGGCCCAGAT | 249 |
| PAstV-R | CCTGTCCATCTGCCTTTCTGT | ||
| PKV-F | 3D | ATGCTGCTTGGTGGACTCATT | 280 |
| PKV-R | ACGTCTCGTTGCCAAAGACAT | ||
| PoSaV-F | ORF1 | CGAGGCTAAAGGGAAAAACAAACGT | 340 |
| PoSaV-R | ACTGTCGTAGGTGTCTGTTTT | ||
| PToV-F | S | CTTTTACACCTTGCCATCC | 421 |
| PToV-R | TGCTTCACCTCTACACTGTT | ||
| PDCoV-F | M | ACTTATTCTGCTTTGGCTGCT | 504 |
| PDCoV-R | GAAGTGGTTATGGTGTGAAGTC | ||
| PSV-F | 5′UTR | GATGTGGCGCATGCTCTT | 624 |
| PSV-R | TGCTGCCTCCTGTGTTGTTAT |
| Score | df | Sig. | ||
|---|---|---|---|---|
| Step 0 Variables | PTV | 2.972 | 1 | 0.085 |
| PAstV | 25.901 | 1 | 0.000 | |
| PKoV | 48.619 | 1 | 0.000 | |
| PoSaV | 14.489 | 1 | 0.000 | |
| PToV | 2.012 | 1 | 0.156 | |
| PDCoV | 6.892 | 1 | 0.009 | |
| PSV | 9.607 | 1 | 0.002 | |
| BVDV | 8.485 | 1 | 0.004 | |
| TGEV | 2.491 | 1 | 0.115 | |
| PEDV | 45.179 | 1 | 0.000 | |
| PRoV | 2.132 | 1 | 0.144 | |
| Overall Statistics | 125.131 | 11 | 0.000 |
| B | S.E. | Wald | df | Sig. | Exp(B) | ||
|---|---|---|---|---|---|---|---|
| Step 1(a) | PAstV | 0.734 | 0.231 | 10.129 | 1 | 0.001 | 2.083 |
| PKoV | 1.554 | 0.286 | 29.625 | 1 | 0.000 | 4.730 | |
| PoSaV | 18.787 | 3503.178 | 0.000 | 1 | 0.996 | 1.443 × 108 | |
| PDCoV | 18.499 | 4793.354 | 0.000 | 1 | 0.997 | 1.082 × 108 | |
| BVDV | 1.747 | 0.724 | 5.820 | 1 | 0.016 | 5.739 | |
| PEDV | 3.709 | 1.006 | 13.603 | 1 | 0.000 | 40.806 | |
| Constant | −46.439 | 5937.043 | 0.000 | 1 | 0.994 | 0.000 |
| ID | PKoV | PoSaV | PDCoV | BVDV | PEDV |
|---|---|---|---|---|---|
| PKoV | 0 | 0.92691 | 0.52696 | 0.00376 | 0.00276 |
| PoSaV | 0.92691 | 0 | 0.00097 | 0.00025 | 1.04 × 10−6 |
| PDCoV | 0.52696 | 0.00097 | 0 | 0.34198 | 0.91645 |
| BVDV | 0.00376 | 0.00025 | 0.34198 | 0 | 0.88815 |
| PEDV | 0.00276 | 1.04 × 10−6 | 0.91645 | 0.88815 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, B.; Tao, J.; Li, X.; Cheng, J.; Shi, Y.; Tang, P.; Liu, H. Relevancy Prediction of the Emerging Pathogens with Porcine Diarrhea by Logistic Regression Model. Microorganisms 2025, 13, 528. https://doi.org/10.3390/microorganisms13030528
Li B, Tao J, Li X, Cheng J, Shi Y, Tang P, Liu H. Relevancy Prediction of the Emerging Pathogens with Porcine Diarrhea by Logistic Regression Model. Microorganisms. 2025; 13(3):528. https://doi.org/10.3390/microorganisms13030528
Chicago/Turabian StyleLi, Benqiang, Jie Tao, Xin Li, Jinghua Cheng, Ying Shi, Pan Tang, and Huili Liu. 2025. "Relevancy Prediction of the Emerging Pathogens with Porcine Diarrhea by Logistic Regression Model" Microorganisms 13, no. 3: 528. https://doi.org/10.3390/microorganisms13030528
APA StyleLi, B., Tao, J., Li, X., Cheng, J., Shi, Y., Tang, P., & Liu, H. (2025). Relevancy Prediction of the Emerging Pathogens with Porcine Diarrhea by Logistic Regression Model. Microorganisms, 13(3), 528. https://doi.org/10.3390/microorganisms13030528




