Abstract
Probiotics are widely used to improve pet health and welfare due to their significant biological activity and health benefits. Lactobacillus acidophilus GLA09 was derived from the intestinal tract of healthy beagles. The safety and suitability evaluation of GLA09 was completed through a combination of whole genome sequence and phenotypic analyses, including tests for the inhibition of harmful bacteria, acid resistance, bile salt tolerance, adhesion, and amine-producing substance content. The findings revealed that GLA09 has good gastrointestinal tolerance, inhibits the growth of pathogenic bacteria, and does not produce toxic biogenic amines. The genome of GLA09 comprises one chromosome and one plasmid, with a genome size of 2.10 M and a Guanine + Cytosine content of 38.71%. It encodes a total of 2208 genes, including 10 prophages, and 1 CRISPR sequence. Moreover, 56 carbohydrate-encoding genes were identified in the CAZy database, along with 11 genes for cold and heat stress tolerance, 5 genes for bile salt tolerance, 12 genes for acid tolerance, and 14 predicted antioxidant genes. Furthermore, GLA09 has one lincosamide resistance gene, but there is no risk of transfer. GLA09 harbors a cluster of Helveticin J and Enterolysin A genes linked to antimicrobial activity. Genomic analysis validated the probiotic attributes of GLA09, indicating its potential utility as a significant probiotic in the pet food industry. In summary, L. acidophilus GLA09 has the potential to be used as a probiotic in pet food and can effectively combat intestinal health in pets.
1. Introduction
Lactobacillus acidophilus is a microaerobic, Gram-positive bacterium classified within the phylum Firmicutes [1]. Since L. acidophilus was first isolated from infant feces in 1900, it has been recognized for its diverse probiotic effects on gastrointestinal health and overall well-being [2]. L. acidophilus can alleviate inflammatory bowel disease by reducing levels of inflammatory cytokines [3,4]. In addition, L. acidophilus serves multiple functional roles, including immune regulation, the enhancement of the intestinal epithelial barrier, the modulation of gut microbiota, cholesterol reduction, and the improvement of the body’s antioxidant capacity [5,6,7]. Recent studies have demonstrated that L. acidophilus can produce active metabolites such as extracellular polysaccharides and bacteriocins, which can directly or indirectly modulate host metabolism and immune responses [8,9]. Although numerous studies have documented the biological functions and probiotic properties of various L. acidophilus strains, most research has focused on physiological aspects. In contrast, relatively few studies have been conducted on the genetic function of L. acidophilus.
With the advancement of high-throughput sequencing technology, many studies have been conducted to analyze the safety and probiotic properties of strains at the genomic level, such as Lacticaseibacillus rhamnosus, Limosilactobacillus reuteri, Lactobacillus johnsonii, Bifidobacterium longum [10,11,12,13]. Whole-genome sequencing (WGS) reveals gene function, metabolic pathways, genetic features, and the genome structure of microorganisms [14]. WGS, along with bioinformatics analysis, has emerged as a crucial methodology. Microorganisms derived from the host are preferred as probiotics over non-host microorganisms because they have co-evolved with their hosts for approximately 10 million years, are better adapted to the host’s gastrointestinal environment, exhibit superior adherence, and provide lasting effects [15,16,17]. Currently, while some studies on probiotics in dogs have been conducted, there remains a scarcity of research specifically focusing on canines, prompting researchers to seek improved probiotic options for the future [18]. In this study, L. acidophilus GLA09 was isolated from the gastrointestinal tract of healthy beagle dogs. The objective was to investigate the potential probiotic effects of GLA09 as a pet food probiotic and to evaluate its safety through whole genome sequencing (WGS), mass spectrometry detection, gene sequence prediction, and functional annotation, thereby providing a theoretical basis for its application.
2. Materials and Methods
2.1. Main Reagents and Instruments
MRS broth medium (MRS, Haibo, Qingdao, China); Luria-Bertani (LB, Haibo, Qingdao, China); DNA extraction kit (Solarbio, Beijing, China); sequencing platform: NovaSeq 6000 (Illumina, San Diego, CA, USA).
2.2. Strain Source and Culture
L. acidophilus GLA09, derived from canine sources, was isolated from the gastrointestinal tract of healthy beagles in our laboratory and preserved at the Chinese Typical Cultures Depository Center in Wuhan, China, under the deposit number CCTCC NO: M2023982. In this study, L. acidophilus GLA09 was cultured in a 10 mL MRS broth at 37 °C with shaking for 24 h to activate the strain. Subsequently, it was passaged three times with a 3.0% (v/v) inoculum before being prepared for use.
The strains were inoculated into MRS liquid medium at an inoculum of 3.0% and incubated at 37 °C for 48 h. The control was a blank MRS liquid medium without strains. During the incubation period, OD600 nm absorbance was measured at 0, 1, 2, 3, 6, 9, 12, 15, 18, 21, 24, 36, and 48 h. The growth curves were plotted with time as the horizontal coordinate and OD600 absorbance as the vertical coordinate (subtracting the OD600 nm of the control group, removing the effect of the base color of the medium).
2.3. WGS, Assembly and Annotation
2.3.1. Strain DNA Extraction and Detection
After a 24-h incubation period, the bacterial solution was centrifuged at 4 °C at 10,621× g for 10 min, and the supernatant was discarded to collect the bacterial pellet. Bacterial DNA was then extracted using a bacterial DNA extraction kit (Qiagen, Beijing, China), followed by a quality assessment of the DNA concentration and purity using an ultra-micro spectrophotometer and agarose gel electrophoresis.
2.3.2. WGS and Genome Assembly
The WGS analysis was conducted using the Oxford PromethION platform and the Illumina NovaSeq 6000 platform. The genome was assembled de novo from the filtered reads using Unicycler software (version 0.5.0) based on the sequencing data. Subsequently, Pilon version 1.24 was employed to further refine the assembled genome using second-generation data [19].
2.3.3. Functional Annotation of Genomes
The coding genes were predicted from the assembled genome using Prokka software (version 1.14.6) to obtain general genomic features. To gather more comprehensive information on gene function, gene function annotation was performed using BLAST+ (version 2.11.0+) via Gene Ontology (GO), Kyoto Encyclopaedia of Genes and Genomes (KEGGs), and Cluster of Orthologous Groups of Proteins (COGs) (version 2.11.0+) comparisons [20,21,22]. A carbohydrase annotation was performed according to the Carbohydrate-Active EnZymes database (CAZy, https://www.cazy.org, accessed on 20 July 2023), and strain safety prediction was conducted using the Pathogen Host Interactions Database (PHI version 4.12, http://www.phi-base.org/, accessed on 20 July 2023) (identity > 40%), Comprehensive Antibiotic Research Database (CARD version 3.2.0, http://arpcard.mcmaster.ca, accessed on 20 July 2023), Pathogenfinder 1.1 (https://cge.food.dtu.dk/services/PathogenFinder/, accessed on 20 July 2023), and Virulence Factor Database (VFDB, http://www.mgc.ac.cn/VFs/, accessed on 20 July 2023, identity > 60%) [23,24,25].
2.3.4. Secondary Metabolites
The predictions of secondary metabolites were performed using antiSMASH 7.0 (https://docs.antismash.secondarymetabolites.org/, accessed on 20 July 2024).
2.3.5. Comparative Genomes
Genome evolutionary trees were constructed using a Type Strain Genome Server (https://tygs.dsmz.de, accessed on 26 June 2024), and comparative genome analysis was performed using TBools v2.056.
2.4. Biogenic Amines Detection
The content of biogenic amines produced by GLA09 was analyzed using LC-MS/MS (Sciex 4500, Shanghai, China). A 100 μL sample was taken, and 100 μL of sodium bicarbonate solution, 10 μL of sodium hydroxide solution, and 100 μL of dansyl chloride solution were added. The mixture was vortexed and mixed thoroughly. Subsequently, it was incubated in a 60 °C water bath for 25 min, cooled to room temperature, and 10 μL of ammonia was added. The sample was then concentrated to dryness and redissolved in 1 mL of acetonitrile. The supernatant was filtered through a 0.22 μm membrane, and the final sample volume was 3 μL. The chromatography column used was a HYPERSIL GOLD C18 column (3 μm, 2.1 mm × 100 mm) with a column temperature of 35 °C and a flow rate of 0.3 mL/min. The criteria for determining Biogenic amines were referenced from Lorencová et al. [26].
2.5. In Vitro Probiotic Potential of Strain
2.5.1. Cell Surface Hydrophobicity and Auto-Aggregation
The auto-aggregation assays referred to the method by Zhao et al. [27]. Following a 24-h growth period, the isolated cultures were washed three times with phosphate-buffered saline (PBS, pH 7.0) and adjusted to an approximate concentration of 1 × 108 CFU/mL. The bacterial suspension was vortexed for 30 s, and the initial absorbance was measured at OD600 nm (A0). Subsequently, the solution was incubated at 37 °C for 2 h. The OD value (A1) was determined at 600 nm using PBS buffer as a control. The aggregation activity was then calculated using the following equation:
The cell surface hydrophobicity assays referred to the method by Chen et al. [28]. GLA09 was cultured for 24 h, then washed three times with PBS (pH 7.0), and adjusted to a concentration of approximately 1 × 108 CFU/mL. The absorbance at OD600 nm was measured (A2). An equal volume of xylene and chloroform was then added, and the mixture was vortexed for 30 s before being incubated at 37 °C for 1 h. The organic phase was removed, and the OD value of the aqueous phase was determined at 600 nm (A3). The cell surface hydrophobicity was then calculated using the following equation:
2.5.2. Antibacterial Activity
The antibacterial activity of GLA09 was detected using the Oxford cup double-layer agar diffusion method [29] with slight modifications. Following incubation, the isolates were harvested from an MRS liquid medium and adjusted to a concentration of approximately 1 × 107 CFU/mL. Moreover, the pathogenic bacteria (Escherichia coli ATCC 25922, Staphylococcus aureus ATCC 25923, and Salmonella typhimurium ATCC 14028) were cultured in an LB liquid medium and adjusted to 1 × 107 CFU/mL. The plates were then incubated at 37 °C for 24 h, after which the inhibition zone diameter (IZD) was measured. A blank MRS medium was used as a negative control and laboratory strain CLP03 as a positive control (previously tested for bacteriostatic properties) [27].
2.5.3. Low pH and Bile Salt Tolerance
Additionally, the growth curve of GLA09 at different pH and bile concentrations was analyzed. To enhance the experimental procedure described by Ramos et al. [30] for determining the acid and bile salt tolerance of strains, bacteria were inoculated into modified pH-MRS (pH = 1.5–4.0) and BS-MRS (MRS medium with bile salt concentrations of 0.1%, 0.2%, and 0.3%) with an inoculum size of 3.0%, and samples were collected at 0, 2, 4, and 8 h. All experiments were conducted in triplicate for each experiment.
2.5.4. Data Analysis
Statistical analysis of the data in vitro probiotic potential was expressed as the mean ± standard deviation (SD). Each experiment was repeated three times independently.
3. Results
3.1. Genomic Characterization of L. acidophilus GLA09
By WGS of L. acidophilus GLA09, 9,487,074 valid reads remained after filtering. The size of the assembled complete genome was 2,102,723 bp, with a Guanine + Cytosine (G + C) content of 38.71%. As shown in Figure 1A, it contained one chromosome and one plasmid with sizes of 2,042,740 bp and 59,983 bp and G + C contents of 34.94% and 33.38%, respectively. The total number of coding genes was 2208, and the total length of coding genes was 1,860,808 bp, accounting for 88.50% of the total genome length, with an average gene length of 843 bp. In addition, the non-coding genome of the whole genome of strain GLA09 contained 64 tRNAs, 5 23S rRNAs, 5 16S rRNAs, 5 5S rRNAs, eight gene islands, 10 prophages, and one CRISPR sequence was predicted (Table 1 and Table S1). Furthermore, the results of strain GLA09 alignment were similar to those of L. acidophilus NBRC 13951 using the Type genome server (Figure 1B).
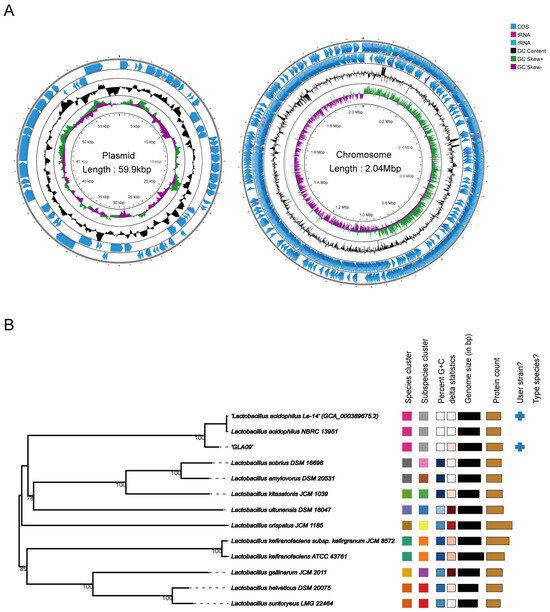
Figure 1.
Genome analysis of L. acidophilus GLA09. (A) Genome map (circles are indicated from outside to inside: circles 1 and 2 (blue) indicate forward and reverse strands, which represent genes for CDS, tRNA, and rRNA. Circle 3 (black) indicates the GC percentage of the genome. Circle 4 (purple and green) represents GC skew). (B) Comparison of the genome of L. acidophilus GLA09 using Type genome server.

Table 1.
Genome components of L. acidophilus GLA09.
3.2. Functional Annotation Analysis of the L. acidophilus GLA09
Prokka is a tool designed for rapid annotation of prokaryotic genomes [31]. The genome-wide coding genes of strain GLA09 were annotated using universal database annotation; the statistical results are shown in Figure 2 and Figure 3. A total of 2076 functional genes were annotated, of which the number of genes that appeared in at least one database was 2062, which accounted for 99.33%.

Figure 2.
Database annotations of L. acidophilus GLA09. (A) General database annotation percentage statistics; (B) COG functional annotation; and (C) GO functional annotation of strain GLA09.

Figure 3.
Proprietary database annotations of L. acidophilus GLA09. (A) KEGG functional annotation of L. acidophilus GLA09; (B) Pathogen Host Interactions annotations; (C) TCDB transporter protein; (D) non-redundant annotations; and (E) carbohydrase annotations of strain GLA09.
3.2.1. COG Annotations
The COG database is a database developed by the NCBI for the annotation of homologous proteins. The strength of the COG database lies in the accuracy and comprehensiveness of its classification. There are 2005 genes in the whole genome of L. acidophilus GLA09 completed protein annotation. The highest number of annotations was for translation, ribosome structure, and biosynthesis with 183 genes, followed by carbohydrate transport and metabolism with 174 genes, major function prediction with 167 genes, amino acid transport metabolism annotated to 160 genes, and transcription annotated to 149 functional genes (Figure 2B).
3.2.2. GO Annotations
GO is a database created by the Gene Ontology Consortium that provides a comprehensive description of the properties of genes and gene products in organisms. A total of 1120 genes were annotated, and the percentage of genes with GO annotation was 53.95% (Figure 2C). Among the three major functional gene categories of GO, the top three functional annotations in biological processes were translation, transposition, DNA-mediated, and DNA integration; the more annotated genes in cellular composition were integrated. Among the cellular components, the most frequently annotated are an integral component of the membrane, cytoplasm, plasma membrane, and ribosome. Among the molecular functions, the most frequently annotated were binding, ATP binding, and DNA binding.
3.2.3. KEGG Annotations
The KEGG database is a systematic analysis of the metabolic pathways of gene products in cells and the functions of these gene products. A total of 1203 functional genes were annotated by KEGG for L. acidophilus GLA09 (Figure 3A). Metabolism had the highest proportion, while cellular processes had the lowest proportion. Among the metabolic processes, 128 functional genes were related to carbohydrate metabolism, 76 genes were associated with amino acid metabolism, and 61 genes were related to nucleotide metabolism. In addition, there were 124 genes in the environmental information processing process related to membrane transport.
3.2.4. TCDB Functional Notes and Pathogen Host Interactions
PHI annotation results show 215 Unaffected pathogenicity and 484 Reduced virulence (Figure 3B). Furthermore, a total of 421 membrane transporter protein-coding genes were annotated in the TCDB database (Figure 3C). Among them, the major active transporter proteins were annotated with 184 coding genes, followed by electrochemical potential-driven transporters with 94 coding genes.
3.2.5. Non-Redundant Protein Database
L. acidophilus GLA09 using the Non-Redundant Protein database, the results show that strain GLA09 with L. acidophilus had the highest rate, at 80.04% (Figure 3D).
3.2.6. CAZy Functional Notes
L. acidophilus GLA09 was annotated with a total of 56 carbohydrate-encoding genes in the CAZy database (Figure 3E). Glycoside hydrolase genes were annotated with 34, with the highest percentage of 60.71%, followed by glycosyltransferase genes with 19, with 33.93%; a carbohydrate-binding module was annotated with 2 genes, representing 3.57%, and carbohydrate lipase was annotated with only 1 gene.
3.3. L. acidophilus GLA09 Tolerance-Related Genes
L. acidophilus GLA09 annotated to 3 stress protein genes, 8 heat tolerance-related genes, 3 cold tolerance-related genes, 5 bile salt tolerance-related genes, 12 acid tolerance-related genes, and 14 antioxidant-related genes (Table 2).

Table 2.
Tolerance-related genes of Lactobacillus acidophilus GLA09.
3.4. L. acidophilus GLA09 Safety-Related Gene
Annotation using the CARD database was performed for L. acidophilus GLA09 with one lincosamide resistance gene (Table 3). Moreover, the number of virulence factors annotated to 8 with a complete match rate greater than 60% (Table 4).

Table 3.
Drug resistance gene prediction of L. acidophilus GLA09.

Table 4.
Prediction of virulence factor to Lactobacillus acidophilus GLA09.
3.5. Biogenic Amine Content of L. acidophilus GLA09
L. acidophilus GLA09 was not detected in the supernatant of biogenic amine, showing that the strain is safe (Table S2).
3.6. Secondary Metabolite Prediction and Comparative Genome
L. acidophilus GLA09 produced three bacteriocins as predicted in Figure 4, along with a comparative genome analysis (Figure S1). The first one was Bacteriocin Helveticin J, with a match of 99.61%, which was in the genome between 498,704~519,466 bp. The second one was Enterolysin A, with a match of 58.39%, which was in the genome. And, the third one is Helveticin J, with a matching degree of 44.66%, which all belong to the class III bacteriocins.

Figure 4.
Bacteriocin prediction of L. acidophilus GLA09. (A–C) Cluster of genes encoding secondary metabolites and protein tertiary structure prediction of (D) Bacteriocin Helveticin J; (E) Enterolysin A; and (F) Helveticin J.
3.7. Results of In Vitro Probiotic Potential Test of the Strain
L. acidophilus GLA09 exhibits an excellent bacteriostatic effect (Table S3) and superior adhesion. Moreover, it can withstand pH levels of ≥2.5 and is resistant to 0.3% (Figure 5).
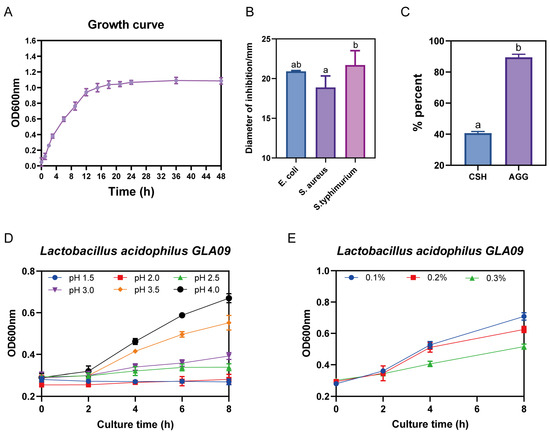
Figure 5.
Growth curve and probiotic potential evaluation of L. acidophilus GLA09. (A) Growth curve; (B) in vitro bacterial inhibition; (C) cell surface hydrophobicity and auto-aggregation ability; (D) acid resistance; and (E) bile salt resistance. Values are displayed as the mean ± SD. The different letters in the figure represent significant differences (p < 0.05), the same letter in the figure indicates no significant difference (p > 0.05).
4. Discussion
In this study, the whole genome of L. acidophilus GLA09 of canine origin was sequenced and mapped. The genome size of L. acidophilus GLA09 was 2.103 M, containing one chromosome and one plasmid, which belonged to the medium-sized genome. The G + C content was 38.71%, slightly higher than the average G+C content of L. acidophilus of 34.66% [1]. In this study, the strains were functionally annotated using GO, KEGG, and COG databases. The most functionally annotated genes in the GO database were integral components of the membrane, and the proteins encoded by these genes may have transmembrane transport abilities. Another significant annotation was ATP binding, which hydrolyzes ATP to provide energy for other biological processes. KEGG is a database that analyzes the metabolic pathways of gene products in cells and their functions of gene products at the molecular level [22]. L. acidophilus GLA09 had the most annotations in the KEGG database for metabolism-related genes. Studies have shown that L. acidophilus can produce large amounts of organic acids such as lactic acid, acetic acid, and butyric acid during fermentation. Additionally, it can metabolize and produce bacteriocins, extracellular polysaccharides, and other substances [32,33,34]. These metabolites contribute to the productivity of L. acidophilus and are utilized in various functional product developments, such as preventing diarrhea, lowering cholesterol, improving immunity, and other health-related products. Furthermore, the strains were determined to contain CRISPR sequences. The CRISPR/Cas system has been extensively utilized for genome editing. In the future, the development of efficient and precise genome editing tools is anticipated to enhance the research and application of gene functions [35].
Carbohydrates play a crucial role in many biological functions, and the CAZy database focuses on analyzing genomic, structural, and biochemical information on carbohydrate enzymes [36]. In this study, the highest number of glycoside hydrolase genes were annotated, demonstrating that glycoside hydrolases promote the hydrolysis of α-1,4 glycosidic bonds present in carbohydrates such as starch and also play a significant role in industrial production [37,38]. This study highlights that glycoside hydrolases can facilitate the hydrolysis of α-1,4 glycosidic bonds in carbohydrates such as starch, and are crucial for industrial production. Therefore, it is suggested that L. acidophilus GLA09 may enhance the digestive utilization of dietary carbohydrates or regulate glucose metabolism in the body.
L. acidophilus GLA09 genomic analysis revealed the presence of genes associated with resistance to cold and heat shock stress, bile salts, pH stress, and antioxidant stress. Firstly, concerning hot and cold stress, L. acidophilus is frequently exposed to high- or low-temperature extremes in industrial production. For example, when used as feed additives, high temperatures are generated during granulation and mixing or when fermented milk is subjected to prolonged low-temperature stress during cold storage [39,40]. The stresses of heat and cold can easily lead to the development of the bacterium. These hot and cold stresses can easily lead to sub-lethal or lethal conditions in the strains, thus reducing the probiotic function of the probiotics. In this study, L. acidophilus GLA09 was predicted to possess eight heat stress-related genes and three cold stress-related genes, suggesting that L. acidophilus GLA09 may exhibit better resistance to hot and cold stress, making it suitable for industrial applications. Secondly, probiotic bacteria can withstand low pH and high bile salt concentrations in the gastrointestinal tract, colonizing the intestine for long-lasting probiotic effects [41]. L. acidophilus GLA09 was predicted to have several genes related to pH tolerance and bile salt tolerance. Among them, the Na+/H+ ion reverse transporter protein and F0F1-ATP synthase have been shown to maintain intracellular pH homeostasis and pump H+ out of the cell to help the strain defend itself against the acidic environment [42]. The bile salt hydrolase gene may help to improve bile salt tolerance and increase the number of strains surviving in the gastrointestinal tract. The primary requirement for probiotics to be effective is their ability to withstand the harsh conditions of the stomach. GLA09 possesses multiple genes associated with resistance to pH stress and bile salts. This finding aligns with an in vitro study that demonstrated the strain’s capacity to tolerate pH levels of 3.0 and a wide range of bile salt concentrations.
In addition, oxidation is a necessary process for cellular metabolism in organisms, and the high levels of oxidative stress produced when organisms are subjected to oxidative stress can lead to aging as well as cause various chronic diseases [43]. L. acidophilus GLA has been reported as a natural antioxidant [44,45]. Several antioxidant-related genes were predicted in the whole genome of L. acidophilus GLA09. Therefore, it was hypothesized that L. acidophilus GLA09 might improve the antioxidant capacity of the body by increasing the activity of oxidative enzymes and so on. Additionally, three class III bacteriocins were predicted by the secondary metabolite database, containing two Helveticin J genes and one Enterolysin A gene. Bacteriocins produced by lactic acid bacteria are usually bacteriostatic, and Helveticin J bacteriocin is a chromosomally encoded bacteriocin with narrow-spectrum bacteriostatic effects. Enterolysin A bacteriocin has broader bacteriostatic effects by breaking down the cell walls of sensitive bacteria, including Streptococcus, Listeria monocytogenes, and Lactococcus lactis [46,47]. Therefore, it is suggested that L. acidophilus GLA09 may likewise be bacteriostatic and thus have the effect of regulating the intestinal flora.
The safety assessment of strains is another important consideration in the selection of potential probiotics. Although one lincosamide resistance gene was predicted, there was no risk of metastasis. Antibiotic resistance can occur through the horizontal transfer of antibiotic-resistance genes [48]. Mobile genetic elements, such as transposon-promoted enzyme expression, integrase, or recombinase, contribute to the initial mobilization. Mobile genetic elements are able to capture antibiotic resistance genes from chromosomes and transfer them horizontally to other bacteria via plasmids or phages [49,50]. In this study, strain GLA09 was predicted to have one lincosamide resistance gene by the CARD database, but it was located on the chromosome and had no repetitive sequences before or after it to constitute a transposon [51]. It was also not located on mobile genetic elements such as phages and gene islands as predicted by the genome, so it did not have the possibility of transfer and was not at risk of antibiotic transfer [52].
The virulence factors are divided into two categories: surface factors involved in host cell colonization and factors that lead to host tissue damage [53]. Classification is ambiguous in probiotics, and a large number of genes found in the VRDB are associated with virulence factors in some pathogens but with adaptive factors in probiotics [52]. The VFBD database is dedicated to collecting information on the virulence factors of bacterial pathogens. There are eight virulence genes with greater than 60% genetic similarity predicted in the whole genome of L. acidophilus GLA09. Of these, EF-Tu and GroEL are adhesion-associated factors that promote the adhesion and colonization of strains within the stomach [54]. The two-component response regulator LisR is ubiquitous in bacteria, facilitating their adaptation to diverse environments and playing a critical role in the regulation of oxidative stress [55]. In addition, the GLA09 genome is predicted to have a UTP, glucose-1-phosphate uridylyltransferase GalU, which enhances the resistance of the strain to freeze-drying [56]. There is a fraction of pathogenic virulence factors, but there is also a fraction of virulence factors that are not involved in pathogenicity but are essential for probiotics and are health factors. Therefore, whether these virulence genes are expressed in L. acidophilus GLA09 needs to be further verified. The PHI database is currently commonly used for the detection of pathogenicity genes, virulence genes, and effector protein genes in bacteria. The L. acidophilus GLA09 genome had the highest number of virulence-reducing genes annotated at 484, followed by 215 genes annotated with no effect on pathogenicity. The presence of many virulence-reducing and non-pathogenicity genes suggests that L. acidophilus GLA09 may have a pathogenicity-suppressing effect. Acceptable reference limits for histamine and tyramine are 0–100 and 100–800 mg/Kg [57,58]. For other amines, no criteria have been established in the literature. According to the Chinese national standard GB/T 5009.208-2016 [59], the minimum limit of BAs is 2–5 ug/mL. In this study, strain GLA09 produced less than 2 ug/mL of biogenic amines in all cases, and the strain is safe [60,61].
Although this study effectively established a connection between the genome and the probiotic properties, further validation of metabolites (bacteriocins), native animal testing, and assessments of product industrialization are necessary to provide substantial technical support for the strain as a probiotic product for pets. To ensure the quality of our probiotic product, GLA09, it is essential to establish standardized production processes. This entails optimizing culture conditions, as well as handling and storage practices, to guarantee the product’s effectiveness and stability.
5. Conclusions
In this study, the whole genome of L. acidophilus GLA09 was sequenced, and its genes were functionally annotated to analyze the probiotic properties and safety of the strain at the genetic level. The genome mainly related to functional genes includes carbohydrate metabolism, amino acid metabolism, nucleotide metabolism, and three predicted genes coding for bacteriocins. Additionally, a significant number of probiotic-related genes were identified in the L. acidophilus GLA09 genome, including trxA, mvaA, trxB, TRR, pcaC, GSR, gor, and fabG. These genes are closely associated with antioxidant functions and contribute to the unique genomic features of this strain. The safety of the strain was elucidated at the gene level. Although one lincosamide resistance gene was predicted, there was no risk of metastasis. Therefore, L. acidophilus GLA09 of canine origin has excellent probiotic properties and safety and can be a candidate strain for probiotics.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/microorganisms13020350/s1, Figure S1: Comparative genomic covariance map of L. acidophilus GLA09. Table S1: Prediction of CRISPRs to L. acidophilus GLA09. Table S2. Biogenic amine content of L. acidophilus GLA09. Table S3. Inhibition test results of L. acidophilus GLA09.
Author Contributions
G.L., conceptualization, funding acquisition; M.Z. and Y.Z., investigation, methodology, visualization, and writing—original draft; Y.L., writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
The manuscript was supported by the High-level Talents Fund of Qingdao Agricultural University (1121021) to G.L.
Institutional Review Board Statement
The laboratory animals needed for this study were approved by the Laboratory Animal Ethics Committee of Qingdao Agricultural University (grant No. DWKJ202304044; Qingdao, China).
Informed Consent Statement
Not applicable.
Data Availability Statement
The whole genome sequencing results have been uploaded to the NCBI database under the project number PRJNA1028138 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1028138/, accessed on 14 October 2023).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| WGS | Whole-genome sequencing |
| MRS | MRS broth medium |
| LB | Luria-Bertani |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopaedia of Genes and Genomes |
| COG | Cluster of Orthologous Groups of Proteins |
| CAZy | Carbohydrate-Active EnZymes database |
| PHI | Pathogen Host Interactions Database |
| CARD | Comprehensive Antibiotic Research Database |
| VFDB | Virulence Factor Database |
| SD | Standard deviation |
| G + C | Guanine + Cytosine |
References
- Huang, Z.; Zhou, X.; Stanton, C.; Ross, R.P.; Zhao, J.; Zhang, H.; Yang, B.; Chen, W. Comparative Genomics and Specific Functional Characteristics Analysis of Lactobacillus Acidophilus. Microorganisms 2021, 9, 1992. [Google Scholar] [CrossRef] [PubMed]
- Altermann, E.; Russell, W.M.; Azcarate-Peril, M.A.; Barrangou, R.; Buck, B.L.; McAuliffe, O.; Souther, N.; Dobson, A.; Duong, T.; Callanan, M.; et al. Complete Genome Sequence of the Probiotic Lactic Acid Bacterium Lactobacillus Acidophilus NCFM. Proc. Natl. Acad. Sci. USA 2005, 102, 3906–3912. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gu, Y.; Fang, K.; Mao, K.; Dou, J.; Fan, H.; Zhou, C.; Wang, H. Lactobacillus Acidophilus and Clostridium Butyricum Ameliorate Colitis in Murine by Strengthening the Gut Barrier Function and Decreasing Inflammatory Factors. Benef. Microbes 2018, 9, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, L.L.; Hoffmann, C.; Lacorte, G.A.; de Melo Franco, B.D.G.; Todorov, S.D. Genomic and Functional Characterization of Bacteriocinogenic Lactic Acid Bacteria Isolated from Boza, a Traditional Cereal-Based Beverage. Sci. Rep. 2022, 12, 1460. [Google Scholar] [CrossRef]
- Yan, F.; Li, N.; Shi, J.; Li, H.; Yue, Y.; Jiao, W.; Wang, N.; Song, Y.; Huo, G.; Li, B. Lactobacillus Acidophilus Alleviates Type 2 Diabetes by Regulating Hepatic Glucose, Lipid Metabolism and Gut Microbiota in Mice. Food Funct. 2019, 10, 5804–5815. [Google Scholar] [CrossRef]
- De Cesare, A.; Sirri, F.; Manfreda, G.; Moniaci, P.; Giardini, A.; Zampiga, M.; Meluzzi, A. Effect of Dietary Supplementation with Lactobacillus Acidophilus D2/CSL (CECT 4529) on Caecum Microbioma and Productive Performance in Broiler Chickens. PLoS ONE 2017, 12, e0176309. [Google Scholar] [CrossRef]
- Song, M.; Park, S.; Lee, H.; Min, B.; Jung, S.; Park, S.; Kim, E.; Oh, S. Effect of Lactobacillus Acidophilus NS1 on Plasma Cholesterol Levels in Diet-Induced Obese Mice. J. Dairy Sci. 2015, 98, 1492–1501. [Google Scholar] [CrossRef]
- Li, L.; Jiang, Y.-J.; Yang, X.-Y.; Liu, Y.; Wang, J.-Y.; Man, C.-X. Immunoregulatory Effects on Caco-2 Cells and Mice of Exopolysaccharides Isolated from Lactobacillus Acidophilus NCFM. Food Funct. 2014, 5, 3261–3268. [Google Scholar] [CrossRef]
- Deepak, V.; Sundar, W.A.; Pandian, S.R.K.; Sivasubramaniam, S.D.; Hariharan, N.; Sundar, K. Exopolysaccharides from Lactobacillus Acidophilus Modulates the Antioxidant Status of 1,2-Dimethyl Hydrazine-Induced Colon Cancer Rat Model. 3 Biotech. 2021, 11, 225. [Google Scholar] [CrossRef]
- Douillard, F.P.; Ribbera, A.; Kant, R.; Pietilä, T.E.; Järvinen, H.M.; Messing, M.; Randazzo, C.L.; Paulin, L.; Laine, P.; Ritari, J.; et al. Comparative Genomic and Functional Analysis of 100 Lactobacillus Rhamnosus Strains and Their Comparison with Strain GG. PLoS Genet. 2013, 9, e1003683. [Google Scholar] [CrossRef]
- Shi, S.; Dong, J.; Cheng, X.; Hu, J.; Liu, Y.; He, G.; Zhang, J.; Yu, H.; Jia, L.; Zhou, D. Biological Characteristics and Whole-Genome Analysis of the Potential Probiotic, Lactobacillus Reuteri S5. Lett. Appl. Microbiol. 2022, 74, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wang, Y.; Gu, L.; Yu, J.; Liu, Q.; Zhang, R.; Liang, G.; Chen, H.; Gu, F.; Liu, H.; et al. Characterization of Probiotic Properties and Whole-Genome Analysis of Lactobacillus Johnsonii N5 and N7 Isolated from Swine. Microorganisms 2024, 12, 672. [Google Scholar] [CrossRef] [PubMed]
- Sundararaman, A.; Bansal, K.; Sidhic, J.; Patil, P.; Halami, P.M. Genome of Bifidobacterium Longum NCIM 5672 Provides Insights into Its Acid-Tolerance Mechanism and Probiotic Properties. Arch. Microbiol. 2021, 203, 6109–6118. [Google Scholar] [CrossRef] [PubMed]
- Kiousi, D.E.; Rathosi, M.; Tsifintaris, M.; Chondrou, P.; Galanis, A. Pro-Biomics: Omics Technologies to Unravel the Role of Probiotics in Health and Disease. Adv. Nutr. 2021, 12, 1802–1820. [Google Scholar] [CrossRef]
- Dowarah, R.; Verma, A.K.; Agarwal, N.; Singh, P.; Singh, B.R. Selection and Characterization of Probiotic Lactic Acid Bacteria and Its Impact on Growth, Nutrient Digestibility, Health and Antioxidant Status in Weaned Piglets. PLoS ONE 2018, 13, e0192978. [Google Scholar] [CrossRef]
- Johnson, A.; Miller, E.A.; Weber, B.; Figueroa, C.F.; Aguayo, J.M.; Johny, A.K.; Noll, S.; Brannon, J.; Kozlowicz, B.; Johnson, T.J. Evidence of Host Specificity in Lactobacillus Johnsonii Genomes and Its Influence on Probiotic Potential in Poultry. Poult. Sci. 2023, 102, 102858. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Han, G.G.; Choi, J.; Jin, G.-D.; Kang, S.-K.; Chae, B.J.; Kim, E.B.; Choi, Y.-J. Pan-Genomic Approaches in Lactobacillus Reuteri as a Porcine Probiotic: Investigation of Host Adaptation and Antipathogenic Activity. Microb. Ecol. 2017, 74, 709–721. [Google Scholar] [CrossRef]
- Casarotti, S.N.; Carneiro, B.M.; Todorov, S.D.; Nero, L.A.; Rahal, P.; Penna, A.L.B. In Vitro Assessment of Safety and Probiotic Potential Characteristics of Lactobacillus Strains Isolated from Water Buffalo Mozzarella Cheese. Ann. Microbiol. 2017, 67, 289–301. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG Database: A Tool for Genome-Scale Analysis of Protein Functions and Evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG Resource for Deciphering the Genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef] [PubMed]
- Saier, M.H.; Tran, C.V.; Barabote, R.D. TCDB: The Transporter Classification Database for Membrane Transport Protein Analyses and Information. Nucleic Acids Res. 2006, 34, D181–D186. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Urban, M.; Cuzick, A.; Seager, J.; Wood, V.; Rutherford, K.; Venkatesh, S.Y.; Sahu, J.; Iyer, S.V.; Khamari, L.; De Silva, N.; et al. PHI-Base in 2022: A Multi-Species Phenotype Database for Pathogen-Host Interactions. Nucleic Acids Res. 2022, 50, D837–D847. [Google Scholar] [CrossRef]
- Lorencová, E.; Buňková, L.; Matoulková, D.; Dráb, V.; Pleva, P.; Kubáň, V.; Buňka, F. Production of Biogenic Amines by Lactic Acid Bacteria and Bifidobacteria Isolated from Dairy Products and Beer. Int. J. Food Sci. Technol. 2012, 47, 2086–2091. [Google Scholar] [CrossRef]
- Zhao, M.; Zhang, Y.; Li, Y.; Liu, K.; Zhang, C.; Li, G. Complete Genome Sequence and Probiotic Properties of Pediococcus Acidilactici CLP03 Isolated from Healthy Felis Catus. Probiotics Antimicrob. Proteins 2023, 1–15. [Google Scholar] [CrossRef]
- Chen, T.; Wang, L.; Li, Q.; Long, Y.; Lin, Y.; Yin, J.; Zeng, Y.; Huang, L.; Yao, T.; Abbasi, M.N.; et al. Functional Probiotics of Lactic Acid Bacteria from Hu Sheep Milk. BMC Microbiol. 2020, 20, 228. [Google Scholar] [CrossRef]
- Liu, X.; Lv, X.; Sun, Y.; Liu, C.; Wang, R.; Liu, R.; Ma, Y.; Li, Q. Probiotic Properties of Lacticaseibacillus Rhamnosus Grx10 Revolved with Complete Genome. Food Biosci. 2023, 51, 102219. [Google Scholar] [CrossRef]
- Ramos, C.L.; Thorsen, L.; Schwan, R.F.; Jespersen, L. Strain-Specific Probiotics Properties of Lactobacillus fermentum, Lactobacillus plantarum and Lactobacillus brevis Isolates from Brazilian Food Products. Food Microbiol. 2013, 36, 22–29. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Liong, M.T.; Shah, N.P. Production of Organic Acids from Fermentation of Mannitol, Fructooligosaccharide and Inulin by a Cholesterol Removing Lactobacillus Acidophilus Strain. J. Appl. Microbiol. 2005, 99, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Anjum, N.; Maqsood, S.; Masud, T.; Ahmad, A.; Sohail, A.; Momin, A. Lactobacillus Acidophilus: Characterization of the Species and Application in Food Production. Crit. Rev. Food Sci. Nutr. 2014, 54, 1241–1251. [Google Scholar] [CrossRef] [PubMed]
- Amiri, S.; Rezaei Mokarram, R.; Sowti Khiabani, M.; Rezazadeh Bari, M.; Alizadeh Khaledabad, M. Exopolysaccharides Production by Lactobacillus acidophilus LA5 and Bifidobacterium animalis Subsp. Lactis BB12: Optimization of Fermentation Variables and Characterization of Structure and Bioactivities. Int. J. Biol. Macromol. 2019, 123, 752–765. [Google Scholar] [CrossRef]
- Sander, J.D.; Joung, J.K. CRISPR-Cas Systems for Editing, Regulating and Targeting Genomes. Nat. Biotechnol. 2014, 32, 347–355. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes Database (CAZy): An Expert Resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Gangoiti, J.; Pijning, T.; Dijkhuizen, L. Biotechnological Potential of Novel Glycoside Hydrolase Family 70 Enzymes Synthesizing α-Glucans from Starch and Sucrose. Biotechnol. Adv. 2018, 36, 196–207. [Google Scholar] [CrossRef]
- Plaza-Vinuesa, L.; Hernandez-Hernandez, O.; Moreno, F.J.; de Las Rivas, B.; Muñoz, R. Unravelling the Diversity of Glycoside Hydrolase Family 13 α-Amylases from Lactobacillus Plantarum WCFS1. Microb. Cell Fact. 2019, 18, 183. [Google Scholar] [CrossRef]
- Belhadj Slimen, I.; Najar, T.; Ghram, A.; Abdrrabba, M. Heat Stress Effects on Livestock: Molecular, Cellular and Metabolic Aspects, a Review. J. Anim. Physiol. Anim. Nutr. 2016, 100, 401–412. [Google Scholar] [CrossRef]
- Zhang, M.; Yao, M.; Lai, T.; Zhao, H.; Wang, Y.; Yang, Z. Response of Lactiplantibacillus Plantarum NMGL2 to Combinational Cold and Acid Stresses during Storage of Fermented Milk as Analyzed by Data-Independent Acquisition Proteomics. Foods 2021, 10, 1514. [Google Scholar] [CrossRef]
- Reuben, R.C.; Roy, P.C.; Sarkar, S.L.; Alam, R.-U.; Jahid, I.K. Isolation, Characterization, and Assessment of Lactic Acid Bacteria toward Their Selection as Poultry Probiotics. BMC Microbiol. 2019, 19, 253. [Google Scholar] [CrossRef] [PubMed]
- Counillon, L.; Bouret, Y.; Marchiq, I.; Pouysségur, J. Na+/H+ Antiporter (NHE1) and Lactate/H+ Symporters (MCTs) in pH Homeostasis and Cancer Metabolism. Biochim. Biophys. Acta BBA—Mol. Cell Res. 2016, 1863, 2465–2480. [Google Scholar] [CrossRef]
- Liguori, I.; Russo, G.; Curcio, F.; Bulli, G.; Aran, L.; Della-Morte, D.; Gargiulo, G.; Testa, G.; Cacciatore, F.; Bonaduce, D.; et al. Oxidative Stress, Aging, and Diseases. Clin. Interv. Aging 2018, 13, 757–772. [Google Scholar] [CrossRef] [PubMed]
- Khalafalla, M.M.; Zayed, N.F.A.; Amer, A.A.; Soliman, A.A.; Zaineldin, A.I.; Gewaily, M.S.; Hassan, A.M.; Van Doan, H.; Tapingkae, W.; Dawood, M.A.O. Dietary Lactobacillus Acidophilus ATCC 4356 Relieves the Impacts of Aflatoxin B1 Toxicity on the Growth Performance, Hepatorenal Functions, and Antioxidative Capacity of Thinlip Grey Mullet (Liza Ramada) (Risso 1826). Probiotics Antimicrob. Proteins 2022, 14, 189–203. [Google Scholar] [CrossRef] [PubMed]
- Xia, M.; Li, C.; Wu, D.; Wu, F.; Kong, L.; Jia, Z.; Han, W.; Chen, S.; Fang, W.; Liu, Y.; et al. Benefits of Heat-Killed Lactobacillus Acidophilus on Growth Performance, Nutrient Digestibility, Antioxidant Status, Immunity, and Cecal Microbiota of Rabbits. Front. Vet. Sci. 2024, 11, 1361908. [Google Scholar] [CrossRef] [PubMed]
- Nilsen, T.; Nes, I.F.; Holo, H. Enterolysin A, a Cell Wall-Degrading Bacteriocin from Enterococcus Faecalis LMG 2333. Appl. Environ. Microbiol. 2003, 69, 2975–2984. [Google Scholar] [CrossRef]
- Tse, T.J.; Shen, J.; Shim, Y.Y.; Reaney, M.J.T. Changes in Bacterial Populations and Their Metabolism over 90 Sequential Cultures on Wheat-Based Thin Stillage. J. Agric. Food Chem. 2020, 68, 4717–4729. [Google Scholar] [CrossRef]
- Sommer, M.O.A.; Munck, C.; Toft-Kehler, R.V.; Andersson, D.I. Prediction of Antibiotic Resistance: Time for a New Preclinical Paradigm? Nat. Rev. Microbiol. 2017, 15, 689–696. [Google Scholar] [CrossRef]
- Jiang, X.; Ellabaan, M.M.H.; Charusanti, P.; Munck, C.; Blin, K.; Tong, Y.; Weber, T.; Sommer, M.O.A.; Lee, S.Y. Dissemination of Antibiotic Resistance Genes from Antibiotic Producers to Pathogens. Nat. Commun. 2017, 8, 15784. [Google Scholar] [CrossRef]
- Bennett, P.M. Plasmid Encoded Antibiotic Resistance: Acquisition and Transfer of Antibiotic Resistance Genes in Bacteria. Br. J. Pharmacol. 2008, 153 (Suppl. S1), S347–S357. [Google Scholar] [CrossRef]
- Yang, Y.; Xie, S.; He, F.; Xu, Y.; Wang, Z.; Ihsan, A.; Wang, X. Recent Development and Fighting Strategies for Lincosamide Antibiotic Resistance. Clin. Microbiol. Rev. 2024, 37, e0016123. [Google Scholar] [CrossRef] [PubMed]
- Senan, S.; Prajapati, J.B.; Joshi, C.G. Feasibility of Genome-Wide Screening for Biosafety Assessment of Probiotics: A Case Study of Lactobacillus Helveticus MTCC 5463. Probiotics Antimicrob. Proteins 2015, 7, 249–258. [Google Scholar] [CrossRef]
- Terzić-Vidojević, A.; Veljović, K.; Popović, N.; Tolinački, M.; Golić, N. Enterococci from Raw-Milk Cheeses: Current Knowledge on Safety, Technological, and Probiotic Concerns. Foods 2021, 10, 2753. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, H.; Ohuchi, S.; Arakawa, K.; Watanabe, M.; Kitazawa, H.; Saito, T. Isolation of Lactic Acid Bacteria Bound to the Porcine Intestinal Mucosa and an Analysis of Their Moonlighting Adhesins. Biosci. Microbiota Food Health 2016, 35, 185–196. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, S.; Du, J.; Li, Y.; Wu, M.; Chen, S.; Jiang, S.; Zhan, L.; Huang, X. LiaSR Two-Component System Modulates the Oxidative Stress Response in Streptococcus mutans. Microb. Pathog. 2023, 185, 106404. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Zeng, X.; Guo, Y.; Wu, Z.; Cai, Z.; Pan, D. Determining the Role of UTP-Glucose-1-Phosphate Uridylyltransferase (GalU) in Improving the Resistance of Lactobacillus Acidophilus NCFM to Freeze-Drying. Foods 2022, 11, 1719. [Google Scholar] [CrossRef]
- Deepika Priyadarshani, W.M.; Rakshit, S.K. Screening Selected Strains of Probiotic Lactic Acid Bacteria for Their Ability to Produce Biogenic Amines (Histamine and Tyramine). Int. J. Food Sci. Technol. 2011, 46, 2062–2069. [Google Scholar] [CrossRef]
- Roselino, M.N.; Maciel, L.F.; Sirocchi, V.; Caviglia, M.; Sagratini, G.; Vittori, S.; Taranto, M.P.; Cavallini, D.C.U. Analysis of Biogenic Amines in Probiotic and Commercial Salamis. J. Food Compos. Anal. 2020, 94, 103649. [Google Scholar] [CrossRef]
- GB/T 5009.208; National Standards for Food Safety—Determination of Biogenic Amines in Food. National Health and Family Planning Commission of the People’s Republic of China and China Food and Drug Administration: Beijing, China, 2017.
- Barbieri, F.; Montanari, C.; Gardini, F.; Tabanelli, G. Biogenic Amine Production by Lactic Acid Bacteria: A Review. Foods 2019, 8, 17. [Google Scholar] [CrossRef]
- Saha Turna, N.; Chung, R.; McIntyre, L. A Review of Biogenic Amines in Fermented Foods: Occurrence and Health Effects. Heliyon 2024, 10, e24501. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).