Abstract
The use of antibiotics has led to the emergence of antibiotic resistance, posing significant challenges in the prevention, control, and treatment of microbial diseases, while threatening public health, the environment, and food safety. In this study, the antibiotic resistance phenotypes and genotypes of 56 endophytic bacteria isolates from three species of wild edible fungi in Yunnan were analyzed using the Kirby-Bauer disk diffusion method and PCR amplification. The results revealed that all isolates were sensitive to ofloxacin, but resistance was observed against 17 other antibiotics. Specifically, 55, 53, and 51 isolates exhibited resistance to amoxicillin, penicillin, and vancomycin, respectively. Antibiotic resistance gene (ARG) detection indicated that the sulfonamide sul1 gene had the highest detection rate (53.57%). Excluding the ARG that was not detected, the lowest detection rates were the sulfonamide sul2 and sul3 genes, both at 1.79%. Among six tetracycline resistance genes, only tetK and tetM were detected. For β-lactam antibiotics, blaTEM, blaVIM, and blaSHV genes were present, while blaOXA was absent. In aminoglycoside resistance genes, aadB was not detected, while detection rates for aac(3′)-IIa, acrB, and aadA1 were 3.57%, 1.79%, and 37.5%, respectively. The chloramphenicol Cat gene was detected at a rate of 14.29%, whereas floR was absent. For polypeptide resistance, VanC was detected at 3.57%, with EmgrB not detected. All three quinolone genes were detected, with detection rates of 8.92% for GyrA, 39.29% for GyrB, and 37.5% for ParC. Through phylogenetic analysis, 12 isolates that are closely related to ten common foodborne pathogenic bacteria were further selected for whole-genome sequencing and assembly. Gene annotations revealed that each isolate contained more than 15 ARGs and over 30 virulence factors. Notably, the detection rate of antibiotic resistance phenotypes was higher than that of genotypes, highlighting the importance of studying phenotypic antibiotic resistance that lacks identifiable ARGs. This study enriches the research on endophytes in wild edible fungi and provides new data for microbial ecology and antibiotic resistance research. It also offers critical insights for monitoring microbial antibiotic resistance in wild edible fungi and potentially other food sources, contributing to more effective strategies for ecological protection, sustainable agricultural development, and public health security.
1. Introduction
Plant endophytes are a group of microorganisms that live in plant tissues, interstitial tissues, and organs at a certain stage or throughout their life cycle. Studies on the diversity of plant endophytes have shown that these species mainly include endophytic fungi, endophytic bacteria, endophytic actinomycetes, and so on [1]. The main members of the plant microbial community are endophytes, which inhabit plant tissues. It has been found that plant endophytes are an integral part of plant microcosm, and most of them assist in plant growth through various pathways [2]. Many studies have revealed beneficial relationships between endophytes and their hosts, promoting plant growth through mechanisms such as nitrogen fixation, phosphorus solubilization, secretion of iron carriers, production of volatile organic substances, enhancement of nutrient uptake, and suppression of pathogenic fungi [3]. Simultaneously, the host plant provides a habitat for endophytes. As the application of toxic chemical pesticides and the production and long-term use of chemical fertilizers in agriculture has led to significant environmental pollution and plant pathogen resistance, research has found that most endophytes can colonize in the internal tissues of plants, and can reduce pests and diseases and promote plant growth, thus circumventing the negative impacts of chemical pesticides and providing an effective and environmentally friendly alternative [4]. In recent years, with the gradual expansion of various cross-disciplinary research areas and the deepening of research methods, advances in molecular techniques have simplified the isolation and characterization of some non-culturable species, and the understanding of endophyte diversity, function, metabolism, and genetics has continued to improve [5,6].
The era of antibiotics arrived with the discovery of penicillin, and an increasing number of antibiotics have been found and used in the treatment of clinical diseases, thus greatly promoting the development and progress of human health and society. The discovery of antibiotics is a great advancement in the history of world medicine and the beginning of another difficult global problem. Research and development of new antibiotics slowed down in the late 1970s, and no new antibiotics with novel structures appeared after 1984 [7]. The alarming rise of bacterial antibiotic resistance has resulted from the widespread and improper use, or even abuse, of antibiotics, with antibiotic residues detected in water, livestock, and food [8].
Bacterial resistance refers to the ability of bacteria to withstand antimicrobial agents [9]. The emergence of bacterial antibiotic resistance is an ancient and natural phenomenon. After penicillin [7], scientists identified resistance to other antibiotics in certain bacteria, such as Streptococcus [10] and Mycobacterium tuberculosis [11]. Bacteria continually exhibit resistance to newly discovered and applied antibiotics, leading to increasing rates of antibiotic resistance [12,13]. The existence of multiple antibiotic-resistant bacteria and super bacteria, such as methicillin-resistant Staphylococcus aureus [14], is an important factor for the increase in patient mortality. The relationship between humans, animals, and the environment facilitates the widespread dissemination of resistant bacteria and resistance genes (ARGs). In particular, food can effectively transfer and spread antibiotic resistance genes to human intestinal microbiota through consumption, allowing additional microorganisms to acquire antibiotic resistance. Consequently, food is an important carrier of ARGs for transmission [15,16]. The types of antimicrobial drugs used by food animals are almost the same as those used in human medicine [17]. In some countries, the amount of antibiotics used for food animals and pets surpasses that used for human medical treatment [18]. This situation broadens the path for ARG transmission. While foodborne pathogens are the primary focus of antibiotic resistance monitoring and research, nonpathogenic bacteria are abundant, widely distributed, and form the backbone of the microbial ecosystem. Given that ARGs in pathogenic bacteria are likely to be transmitted by nonpathogenic bacteria as their original carriers [19], understanding and monitoring the antibiotic resistance of both pathogenic and nonpathogenic bacteria is essential.
The province of Yunnan is abundant in wild edible fungi, which thrive in rich environments that also support a diverse range of endophytic bacteria. As an integral part of the ecosystem, the endophytic communities associated with these wild edible fungi play a crucial role in maintaining ecological balance and biodiversity. However, there are few reports on endophytic bacteria pathogenicity and their antibiotic resistance in wild edible fungi [20]. If these endophytes exhibit antibiotic resistance, they could pose a potential threat to human health. By investigating their resistance mechanisms, we can assess and monitor the safety of consuming wild edible fungi, thereby reducing the risk of foodborne diseases. Therefore, studying the antibiotic resistance of endophytic bacteria in wild edible fungi is necessary to provide a reference and basis for clinical antibiotic use. At present, the most commonly used method to study bacterial resistance is to investigate the molecular mechanism of ARGs after bacterial isolation through traditional culture methods. In this study, endophytic bacteria isolated from 3 kinds of wild edible fungi were tested for antibiotic resistance, and their phenotypes were observed and analyzed through an antibiotic sensitivity test. At the same time, the results of PCR were combined with the results of ARG annotation to analyze antibiotic resistance genotypes.
2. Materials and Methods
2.1. Materials
A total of 56 isolates were isolated from 8 groups of 3 wild edible fungi from Yunnan, namely Lactarius deliciosus (5 samples), Russula virescens (2 samples), and Cantharellus cibarius (1 sample). Numbered H1, H2, H3, H4, H5, Q1, Q2, and J, these samples were procured from different wild fungus stores in Yunnan Province.
The following antibiotics were used: penicillin 10 µg, amoxicillin 10 µg, ampicillin 10 µg, ticarcillin 30 µg, ceftazidime 30 µg, cefotaxime 10 µg, gentamicin 10 µg, streptomycin 10 µg, neomycin 30 µg, tetracycline 30 µg, minocycline 30 µg, doxycycline 30 µg, vancomycin 30 µg, compound sulfamethoxazole 25 µg, chloramphenicol 30 µg, ofloxacin 5 µg, ciprofloxacin 5 µg, and imipenem 30 µg. All antibiotic disks were purchased from Oxoid, UK.
The following instruments and equipment were utilized: electronic balance (JD200-3, Shenyang Longing Electronics Co., Ltd., Shenyang, China); high-speed desktop centrifuge (H318K, Hunan Kuchen Equipment Co., Ltd., Changsha, China); AB Simplify PCR instrument (Applied Biosystems Co., Ltd., Mumbai, India); Namedrop 2000 ultra-micro UV spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA); Gel DOC ZR gel imaging system (Bio-Rad Laboratories, Thane, India).
2.2. Isolation of Endophytic Bacteria from Wild Edible Fungi
The soil was rinsed off the surface of the samples with tap water, and roots that had come into contact with the soil were removed with a sterile scalpel. Sterilization of samples was carried out on an ultra-clean bench as follows: the sample surface was first sterilized with 75% ethanol and rinsed with sterile water three times; the filter paper was dried and then soaked in 75% ethanol for 2 min, rinsed with sterile water three to five times, and then washed with 2.5% sodium hypoammonate for 3 min; the samples were rinsed with sterile water three to five times to ensure that any microbial residues on the surface were thoroughly washed away [21].
A 10 g sample was weighed from each of the sterilized samples. Two methods [22] were employed for the isolation of endophytes: (1) tissue isolation method: the epidermis of the samples was removed with a sterile scalpel, and the internal tissues were extracted; this tissue was cut into 0.5 cm square pieces and placed on LB agar plate medium (Hope Bio-Technology Co., Ltd., Qingdao, China) at three equidistant points from the center using sterile tubes; (2) grinding isolation method: place the 0.5 cm square piece block into a sterilized mortar, add quartz sand and sterile water, grind thoroughly, and let it stand for 3 min, then use a pipette to extract 100 μL of supernatant, and spread onto LB agar plate medium, finally, use the las cleaned samples of sterile water as a control, with three parallels for both the control and experimental groups.
Each plate was incubated at a constant temperature of 37 °C. After bacterial growth, compared with the control group, according to the morphology of the colony, a small amount of bacteria from the edge of individual colonies was taken using a sterile inoculation loop for isolation and purification. Finally, isolated bacteria were preserved in 30% glycerin (v/v = 1:1) at −80 °C. The sample numbers were as follows: Huanggu ripe H11, H12, H13… H21, H22, H23…; Qingtou bacteria Q11, Q12, Q13… Q21, Q22, Q23…; and chicken oil fungus J1, J2…
2.3. Identification of Isolates Using 16S rRNA Sequencing
A bacterial genome extraction kit (TaKaRa MiniBEST Bacteria Genomic DNA Extraction Kit Ver.3.0, Dalian, China) was utilized to extract the DNA of isolated bacterial strains. The concentration and purity of the bacterial DNA were assessed using NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA)and electrophoresis. The samples with high-quality DNA were subjected to PCR amplification with the universal 16S rRNA bacterial primer (Table 1). PCR was performed by using an ABI SimpliAmp thermal cycler (Applied Biosystems) with a 25 µL reaction system, containing 2.5 µL of 10× PCR buffer (Mg2+ plus), 2 µL of dNTP mixture (2.5 mM), 1 μL of each primer (10 µM), 0.2 μL of rTaq DNA polymerase (5 U/µL) (TaKaRa, Biotechnology, Dalian, China), 2 µL of the DNA template, and 16.3 µL of ultrapure water. The 16S rRNA PCR amplification was carried out with the following program: initial denaturation at 94 °C for 2 min; 30 cycles of denaturation at 94 °C for 30 s, primer annealing at 54 °C for 30 s, and primer extension at 72 °C for 1 min; and followed by a final extension at 72 °C for 10 min.

Table 1.
16S rRNA PCR universal primer amplification sequence.
The amplified products were analyzed on 2% agarose gel electrophoresis with ethidium bromide. The electrophoresis buffer was 1× TAE with the DL2000 marker (TsingKe, Beijing, China) used as the molecular weight standard. PCR products were sent to Shanghai Sangon Co., Ltd., Shanghai, China, for sequencing. Finally, the 16S rRNA sequences of the sequenced samples were uploaded to the NCBI database and compared using the BLAST tool. Moreover, isolates with a homology rate exceeding 99% were considered to belong to the same species.
2.4. Phylogenetic Relationships Between Isolates and Common Foodborne Pathogens
A phylogenetic tree was established by comparing the 16S rRNA sequences of bacteria isolated from wild edible fungi with those of 10 common foodborne pathogens. MEGA7 software (Ver.7.0) was used to build the phylogenetic tree, and isolates with close phylogenetic relationships to foodborne pathogens were selected. The selected bacterial isolates were activated in LB liquid medium, centrifuged at 12,000 r/min for 10 min, and sent to Genesky Biotechnologies Inc. (Shanghai, China) for whole genome sequencing, gene assembly, and bioinformation analysis, including ARGs and virulence factors.
The sequences of foodborne pathogens were obtained and downloaded from GenBank database as follows: Bacillus cereus (NCBI login No.: ABDM0200053.1), Escherichia coli O157: H7 (NZ_JHNG1000174.1), Listeria monocytogenes (AARY02000494.1), Salmonella (KF509913.1), Shigella (HM565981.1), S. aureus (JJEZ010000047.1), Vibrio parahaemolyticus (NZ_AOMN01000024.1), Campylobacter jejuni (NZ_KK365725.1), Cronobacter sakazakii (NZ_AJKT01000053.1), and Vibrio vulnificus (NZ_LBNN01000067.1).
2.5. Antibiotic Sensitivity Tests of 56 Isolates
The antibiotic resistance phenotypes of the isolates were tested using the Kirby–Bauer disk diffusion method of the Clinical and Laboratory Standards Institute (CLSI) [24]. The types and concentrations of antibiotic-sensitive disks used were mentioned in Section 2.1. The mediums with the antibiotic-sensitive disks were inverted and incubated at 37 °C for 18 h within 15 min. The diameter of the bacteriostatic circle was measured and evaluated according to the Enterobacteriaceae standard established by CLSI for each antibiotic susceptibility test. This standard categorizes results into sensitive, intermediate, and resistant [25].
2.6. ARGs Detections of 56 Isolates
We searched for suitable ARGs based on the information provided by the Antibiotic Resistance Gene Database (ARDB). Some primers for ARGs were designed using Vector NTI software (v11.5.2), while others were obtained from references (Table S1) [26,27,28,29,30,31,32,33]. All primers were synthesized by Shanghai SANGON Biotechnology Co., Ltd., Shanghai, China. The PCR reaction system was the same as used in Section 2.3. The PCR products were detected via 2% agarose gel electrophoresis and sent to Shanghai SANGON Biotech Co., Ltd. for sequencing.
3. Results
3.1. DNA Extraction and 16S rRNA Amplification of 56 Isolates
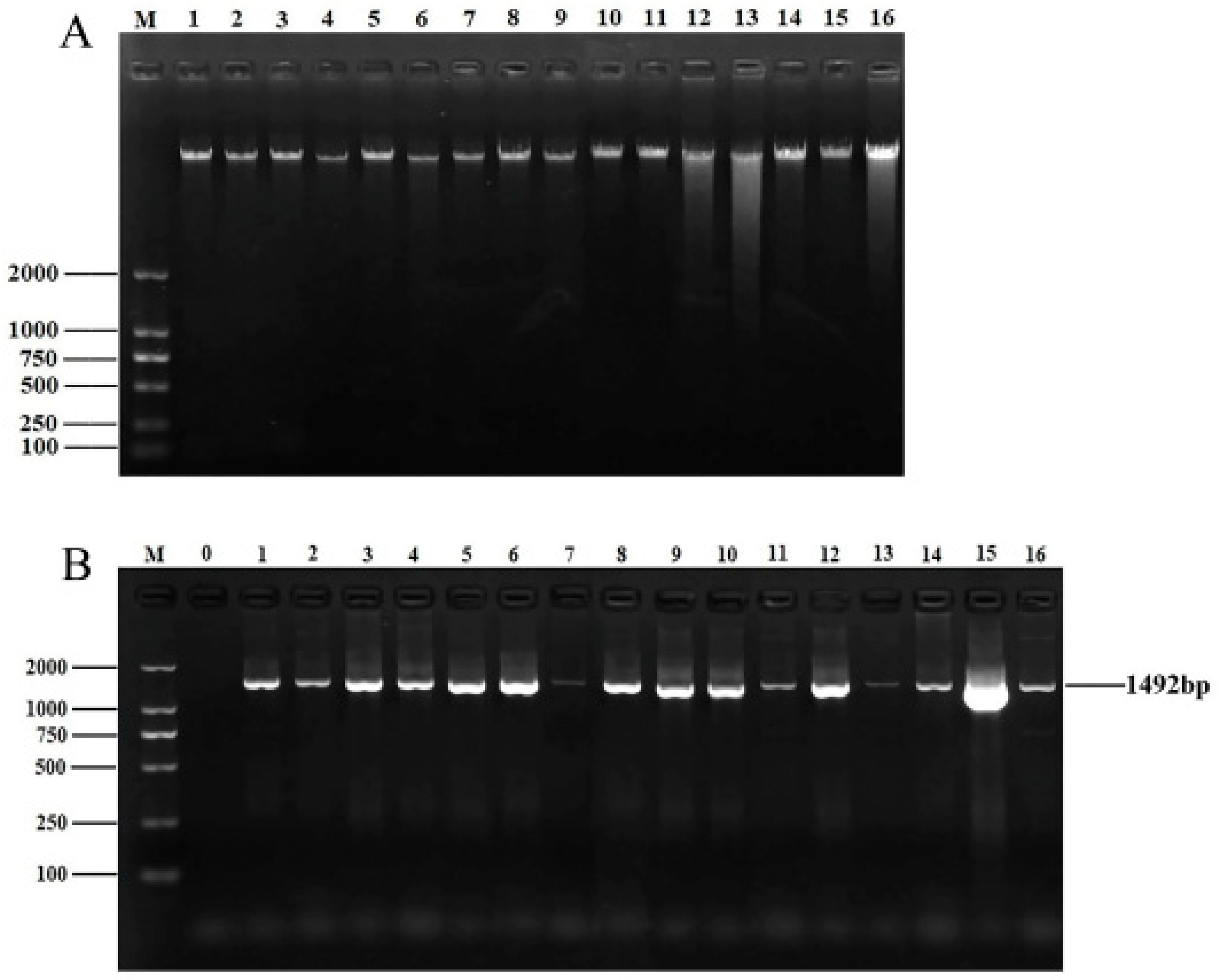
After culture, isolation, and purification, a total of 56 endophytic bacterial isolates were obtained. DNA quality affects subsequent experiments. A bacterial genomic kit was used to extract and isolate the genomic DNA of the isolated strains. The concentration and purity of the strains were measured with an ultra–micro spectrophotometer. The OD260/280 ratios for all DNA ranged from 1.7 to 2.0, indicating that the purity of the extracted DNA was satisfactory. DNA quality was then analyzed via 1% agarose gel electrophoresis (Figure 1A). The DNA of all samples displayed clear and distinct bands, making it suitable for subsequent PCR amplification.
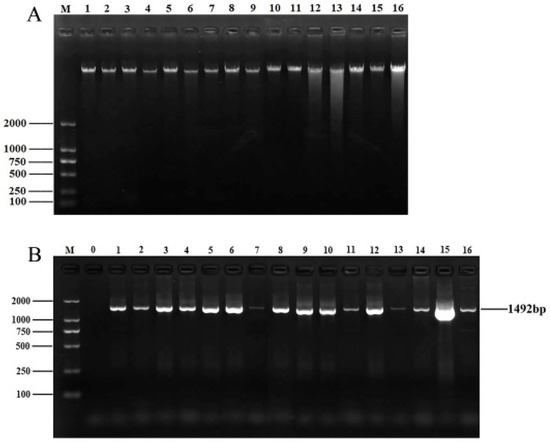
Figure 1.
DNA extraction and 16S rRNA amplification of isolates. (A) Electrophoretic map of genomic DNA of partial isolates. M: DNA marker DL2000; lanes 1–16: genomic DNA of 16 isolates. (B) Electrophoretic map of 16S rRNA amplification products of partial isolates. M: DNA marker DL2000; lane 0: negative control; lanes 1–16: 16S rRNA amplification products of partial isolates.
As shown in Figure 1B, all PCR bands of the isolates were clear and bright, with a length of approximately 1492 bp, which is consistent with the expected product size. After sequencing the PCR products, the 16S rRNA sequences of 56 isolates were uploaded to NCBI for BLAST comparison. Finally, these 56 bacterial isolates belonged to 14 genera, with Pseudomonas, Cedecea, Serratia, and Lelliottia being the dominant groups.
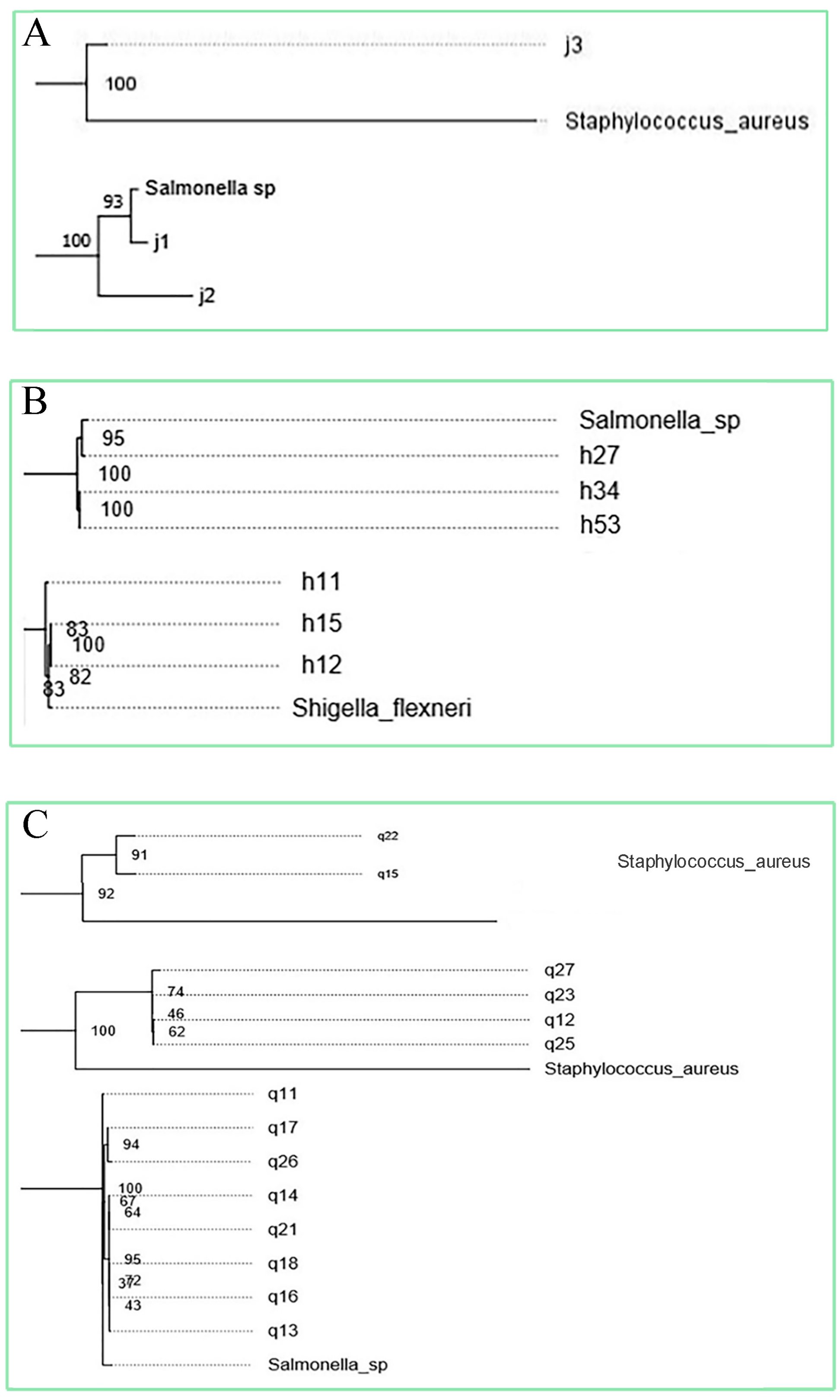
3.2. Phylogenetic Relationships Between Isolates and Common Foodborne Pathogens
A phylogenetic tree was established between 56 isolates and the 10 subspecies of foodborne pathogens, and the relationship between them was determined. Isolates that have a phylogenetic relationship with foodborne pathogenic bacteria greater than 80 and have been reported in the reference as having pathogenic cases were selected. Finally, 12 strains were selected (Figure 2) and sent for whole genome sequencing, including J1, Cedecea; J2, Enterococcus faecalis; J3, Cedecea; H11, Bacillus; H15, Pseudomonas; H27, Enterobacter; H34, Enterobacter; H53, Bacillus; Q11, Enterobacter; Q15, Serratia; Q22, Enterococcus; Q23, Cedecea.
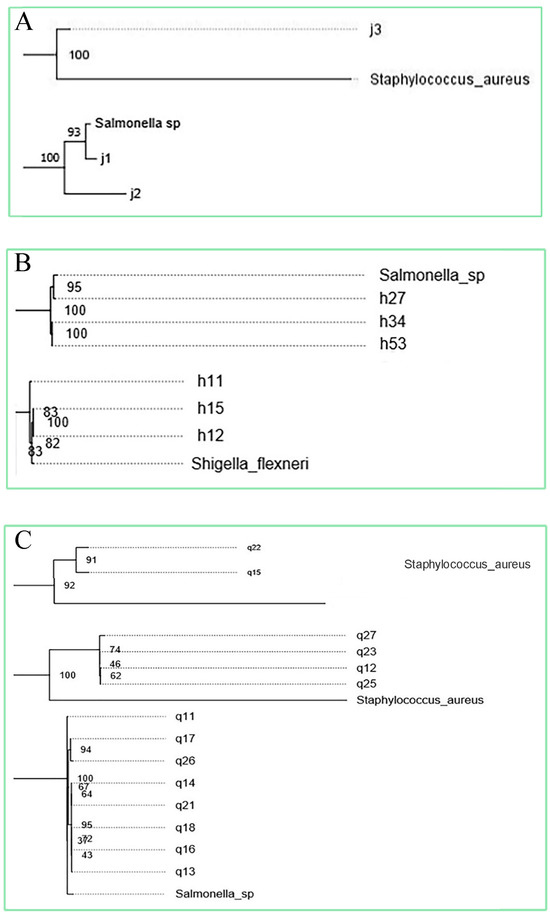
Figure 2.
Phylogenetic tree among solates and common foodborne pathogens (A) Relationship between endophytic isolates of Cantharellus cibarius and foodborne pathogens. (B) Relationship between endophytic isolates of Lactarius deliciosus and foodborne pathogens. (C) Relationship between endophytic isolates of Russul avirescens and foodborne pathogens.
3.2.1. Genome Assembly and Annotation
Trim Galore was used for quality assessment and filtering of the raw data, gene assembly was performed with SPAdes assembly software (4.0), and the GapCloser software (SOAPdenovo2-src-r240) was utilized to fill the local internal holes and correct the bases of the assembly results. The final assembly results are shown in Table S2. The assembly results were subjected to rRNA/tRNA gene prediction, Clusters of Orthologous Groups of proteins function analysis, and Kyoto Encyclopedia of Genes and Genomes pathway analysis.
3.2.2. Virulence Factor Analysis
Based on the genome assembly and annotation, the predicted genes were compared with genes in the VFDB, an integrated and comprehensive database of virus factors for bacterial pathways, using blastp to identify the genes associated with virulence factors. VFDB contains two databases, setA and setB. SetA has been validated through experiments, making its results reliable. Consequently, we used the results of setA for virulence factor analysis. More than 30 kinds of virulence genes were found in the 12 isolates as shown in Table S3 (only 7 kinds of virulence factors represented a significant proportion). However, their mechanisms of action remain unclear and mainly include the formation of oligomer structure of hemolysin e; driven by an enzyme encoded by the iucbcd gene; binding with specific receptors; and other mechanisms. Additional mechanisms included antiphagocytosis, serum resistance, secretion system, effect on cell memory activity, and magnesium absorption.
3.2.3. ARGs Analysis
The genes and mechanisms for antibiotic resistance were identified using blastp to compare predicted genes with those in the ARDB as shown in Table S4. The 12 isolates contained more than 15 kinds of ARGs, with the most prevalent resistance mechanisms being the efflux pump system and inactivated enzymes. Other resistance mechanisms included the resistance operon gene, superfamily transport, pentapeptide repeat family, and ribosome protection protein. These mechanisms can confer resistance to 15 kinds of antibiotics, including fosfomycin, macrolides, vancomycin, tetracycline, fluoroquinolone, penicillin, chloramphenicol, polymyxin, aminoglycoside, cephalosporin, Bacillus subtilis peptide, fosfomycin, and streptomycin.
3.3. Antibiotic Sensitivity Test Results of 56 Isolates
The diameter of the bacteriostatic circle did not exceed 1/3 of the culture dish. The edges of the bacteriostatic circle in the same culture medium with 3 different antibiotics-sensitive disks did not overlap. The bacteriostatic circles did not interpret each other or the interpretation results. Three disks were placed on 1 plate. After BLAST comparison of the 56 isolated, there was no accurate corresponding antimicrobial circle standard for the isolates in the CLSI standard. Therefore, the determination of the antimicrobial circle in this study was based on the CLSI standard for those of Enterobacteriaceae.
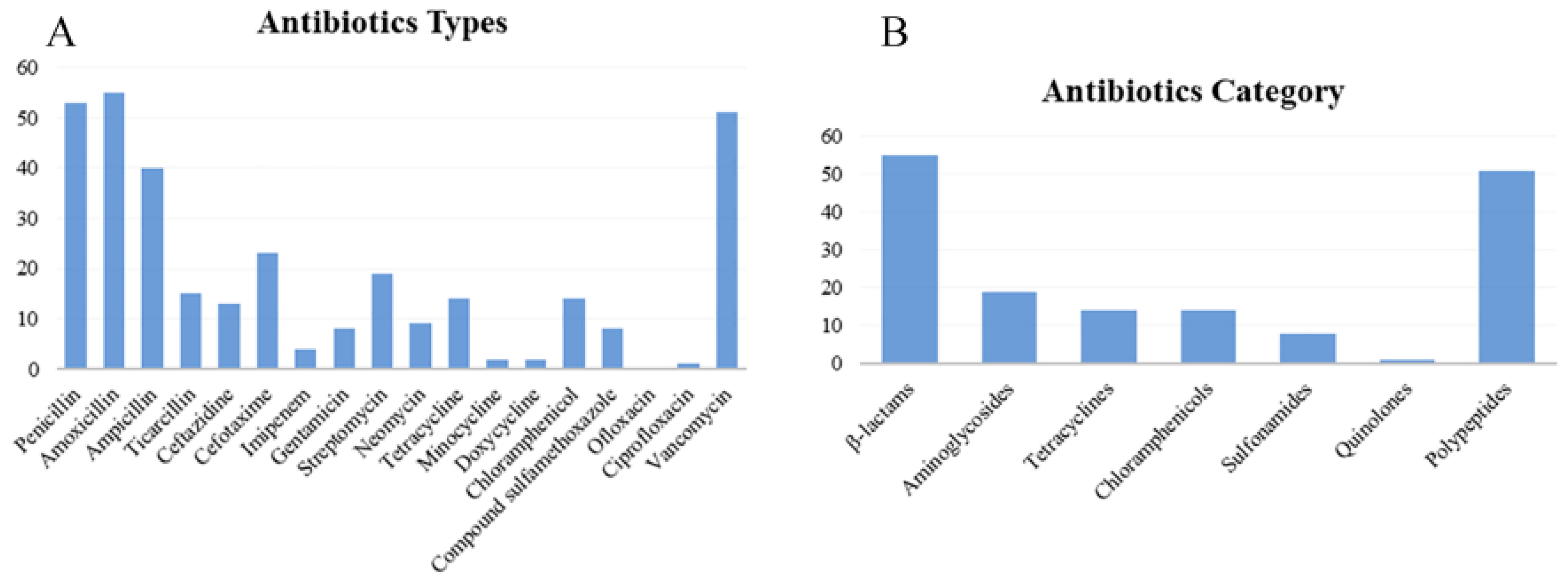
In accordance with the criteria of the antimicrobial circle of Enterobacteriaceae, the resistance phenotypes of 56 isolates were statistically analyzed (Table S5, Figure 3). Table S5 and Figure 3A showed that among the 18 antibiotics, amoxicillin (98.21%), penicillin (94.64%), and vancomycin (91.07%) exhibited the highest resistance rates, while ofloxacin (0), ciprofloxacin (1.69%), minocycline (3.57%), and doxycycline (3.57%) had the lowest resistance rates. The highest sensitivity was observed with ofloxacin (100%), followed by ciprofloxacin (98.21%), and doxycycline (91.07%); the lowest sensitivity was recorded for amoxicillin (1.69%), followed by penicillin (5.36%) and vancomycin (8.93%). The highest intermediate antibiotic resistance rate was demonstrated by neomycin (30%), followed by streptomycin (16.07%) and imipenem (12.5%). The intermediate rates for sulfamethoxazole, ofloxacin, ciprofloxacin, and vancomycin were all 0. Figure 3B shows that 55 isolates were most resistant to β lactams, while 1 isolate exhibited the least resistance to quinolones. The category with the highest number of antibiotic-sensitive isolates is fluoroquinolone, with 56 isolates, while that with the fewest is the polypeptide, with 5 isolates. The highest number of isolates with intermediate-sensitive was 17, for aminoglycosides, and as to sulfonamides, quinolones, and polypeptides were 0. The strains that were sensitive to tetracycline were also sensitive to doxycycline and minocycline. More strains were sensitive to doxycycline and minocycline than to tetracycline. CLSI analysis showed that the isolates with intermediate resistance to tetracycline might be sensitive to doxycycline, minocycline, or both.
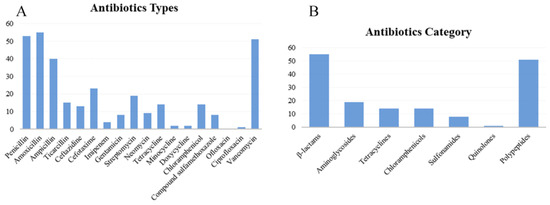
Figure 3.
Distribution of antibiotics resistance phenotype of 56 isolates. (A) Antibiotics types; (B) antibiotics category.
3.4. ARG Detection and Sequencing Comparison Results of 56 Isolates
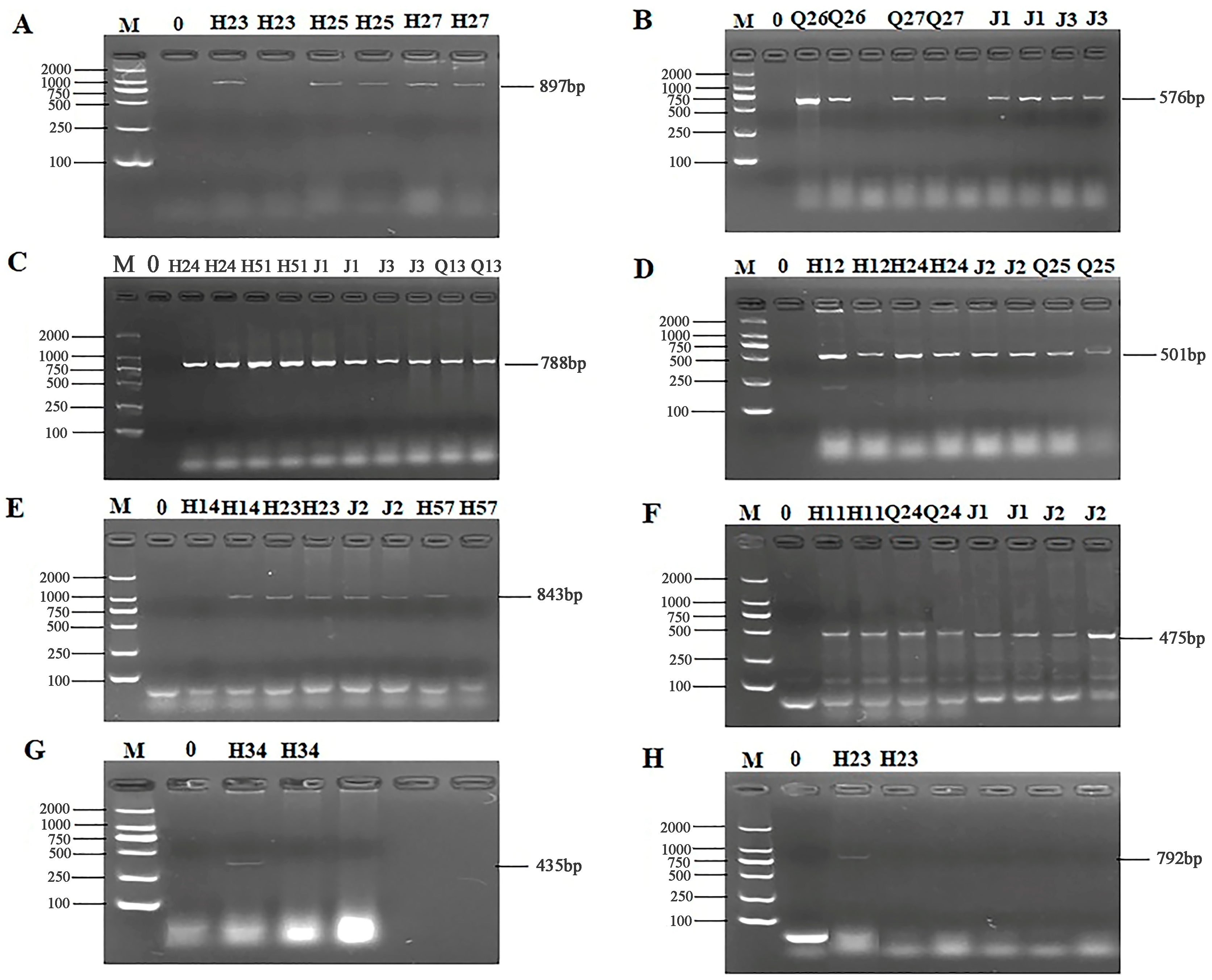
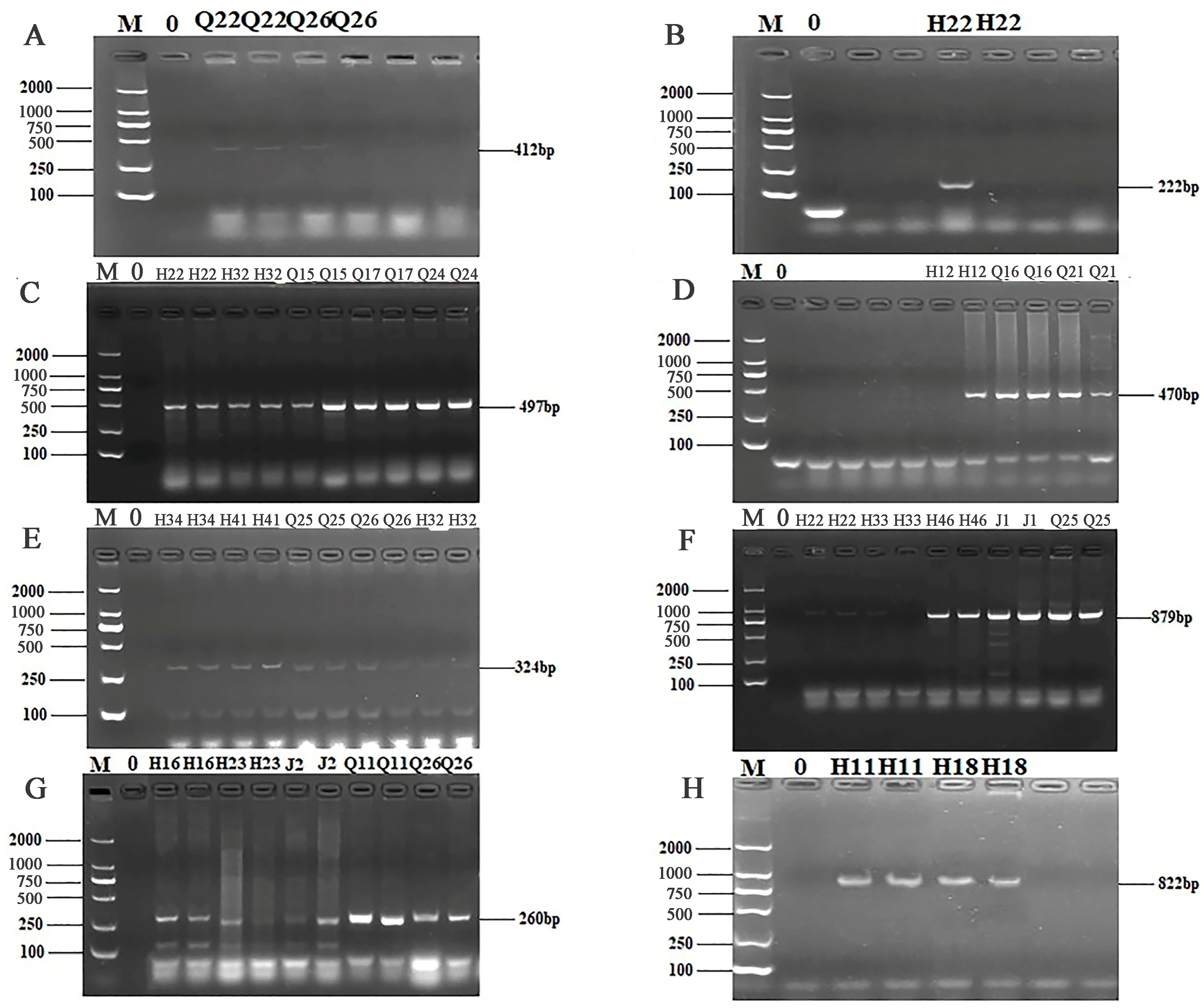
The 24 primers for ARGs were selected from 7 categories of antibiotics for PCR detection. The results of electrophoresis detection are shown in Figure 4 and Figure 5. Table S6 indicated that the highest detection rate was observed for the sul1 (53.57%) gene in sulfonamides, while the lowest detection rate of 1.79% was found for the sul2 and sul3 genes, excluding isolates in which no PCR product was amplified. Among the 6 tetracycline resistance genes, only tetK (8.92%) and tetM (23.21%) were detected. The β-lactam genes blaTEM, blavim, and blaSHV were detected at the rates of 25%, 21.42%, and 23.21%, respectively. Among the 4 aminoglycoside resistance genes, aadB was not detected, while the detection rate for aac (3′)-IIa, acrB, and aadA1 were 3.57%, 1.79%, and 37.5%, respectively. The detection rate of the chloramphenicol gene Cat was 14.29%, and floR was not detected. The peptide VanC gene was detected at the rate of 3.57%, and EmgrB was not detected. Three genes of quinolones, namely GyrA (8.92%), GyrB (39.29%), and ParC (37.5%), were detected. The PCR products also underwent sequencing, the sequences of gel-recovered PCR products were compared with the reference sequences in Genbank using blast, and the accession numbers of sequences with 99% similarity were recorded (Table S7).
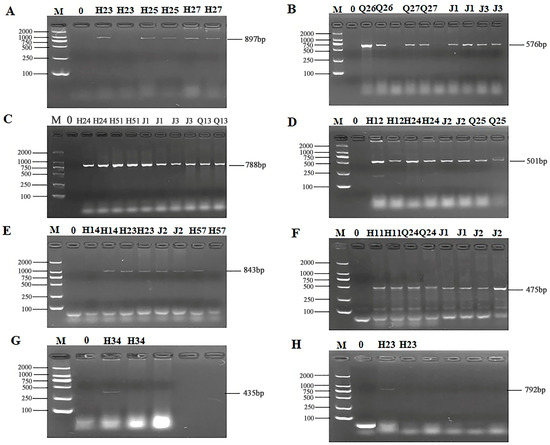
Figure 4.
PCR results of some resistant genes. M: DL2000 marker; 0: negative control; (A) partial electrophoresis of tetK gene; (B) partial electrophoresis of tetM gene; (C) partial electrophoresis of blaTEM gene; (D) partial electrophoresis of bla-vim gene; (E) partial electrophoresis of blaSHV gene; (F) partial electrophoresis of sul1 gene; (G) partial electrophoresis of sul2 gene; (H) partial electrophoresis of sul3 gene.
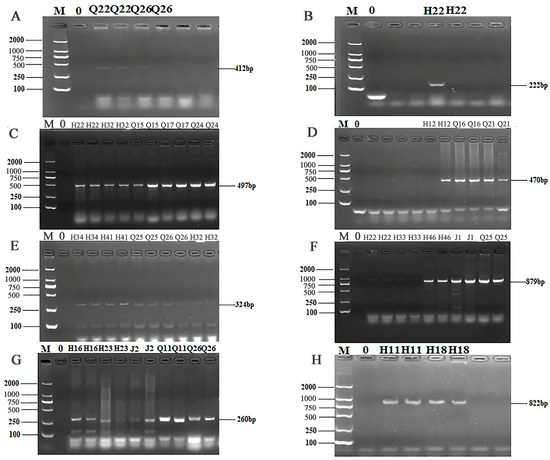
Figure 5.
PCR results of some resistant genes. M: DL2000 marker; 0: negative control; (A) partial electrophoresis of aac(3′)—IIa gene; (B) partial electrophoresis of acrB gene; (C) partial electrophoresis of aadA1 gene; (D) partial electrophoresis of Cat gene; (E) partial electrophoresis of GyrA gene; (F) partial electrophoresis of GyrB gene; (G) partial electrophoresis of ParC gene; (H) partial electrophoresis of VanC gene.
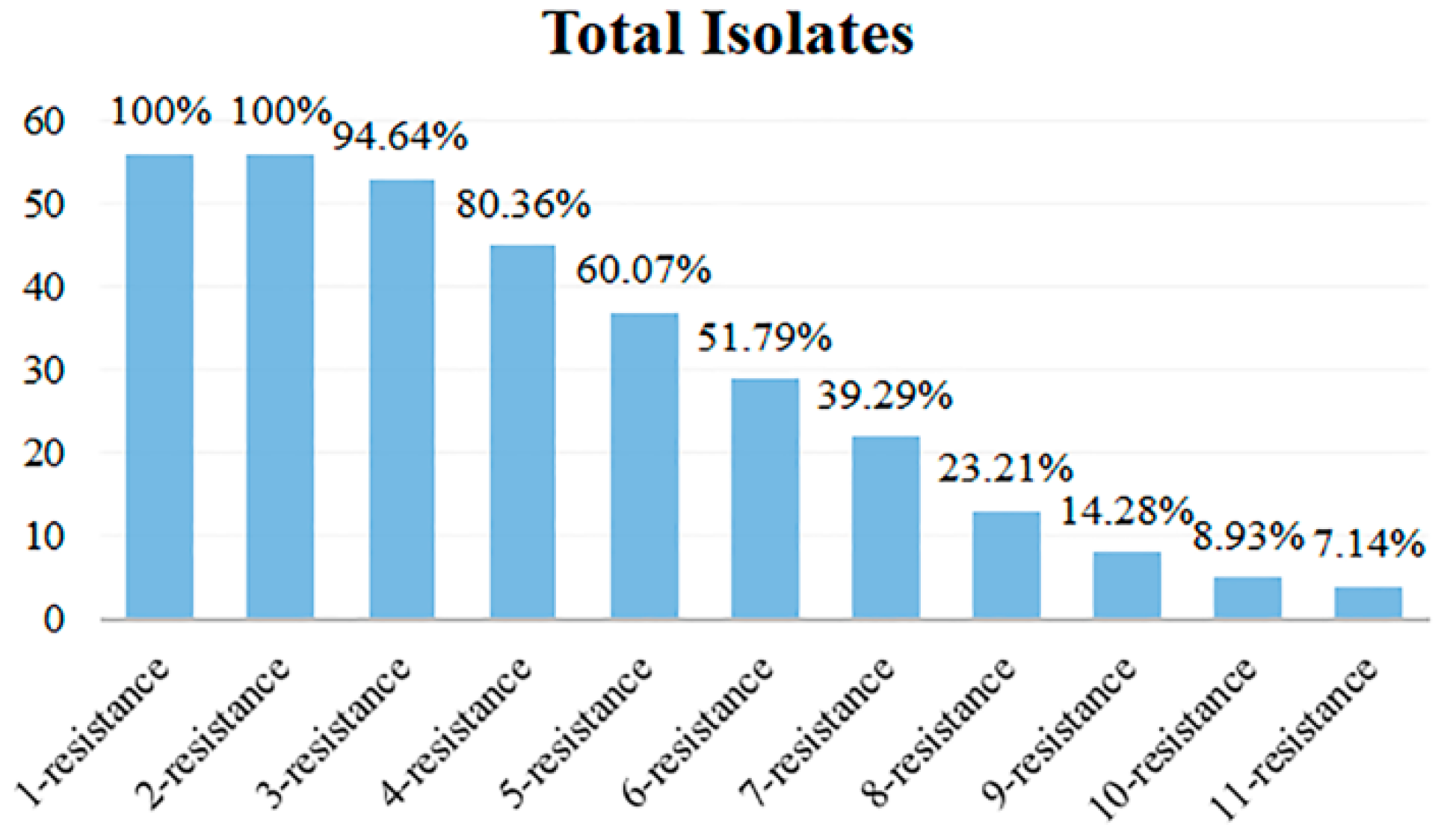
3.5. Analysis of Multi-Antibiotics Resistance Spectrum
The antibiotic resistance spectrum of 56 isolates exhibiting various antibiotic resistance phenotypes was quantified, and the multi-antibiotic-resistant isolates and their types were analyzed. The results are shown in Table 2 and Figure 6. Among the 56 isolates, all isolates exhibited antibiotic resistance, with no cases of 0 resistance, while the others displayed a highly concentrated resistance profile ranging from 1 to 11. The most prevalent resistant types were 1-resistant and 2-resistant, with all 56 isolates exhibiting resistance, which accounted for 100%. Conversely, the isolates categorized as 11-resistance were the fewest, with only 4 isolates accounting for 7.14% of the total. Figure 6 illustrates that isolates with resistance to multiple antibiotics predominantly fell into the categories of 1- to 6-resistance, with their antibiotic resistance isolates exceeding 50%. The multi-antibiotic resistance spectrum with the most forms was 6-resistance, which encompassed 6 types. In contrast, the 3-, 9-, and 10-resistance had only 2 types. The 2-, 4-, and 11-resistance had 3 types, while 5- and 7-resistance exhibited 5 types, and the 8-resistance contained 4 types.

Table 2.
Multi-antibiotic resistance types of 56 isolates.
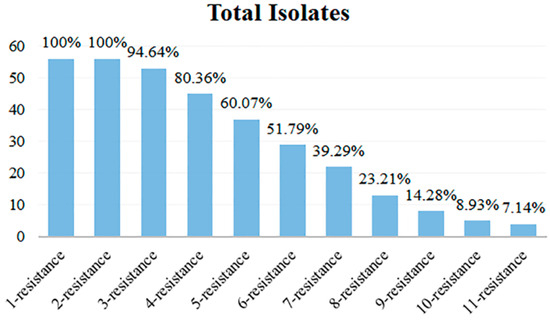
Figure 6.
Multi-antibiotic resistance numbers of 56 isolates.
3.6. Analysis of the Coincidence Rate Between the Antibiotic Resistant Phenotype and Genotype of the 56 Isolates
The antibiotic resistance phenotypes and genotypes of the 56 isolates were compared, and the coincidence rate was analyzed (Table S8). The highest coincidence rate was 90.48% for aminoglycosides, while the lowest was 2.94% for quinolones. Antibiotics exhibiting more phenotypic isolates than genotypes included β-lactams, chloramphenicol, and peptides, whereas those with more genotypes than phenotypes included tetracyclines, sulfonamides, aminoglycosides, and quinolones.
3.7. Gene Annotation Result Matching of Antibiotic Resistance Gene Annotation of the 12 Isolates That Closely Related with the Foodborne Pathogens
As to the 12 isolates that closely related to the foodborne pathogens that were reported to be pathogenic, according to the antibiotic resistance genes annotation and antibiotic resistance genotype and phenotype comparison results (Table S9), no resistant genes for β-lactams or sulfonamides were found in the gene annotation. Additionally, the sulfonamide resistance genes sul1 and sul2, as well as the aminoglycoside resistance gene aadA1, were not present in the resistance gene annotations. However, most other genes were all matched, with the same antibiotic resistance gene corresponding to multiple resistance mechanisms.
4. Discussion
There are various relationships between endophytic bacteria and their hosts, with the most researched being the positive effects they confer on the host plants, some even influence the quality of the host. The results of this study showed that endophytic bacteria isolated from wild edible fungi of the same host exhibit rich diversity. Furthermore, for wild edible fungi of the same species found in different locations, there are both similarities and differences among the endophytic bacteria. A total of 56 isolates were isolated and purified, identified through BLAST comparison, revealing 14 genera, with Pseudomonas, Cedecea, Serratia, and Lelliottia as the dominant bacterial groups. This demonstrates that the endophytes possess significant diversity, and their distribution and colony structure is influenced by the growth stage of the host plant and environmental conditions.
The resistance of bacteria to antibiotics is a common phenomenon in nature, but the abuse of antibiotics leads to bacterial tolerance. Consequently, the issue of acquired resistance to antibiotics, including new ones, is becoming increasingly serious. ARGs are transmitted both horizontally and vertically, resulting in the emergence of superbacteria resistant to multiple antibiotics [34,35]. In this study, 56 isolates of endophytic bacteria isolated from wild edible fungi were tested for antibiotic resistance. The antibiotic sensitivity test was performed with 18 antibiotics across 7 categories. The results indicated that all isolates were sensitive to ofloxacin, while the sensitivity rates for penicillin (the earliest used antibiotic for disease treatment), as well as the commonly used antibiotics amoxicillin and vancomycin, were 5.36%, 1.69%, and 8.93% respectively. The corresponding antibiotic resistance rates were 94.64%, 98.21%, and 91.07%. The resistance rate to ampicillin exceeded 70%; that to cefotaxime surpassed 40%; to streptomycin, it exceeded 30%; and to ticarcillin, ceftazidime, tetracycline, and chloramphenicol, they all exceeded 20%. Currently, carbapenems are effective in the treatment of Gram-negative bacterial infections [36] and are particularly effective against multidrug-resistant Enterobacteriaceae infections [37]. Imipenem is also a new carbapenem antibiotic, but bacteria have developed resistance to it due to its widespread clinical use. Monitoring network data on antibiotic-resistant bacteria in China indicated that the resistance rate of imipenem-resistant bacteria has gradually increased in recent years, a phenomenon that warrants attention. The endophytic bacteria examined in this study are sensitive to ofloxacin and exhibit varying degrees of resistance to 17 kinds of antibiotics. The prolonged misuse of antibiotics and the presence of antibiotic residues in food have impacted environmental microorganisms and the natural growth of plant tissue endophytes through the transfer of ARGs. The rise of antibiotic-resistant microorganisms has resulted in food safety and public health issues, further complicating clinical treatment.
In this study, 24 common ARGs were tested. The results indicated that 12 ARGs were detected among the 56 isolates. More phenotypes than genotypes were identified for β-lactams, chloramphenicol, and polypeptides, likely due to differing biochemical and genetic mechanisms of antibiotic resistance [38]. The β-lactams are the most commonly used antibiotics for treating bacterial infection. Their resistance mechanisms mainly involve the production of β-lactamases, efflux pump systems, and alterations in penicillin-binding proteins. The former can confer antibiotic resistance by hydrolyzing specific β-lactamases [39], such as metal β-lactamases and serine β-lactamases [40]. The mexab-OprM efflux pump system is the primary contributor to the inherent antibiotic resistance of bacteria [41] and enhances resistance against penicillin and cephalosporin. The chloramphenicol resistance mechanism mainly involves the active efflux system and the production of chloramphenicol acetyltransferase [42], with the acetyltransferase mechanism being the principal mechanism of chloramphenicol resistance [43].
Research on the mechanisms underlying vancomycin resistance in China has primarily focused on Enterococcus and S. aureus. Most Enterococcus bacteria develop resistance to vancomycin through the acquisition of ARGs and interspecific transmission [44,45]. The high level of resistance in S. aureus aligns with that of Enterococcus, while the low level of vancomycin resistance may result from multiple chromosomal site mutations [46] or regulation of protein expression [45]. Additionally, cell membranes and active efflux systems are prevalent in both Gram-negative and Gram-positive bacteria. Consequently, due to the biochemical mechanism of bacterial resistance, β lactams, chloramphenicol, and polypeptides exhibited more resistant genotypes than resistant phenotypes in this study. Furthermore, if the strain has an antibiotic-resistant phenotype but lacks the corresponding antibiotic-resistant genotype, it is likely inherently resistant to an antibiotic [47]. ARGs for tetracyclines, sulfonamides, aminoglycosides, and quinolones were detected, but few phenotypes were detected. ARGs may be present but silenced, not expressed, or not fully expressed. Qin et al. [26] conducted antibiotic resistance experiments on lactic acid bacteria isolated from commercial yogurt and found isolates carrying erythromycin and tetracycline ARGs but exhibiting sensitive phenotypes, demonstrating that no antibiotic resistance phenotype carried ARGs.
The analysis of the multi-antibiotics resistance spectrum among the 56 isolates reveals significant insights into the prevalence and distribution of antibiotic resistance phenotypes. The findings indicate that a substantial proportion of the strains exhibit high levels of resistance, particularly in the 1-resistance and 2-resistance categories, which accounted for all strains analyzed. This suggests a concerning trend in antibiotic resistance that could have implications for treatment strategies and public health. The concentration of isolates within the lower resistance categories (1 to 6) indicates that these isolates may be more adaptable or prevalent in the studied environment, potentially due to selective pressures from antibiotic use. The identification of only a few isolates in the higher resistance categories (such as 11-resistance) highlights the need for further investigation into the mechanisms driving such resistance and the factors contributing to the emergence of highly resistant strains. Moreover, the data suggest that multi-antibiotic resistance is a significant issue, with over 50% of strains exhibiting resistance across multiple antibiotic classes. This multi-antibiotic resistance complicates treatment options and underscores the urgency for developing new therapeutic strategies and implementing effective antibiotic stewardship programs.
Although foodborne pathogens are the primary focus of antibiotic resistance monitoring and research, nonpathogenic bacteria are abundant, widely distributed, and constitute the core of microbial ecosystems. Since the ARGs identified in pathogenic bacteria are likely transmitted by nonpathogenic bacteria as their original carriers [19], understanding and monitoring the antibiotic resistance of both pathogenic and nonpathogenic bacteria is crucial. Currently, a key index for assessing the safety of strains is the presence of ARGs. Research on the mechanism of antibiotic resistance primarily encompasses two aspects: biochemical and genetic mechanisms. In this study, the coincidence rates of antibiotic resistance phenotypes and genotypes were inconsistent. The main reason for these findings we speculated was the differing mechanisms of antibiotic resistance. Therefore, a thorough investigation into the resistance mechanisms of various isolates against different antibiotics, as well as exploring the reasons behind the prevalence of phenotype isolates over genotypes, is of great significance and provides valuable insights for studying antibiotic resistance and reducing antibiotic resistance rates.
5. Conclusions
In conclusion, the study highlights the alarming prevalence of multidrug-resistant strains among the analyzed samples. The findings demonstrate that all 56 strains exhibited some level of resistance, with a notable concentration in the lower resistance categories. The results emphasize the critical need for ongoing surveillance of antibiotic resistance patterns and the development of targeted interventions to combat the rise of multidrug-resistant infections. Addressing this issue is essential for ensuring effective treatment options and safeguarding public health.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms13020361/s1, Table S1: Primer sequences of drug resistance gene; Table S2: Assembly results; Table S3: Analysis of virulence factors; Table S4: Antibiotics resistance gene analysis; Table S5: Statistical results of antibiotics resistance phenotype of 56 strains; Table S6: Detection rate of drug resistance genes in 56 isolates; Table S7: Comparison of gel recovery and sequencing results; Table S8: Analysis of the coincidence rate between drug resistance phenotype and genotype; Table S9: Result matching of antibiotic resistance gene annotation of the 12 isolates that closely related with the foodborne pathogens.
Author Contributions
Conceptualization, S.Y. and H.W.; data curation, S.Y. and H.W.; investigation, S.Y. and H.W.; methodology, S.Y. and H.W.; software, Y.L. and X.C.; validation, Y.L. and X.C.; Formal analysis, Y.L. and X.C.; writing—original draft, S.Y. and H.W.; Supervision, Y.S.; writing—review and editing, Y.S.; funding acquisition, Y.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32260624) and Yunnan Major Scientific and Technological Projects (202202AG050009).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Huang, X.Z.; Zhao, F.L. Mechanism of endophytes of medicinal plants in promoting the growth of host plants. Microbiol. China 2023, 50, 1653–1665. [Google Scholar] [CrossRef]
- Verma, S.K.; Sahu, P.K.; Kumar, K.; Pal, G.; Gond, S.K.; Kharwar, R.N.; White, J.F. Endophyte roles in nutrient acquisition, root system architecture development and oxidative stress tolerance. J. Appl. Microbiol. 2021, 131, 2161–2177. [Google Scholar] [CrossRef] [PubMed]
- Vandana, U.K.; Rajkumari, J.; Singha, L.P.; Satish, L.; Alavilli, H.; Sudheer, P.D.; Chauhan, S.; Ratnala, R.; Satturu, V.; Mazumder, P.B. The endophytic microbiome as a hotspot of synergistic interactions, with prospects of plant growth promotion. Biology 2021, 10, 101. [Google Scholar] [CrossRef]
- Liu, M.J.; Xu, Y.J.; Huang, X.Z.; Zhao, F.L. Advances in research methods for plant endophytes. Microbiol. China 2024, 51, 1887–1897. [Google Scholar] [CrossRef]
- Tang, M.-J.; Lu, F.; Yang, Y.; Sun, K.; Zhu, Q.; Xu, F.-J.; Zhang, W.; Dai, C.-C. Benefits of endophytic fungus Phomopsis liquidambaris inoculation for improving mineral nutrition, quality, and yield of rice grains under low nitrogen and phosphorus condition. J. Plant Growth Regul. 2022, 41, 2499–2513. [Google Scholar] [CrossRef]
- Lastochkina, O.; Yuldashev, R.; Avalbaev, A.; Allagulova, C.; Veselova, S. The Contribution of Hormonal Changes to the Protective Effect of Endophytic Bacterium Bacillus subtilis on Two Wheat Genotypes with Contrasting Drought Sensitivities under Osmotic Stress. Microorganisms 2023, 11, 2955. [Google Scholar] [CrossRef] [PubMed]
- Elder, D.P.; Kuentz, M.; Holm, R. Antibiotic resistance: The need for a global strategy. J. Pharm. Sci. 2016, 105, 2278–2287. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.L. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 2009, 157, 2893–2902. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, G.; Midiri, A.; Gerace, E.; Biondo, C. Bacterial Antibiotic Resistance: The Most Critical Pathogens. Pathogens 2021, 10, 1310. [Google Scholar] [CrossRef] [PubMed]
- Bloch, S.; Hager-Mair, F.F.; Andrukhov, O.; Schäffer, C. Oral streptococci: Modulators of health and disease. Front. Cell. Infect. Microbiol. 2024, 14, 1357631. [Google Scholar] [CrossRef]
- Jeon, S.-M.; Park, S.; Kim, S. Whole genome sequence of a Mycobacterium tuberculosis M64 isolate harboring extensive deletions, including the katG-containing region. Microbiol. Resour. Announc. 2024, 14, e00745-24. [Google Scholar] [CrossRef]
- Roca, I.; Akova, M.; Baquero, F.; Carlet, J.; Cavaleri, M.; Coenen, S. Corrigendum to “The global threat of antimicrobial resistance: Science for intervention”. New Microbes New Infect 2015, 8, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Guoxiu, L.; Liliuj, C.; Yingsha, L. Evaluation of Antibiotic Pollution in Soil of Vegetable Base in Yangling District, Shaanxi Province. Asian Agric. Res. 2021, 11, 41–46. [Google Scholar]
- Payne, D.J. Desperately seeking new antibiotics. Science 2008, 321, 1644–1645. [Google Scholar] [CrossRef] [PubMed]
- Sørum, H.; L’Abée-Lund, T.M. Antibiotic resistance in food-related bacteria—A result of interfering with the global web of bacterial genetics. Int. J. Food Microbiol. 2002, 78, 43–56. [Google Scholar] [CrossRef]
- Pesavento, G.; Ducci, B.; Comodo, N.; Nostro, A.L. Antimicrobial resistance profile of Staphylococcus aureus isolated from raw meat: A research for methicillin resistant Staphylococcus aureus (MRSA). Food Control 2007, 18, 196–200. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control; European Food Safety Authority; European Medicines Agency and European Commission’s Scientific Committee on Emerging and Newly Identified Health Risks. Joint Opinion on antimicrobial resistance focused on zoonotic infections. EFSA J. 2009, 7, 1372. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Antimicrobials Sold or Distributed for Use in Food-Producing Animals. 2017. Available online: https://www.fda.gov/media/109457/download (accessed on 14 December 2024).
- Zhou, Y.; Zhang, L.; Wang, Z.H.; Zhong, J.; Wang, H. Key risk factors of antibiotic resistance ecology and targeted control strategies. Acta Microbiol. Sin. 2018, 58, 2040–2050. [Google Scholar] [CrossRef]
- Ahmad, N.; Ali, S.M.; Khan, A.U. First reported new delhi metallo-β-lactamase-1-producing Cedecea lapagei. Int. J. Antimicrob. Agents 2017, 49, 118–119. [Google Scholar] [CrossRef]
- Wu, J.F. Isolation and Identification of the Endophytes from Atractylodes macrocephala Koidz, and Anti-Fungal Activities of Its Fermentation Products. Master’s Thesis, Anhui Agricultural University, Hefei, China, 2020. [Google Scholar]
- Liu, Y.Q.; Luo, Y.L.; Liu, X.R.; Bai, L. Isolation and Identification of Endophytes and Actinomycetes from Underwood Planting Ganoderma lucidum. North. Hortic. 2023, 3, 120–126. [Google Scholar]
- Li, X.; Wu, J.R.; Tian, T.; Yu, X.Q. Isolation, Identification and Preliminary Screening of Acid-Tolerant Lactic Acid Bacteria from Naturally Fermented Pickle Juices from Daqing. Food Sci. 2014, 35, 150–154. [Google Scholar]
- Fraccalvieri, R.; Bianco, A.; Difato, L.M.; Capozzi, L.; Del Sambro, L.; Simone, D.; Catanzariti, R.; Caruso, M.; Galante, D.; Normanno, G.; et al. Toxigenic Genes, Pathogenic Potential and Antimicrobial Resistance of Bacillus cereus Group Isolated from Ice Cream and Characterized by Whole Genome Sequencing. Foods 2022, 11, 2480. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 31st ed.; CLSI Supplement M100; Clinical Laboratory Standards Institute: Wayne, PA, USA, 2021. [Google Scholar]
- Qin, Y.X.; Li, J.; Wang, Q.Y.; Gao, K.X.; Zhu, B.L.; Lv, N. Identification of lactic acid bacteria in commercial yogurt and their antibiotic resistance. Acta Microbiol. Sin. 2013, 53, 889–897. [Google Scholar]
- Li, Y.J.; Shan, L.Y.; Zhang, C.; Lei, Z.; Shang, Y. Isolation and antibiotic resistant research of Tetragenococcus halophilus from Xuanwei ham, a China high-salt-fermented meat products. Antibiotics 2019, 8, 151. [Google Scholar] [CrossRef] [PubMed]
- Ma, C. Detection and Sequence Analysis of Correlative Virulence Genesand Drug Resistance Genes of Superiority Escherichia coli Serotypes Isolated from Cow Mastitis. Master’s Thesis, Ningxia University, Ningxia, China, 2014. [Google Scholar]
- Yue, S. Detection of Drug Resistance Genes and Virulence Genes of Escherichia coli in Beef Cattle in Some Areas of Heilongjiang and Analysis of Drug Resistance. Master’s Thesis, Heilongjiang Bayi Agricultural University, Yinchuan, China, 2017. [Google Scholar]
- Ma, M.G.; Wang, H.Y.; Yu, Y.; Li, C.Z.; Zhang, D.; Yang, Y.F.; Liu, S.G. Detection of Antimicrobial Resistance Genesof Pathogenic Salmonella from Swine. Acta Vet. Zootech. Sin. 2006, 37, 65–70. [Google Scholar]
- Li, L.; Shen, J.Z.; Wu, Y.N.; Wang, Y.P.; Wu, C.M. Expression of marA, soxS and ramA genes and efflux pump genes acrA and acrB in ciprofloxacin-resistant Salmonella. Chin. Vet. Sci. 2009, 39, 928–932. [Google Scholar] [CrossRef]
- Li, C.Y.; Yang, Q. Surveillance of antimicrobial resistance and vancomycin resistance genes in Enterococcus species. Chin. J. Health Lab. 2007, 12, 2244–2246. [Google Scholar]
- Yi, L.X.; Liu, Y.Y.; Wu, R.J.; Liang, Z.S.; Liu, J.H. Research progress on the plasmid-mediated colistin resistancegene mcr-1. Hereditas 2017, 39, 110–126. [Google Scholar] [CrossRef]
- Hou, X.; Wu, R.; Pu, W.X. Research Direction of New Drugs for Superbacteria. Prog. Vet. Med. 2018, 39, 102–106. [Google Scholar] [CrossRef]
- Liu, D.H.; Zhang, X.W.; Zhang, C. Antibiotic abuse and superbugs. World Notes Antibiot. 2019, 40, 1–4. [Google Scholar] [CrossRef]
- Fang, Y.P.; Liu, Z.; Tong, Y.; Zhou, Q.; Guan, S.H.; Shen, J.L. Analysis of antibiotic resistance in 1580 clinical strains of Enterobacteriaceae. Chin. J. Infect. Chemother. 2015, 15, 171–174. [Google Scholar] [CrossRef]
- Wang, J.G.; Yu, F.Y. Prevalence of antimicrobiol-resistant to impenem in Gram-negative bacilli. Chin. J. Antibiot. 2008, 5, 300–302. [Google Scholar]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Bush, K. Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr. Opin. Microbiol. 2010, 13, 558–564. [Google Scholar] [CrossRef]
- Rawat, D.; Nair, D. Extended-spectrum AY-lactamases in Gram Negative Bacteria. J. Glob. Infect. Dis. 2010, 2, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Z.; Ma, D.; Livermore, D.M.; Nikaido, H. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: Active efflux as a contributing factor to beta-lactam resistance. Antimicrob. Agents Chemother. 1994, 38, 1742–1752. [Google Scholar] [CrossRef]
- Zhao, F.J.; Guan, M.; Li, Q.Z.; Li, J.C.; Liang, Q.; Gu, G.B. Detection and Analysis of Chloramphenicols Resistance Genes in Escherichia coli from Swine. Prog. Vet. Med. 2017, 38, 49–51. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Lang, X.Y.; Jiang, H.T.; Liu, J.; Bi, Y.; Zhang, J.J.; Chang, C.L.; Ni, H.B. Detection and analysis of virulence and drug resistance genes of bovine Escherichia coli in Heilongjiang area. Heilongjiang Anim. Sci. Vet. Med. 2015, 1, 137–139. [Google Scholar] [CrossRef]
- Gardete, S.; Tomasz, A. Mechanisms of vancomycin resistance in Staphylococcus aureus. J. Clin. Investig. 2014, 124, 2836–2840. [Google Scholar] [CrossRef]
- Wang, L.P.; He, Y.Y.; Yan, L.; Wang, H.J.; Liang, S.M.; Xia, Y. Investigation on the drug-resistant genes and molecular epidemiology of vancomycin-resistant Enterococcus. Chin. J. Clin. Lab. Sci. 2014, 32, 136–139. [Google Scholar] [CrossRef]
- Dai, Y.Y.; Zhang, T.; Ma, X.L. Research progress on the mechanism of vancomycin resistance in Staphylococcus aureus. Int. J. Lab. Med. 2005, 26, 798–800. [Google Scholar]
- Song, X.M.; Li, S.Y.; Ma, Q.Y.; Li, Z.; Li, S.F. Antibiotic resistance in fermented food lactic acid bacteria—A review. Microbiol. China 2015, 42, 207–213. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).