Impact of Feed Composition on Rumen Microbial Dynamics and Phenotypic Traits in Beef Cattle
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Study
2.2. Rumen Fluid Sampling
2.3. Feed Chemical Analysis
2.4. DNA Extraction
2.5. Quantitative Real-Time PCR Analysis
2.6. Statistical Analysis
3. Results
3.1. Effect of Diet on Phenotypic Traits and Rumen Microbial Counts
3.2. Associations Between Phenotypic Traits and Rumen Microbial Counts Across Feeding Periods
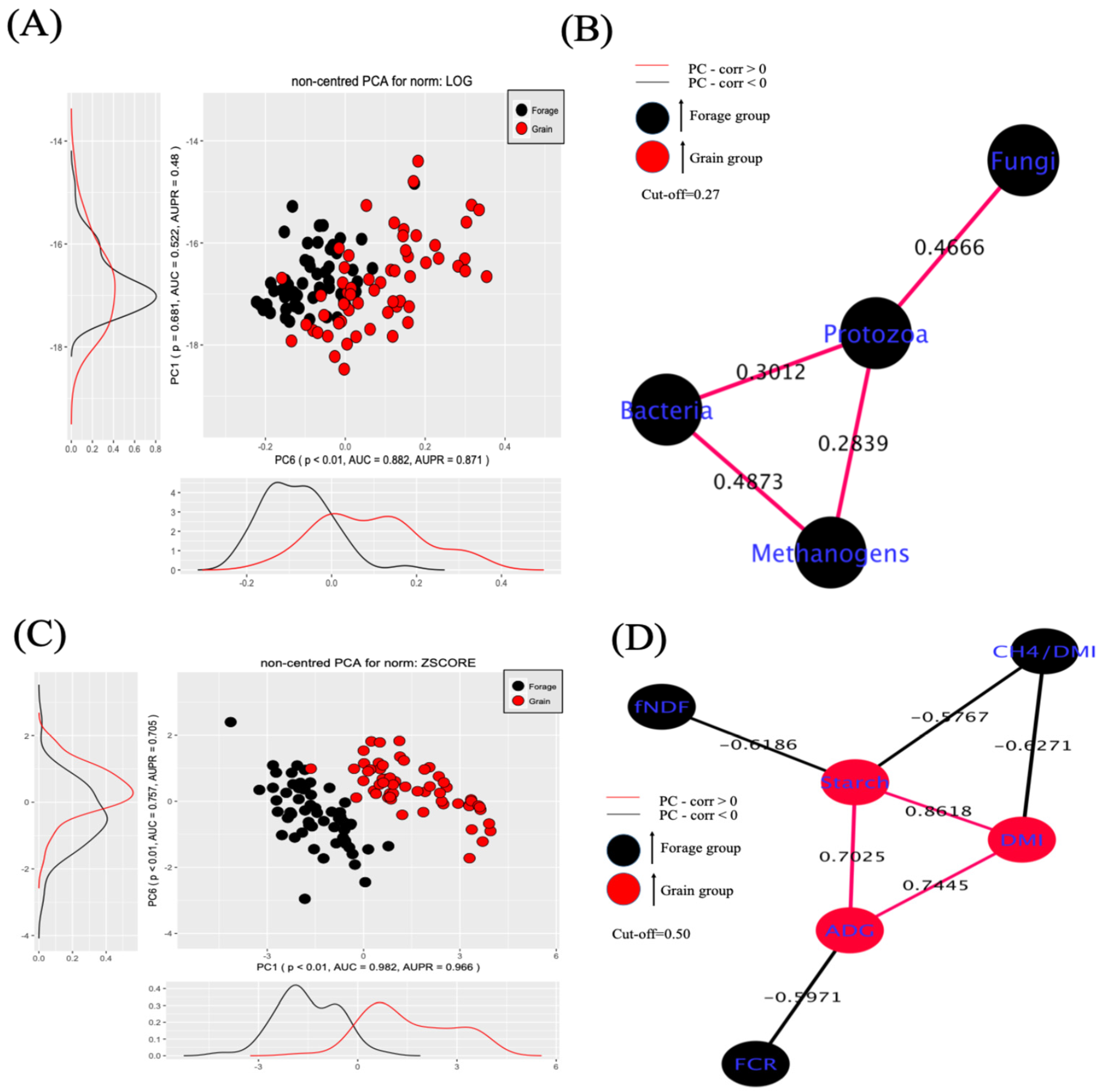
3.3. PC-Corr Analysis of Microbial Counts and Phenotypic Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McAllister, T.A.; Bae, H.D.; Jones, G.A.; Cheng, K.J. Microbial attachment and feed digestion in the rumen. J. Anim. Sci. 1994, 72, 3004–3018. [Google Scholar] [CrossRef] [PubMed]
- Hook, S.E.; Wright, A.-D.G.; McBride, B.W. Methanogens: Methane producers of the rumen and mitigation strategies. Archaea 2010, 2010, 945785. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.A.; Johnson, D.E. Methane emissions from cattle. J. Anim. Sci. 1995, 73, 2483–2492. [Google Scholar] [CrossRef] [PubMed]
- Morgavi, D.P.; Forano, E.; Martin, C.; Newbold, C.J. Microbial ecosystem and methanogenesis in ruminants. Animal 2010, 4, 1024–1036. [Google Scholar] [CrossRef] [PubMed]
- Huws, S.A.; Creevey, C.J.; Oyama, L.B.; Mizrahi, I.; Denman, S.E.; Popova, M.; Muñoz-Tamayo, R.; Forano, E.; Waters, S.M.; Hess, M.; et al. Addressing global ruminant agricultural challenges through understanding the rumen microbiome: Past, present, and future. Front. Microbiol. 2018, 9, 2161. [Google Scholar] [CrossRef]
- Dehority, B.A. Rumen Microbiology; Nottingham University Press: Nottingham, UK, 2003; Volume 113. [Google Scholar]
- Russell, J.B.; Rychlik, J.L. Factors that alter rumen microbial ecology. Science 2001, 292, 1119–1122. [Google Scholar] [CrossRef] [PubMed]
- Grubb, J.A.; Dehority, B.A. Effects of an abrupt change in ration from all roughage to high concentrate upon rumen microbial numbers in sheep. Appl. Microbiol. 1975, 30, 404–412. [Google Scholar] [CrossRef]
- Janssen, P.H. Influence of hydrogen on rumen methane formation and fermentation balances through microbial growth kinetics and fermentation thermodynamics. Anim. Feed Sci. Technol. 2010, 160, 1–22. [Google Scholar] [CrossRef]
- Beauchemin, K.A.; Kreuzer, M.; O’Mara, F.; McAllister, T.A. Nutritional management for enteric methane abatement: A review. Aust. J. Exp. Agric. 2008, 48, 21–27. [Google Scholar] [CrossRef]
- Ramos, S.C.; Jeong, C.D.; Mamuad, L.L.; Kim, S.H.; Kang, S.H.; Kim, E.T.; Cho, Y.I.; Lee, S.S.; Lee, S.S. Diet transition from high-forage to high-concentrate alters rumen bacterial community composition, epithelial transcriptomes and ruminal fermentation parameters in dairy cows. Animals 2021, 11, 838. [Google Scholar] [CrossRef] [PubMed]
- Petri, R.M.; Schwaiger, T.; Penner, G.B.; Beauchemin, K.A.; Forster, R.J.; McKinnon, J.J.; McAllister, T.A. Changes in the Rumen Epimural Bacterial Diversity of Beef Cattle as Affected by Diet and Induced Ruminal Acidosis. Appl. Environ. Microbiol. 2013, 79, 3744–3755. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, H.; Wang, Y.; Li, S.; Cao, Z.; Ji, S.; He, Y.; Zhang, H. Effect of Dietary Forage to Concentrate Ratios on Dynamic Profile Changes and Interactions of Ruminal Microbiota and Metabolites in Holstein Heifers. Front. Microbiol. 2017, 8, 2206. [Google Scholar] [CrossRef] [PubMed]
- Olfert, E.; Cross, B.; McWilliams, A. Guide to the Care and Use of Experimental Steers; Canadian Council on Animal Care: Ottawa, ON, Canada, 1993. [Google Scholar]
- Montanholi, Y.R.; Swanson, K.C.; Palme, R.; Schenkel, F.S.; McBride, B.W.; Lu, D.; Miller, S.P. Assessing feed efficiency in beef steers through feeding behavior, infrared thermography and glucocorticoids. Animal 2010, 4, 692–701. [Google Scholar] [CrossRef] [PubMed]
- Koch, R.M.; Swiger, L.A.; Chambers, D.; Gregory, K.E. Efficiency of feed use in beef cattle. J. Anim. Sci. 1963, 22, 486–494. [Google Scholar] [CrossRef]
- Basarab, J.A.; Price, M.A.; Aalhus, J.L.; Okine, E.K.; Snelling, W.M.; Lyle, K.L. Residual feed intake and body composition in young growing cattle. Can. J. Anim. Sci. 2003, 83, 189–204. [Google Scholar] [CrossRef]
- Berry, D.P.; Crowley, J.J. Residual intake and body weight gain: A new measure of efficiency in growing cattle. J. Anim. Sci. 2012, 90, 109–115. [Google Scholar] [CrossRef]
- NRC. Nutrient Requirements of Beef Cattle, 8th Revised ed.; National Academies of Sciences, Engineering, and Medicine, Ed.; The National Academies Press: Washington, DC, USA, 2016. [Google Scholar]
- Berndt, A.; Boland, T.; Deighton, M.; Gere, J.; Grainger, C.; Hegarty, R.; Iwaasa, A.; Koolaard, J.; Lassey, K.; Luo, D. Guidelines for Use of Sulphur Hexafluoride (SF6) Tracer Technique to Measure Enteric Methane Emissions from Ruminants; Ministry of Primary Industries: Wellington, New Zealand, 2014; pp. 1–66.
- Deighton, M.H.; Williams, S.R.O.; Hannah, M.C.; Eckard, R.J.; Boland, T.M.; Wales, W.J.; Moate, P.J. A modified sulphur hexafluoride tracer technique enables accurate determination of enteric methane emissions from ruminants. Anim. Feed Sci. Technol. 2014, 197, 47–63. [Google Scholar] [CrossRef]
- Duffield, T.; Plaizier, J.C.; Fairfield, A.; Bagg, R.; Vessie, G.; Dick, P.; Wilson, J.; Aramini, J.; McBride, B. Comparison of techniques for measurement of rumen pH in lactating dairy cows. J. Dairy Sci. 2004, 87, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Thompson, S. The Effect of Diet Type on Residual Feed Intake and the Use of Infrared Thermography as a Method to Predict Efficiency in Beef Bulls; University of Manitoba: Winnipeg, MB, Canada, 2015. [Google Scholar]
- AOAC. Official Methods of Analysis (17th); Association of Official Analytical Chemists: Gaithersburg, MD, USA, 2000. [Google Scholar]
- Van Soest, P.J.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.B. Determination of Starch, Including Maltooligosaccharides, in Animal Feeds: Comparison of Methods and a Method Recommended for AOAC Collaborative Study. J. Aoac Int. 2009, 92, 42–49. [Google Scholar] [CrossRef]
- Li, M.; Penner, G.B.; Hernandez-Sanabria, E.; Oba, M.; Guan, L.L. Effects of sampling location and time, and host animal on assessment of bacterial diversity and fermentation parameters in the bovine rumen. J. Appl. Microbiol. 2009, 107, 1924–1934. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, D.M.; Weimer, P.J. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl. Microbiol. Biotechnol. 2007, 75, 165–174. [Google Scholar] [CrossRef]
- Zhou, M.; Hernandez-Sanabria, E.; Guan, L.L. Assessment of the microbial ecology of ruminal methanogens in cattle with different feed efficiencies. Appl. Environ. Microbiol. 2009, 75, 6524–6533. [Google Scholar] [CrossRef] [PubMed]
- Romero-Perez, A.; Okine, E.K.; McGinn, S.M.; Guan, L.L.; Oba, M.; Duval, S.M.; Kindermann, M.; Beauchemin, K.A. The potential of 3-nitrooxypropanol to lower enteric methane emissions from beef cattle. J. Anim. Sci. 2014, 92, 4682–4693. [Google Scholar] [CrossRef] [PubMed]
- Denman, S.E.; McSweeney, C.S. Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. Fems Microbiol. Ecol. 2006, 58, 572–582. [Google Scholar] [CrossRef]
- Malmuthuge, N.; Li, M.; Chen, Y.; Fries, P.; Griebel, P.J.; Baurhoo, B.; Zhao, X.; Guan, L.L. Distinct commensal bacteria associated with ingesta and mucosal epithelium in the gastrointestinal tracts of calves and chickens. Fems Microbiol. Ecol. 2012, 79, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.-A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]
- Ciucci, S.; Ge, Y.; Durán, C.; Palladini, A.; Martínez-Sánchez, L.M.; Cannistraci, C.V.; Gerl, M.J.; Jiménez-Jiménez, V.; Wang, Y.; Sales, S.; et al. Enlightening discriminative network functional modules behind principal component analysis separation in differential-omic science studies. Sci. Rep. 2017, 7, srep43946. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Leng, R. Quantitative ruminant nutrition—A green science. Aust. J. Agric. Res. 1993, 44, 363–380. [Google Scholar] [CrossRef]
- Hackmann, T.J.; Firkins, J.L. Maximizing efficiency of rumen microbial protein production. Front. Microbiol. 2015, 6, 465. [Google Scholar] [CrossRef] [PubMed]
- Van Soest, P.J. Nutritional Ecology of the Ruminant; Cornell University Press: Ithaca, NY, USA, 1994. [Google Scholar]
- Varga, G.A.; Kolver, E.S. Microbial and animal limitations to fiber digestion and utilization. J. Nutr. 1997, 127, 819S–823S. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.W.; Abdullah, N.; Jalaludin, S. Penetrating structures of anaerobic rumen fungi in cattle and swamp buffalo. J. Gen. Microbiol. 1988, 134, 177–182. [Google Scholar] [CrossRef][Green Version]
- Lee, S.S.; Ha, J.K.; Cheng, K.J. Relative contributions of bacteria, protozoa, and fungi to in vitro degradation of orchard grass cell walls and their interactions. Appl. Environ. Microbiol. 2000, 66, 3807–3813. [Google Scholar] [CrossRef] [PubMed]
- McAllister, T.A.; Thomas, K.D.; Gruninger, R.J.; Elshahed, M.; Li, Y.; Cheng, Y. Rumen fungi, archaea and their interactions. J. Dairy Sci. (Press) 2024, 151, 41–61. [Google Scholar]
- Finlay, B.J.; Esteban, G.; Clarke, K.J.; Williams, A.G.; Embley, T.M.; Hirt, R.P. Some rumen ciliates have endosymbiotic methanogens. FEMS Microbiol. Lett. 1994, 117, 157–161. [Google Scholar] [CrossRef]
- Zhou, M.; Chung, Y.H.; Beauchemin, K.A.; Holtshausen, L.; Oba, M.; McAllister, T.A.; Guan, L.L. Relationship between rumen methanogens and methane production in dairy cows fed diets supplemented with a feed enzyme additive. J. Appl. Microbiol. 2011, 111, 1148–1158. [Google Scholar] [CrossRef]
- Newbold, C.J.; Ushida, K.; Morvan, B.; Fonty, G.; Jouany, J.P. The role of ciliate protozoa in the lysis of methanogenic archaea in rumen fluid. Lett. Appl. Microbiol. 1996, 23, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Duin, E.C.; Wagner, T.; Shima, S.; Prakash, D.; Cronin, B.; Yáñez-Ruiz, D.R.; Duval, S.; Rümbeli, R.; Stemmler, R.T.; Thauer, R.K.; et al. Mode of action uncovered for the specific reduction of methane emissions from ruminants by the small molecule 3-nitrooxypropanol. Proc. Natl. Acad. Sci. USA 2016, 113, 6172–6177. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, E.; Neves, A.L.A.; Song, Y.; Guan, L.L. The Role of the Gut Microbiome in Cattle Production and Health: Driver or Passenger? Annu. Rev. Anim. Biosci. 2020, 8, 199–220. [Google Scholar] [CrossRef] [PubMed]
- Louca, S.; Polz, M.F.; Mazel, F.; Albright, M.B.N.; Huber, J.A.; O’Connor, M.I.; Ackermann, M.; Hahn, A.S.; Srivastava, D.S.; Crowe, S.A.; et al. Function and functional redundancy in microbial systems. Nat. Ecol. Evol. 2018, 2, 936–943. [Google Scholar] [CrossRef] [PubMed]
- Shabat, S.K.B.; Sasson, G.; Doron-Faigenboim, A.; Durman, T.; Yaacoby, S.; Berg Miller, M.E.; White, B.A.; Shterzer, N.; Mizrahi, I. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 2016, 10, 2958–2972. [Google Scholar] [CrossRef]
| Diets (Mean ± SE) | |||
|---|---|---|---|
| Predictors | Forage (Upper–Lower Limits) | Grain (Upper–Lower Limits) | p-Value |
| (Unit) | |||
| (kg/day) | 10.6 ± 0.11 (10.1–11.1) | 12.8 ± 0.11 (12.3–13.3) | 0.005 |
| Forage NDF intake, (kg/day) | 4.4 ± 0.06 (4.2–4.5) | 3.5 ± 0.06 (3.3–3.6) | <0.001 |
| intake, (kg/day) | 2.3 ± 0.06 (2.2–2.4) | 4.0 ± 0.06 (3.9–4.1) | <0.001 |
| Feed conversion ratio, (kg feed/kg gain) | 6.3 ± 0.13 (6.0–6.7) | 5.8 ± 0.12 (5.5–6.1) | <0.001 |
| CH4/DMI ratio, (Liters CH4/kg DMI) | 18.0 ± 0.45 (17.1–18.9) | 14.8 ± 0.45 (13.9–15.7) | <0.001 |
| Average daily gain, (kg/day) | 1.5 ± 0.04 (1.3–1.6) | 1.8 ± 0.04 (1.7–2.0) | <0.001 |
| 1 | 2.8 ± 1.82 × 1011 (3.9 × 1010–2.0 × 1012) | 1.1 ± 0.7 × 1011 (1.6 × 1010–8.3 × 1011) | <0.001 |
| 2 | 3.5 ± 2.22 × 104 (5.4 × 103–2.3 × 105) | 1.5 ± 9.9 × 103 (2.4 × 103–1.0 × 105) | 0.026 |
| 3 | 4.5 ± 1.57 × 107 (1.7 × 105–1.1 × 108) | 5.3 ± 1.85 × 107 (1.7 × 107–1.1 × 108) | 0.668 |
| 4 | 4.2 ± 9.7 × 108 (2.1 × 108–8.3 × 108) | 3.5 ± 8.06 × 108 (1.8 × 108–6.9 × 108) | 0.247 |
| Predictors | Estimates | 95% Confidence Intervals (Upper–Lower Limits) | p-Value |
|---|---|---|---|
| —Dry matter intake | |||
| 28.4 ± 0.96 | 26.4–30.5 | 0.005 | |
| −0.2 ± 0.07 | −0.3–[−0.09] | ||
| −0.1 ± 0.05 | −0.3–[−0.08] | ||
| —Forage NDF intake | |||
| 25.3 ± 0.84 | 23.3–27.2 | 0.002 | |
| 0.6 ± 0.18 | 0.2–1.02 | ||
| 0.1 ± 0.14 | −0.1–0.44 | ||
| intake | |||
| 27.4 ± 0.72 | 25.6–29.2 | 0.001 | |
| −0.5 ± 0.14 | −0.7–[−0.24] | ||
| −0.4 ± 0.11 | −0.6–[−0.19] | ||
| —Feed conversion ratio | |||
| 11.1 ± 1.18 | 8.7–13.5 | 0.508 | |
| −0.1 ± 0.18 | −0.5–0.17 | ||
| −0.1 ± 0.17 | −0.4–0.19 | ||
| —Residual feed intake | |||
| 25.9 ± 0.56 | 24.1–27.7 | 0.160 | |
| −0.2 ± 0.23 | −0.7–0.22 | ||
| −0.2 ± 0.14 | −0.5–0.05 | ||
| CH4/DMI ratio | |||
| 24.9 ± 0.74 | 23.2–26.6 | 0.084 | |
| 0.0 ± 0.02 | 0.0–0.10 | ||
| 0.0 ± 0.03 | 0.0–0.13 | ||
| ADG—Average daily gain | |||
| 25.9 ± 0.99 | 23.8–27.9 | 0.310 | |
| −0.7 ± 0.75 | −2.2–0.72 | ||
| 0.0 ± 0.00 | 0.0–0.0 |
| Predictors | Estimates | 95% Confidence Intervals (Upper–Lower Limits) | p-Value |
|---|---|---|---|
| —Dry matter intake | |||
| 12.7 ± 1.25 | 10.2–15.25 | 0.037 | |
| −0.2 ± 0.10 | −0.4–[−0.05] | ||
| −0.1 ± 0.08 | −0.3–[−0.01] | ||
| —Forage NDF intake | |||
| 9.7 ± 1.03 | 7.5–11.8 | 0.146 | |
| 0.5 ± 0.29 | 0.0–1.16 | ||
| 0.1 ± 0.22 | −0.3–0.58 | ||
| intake | |||
| 11.4 ± 0.81 | 9.6–13.2 | 0.027 | |
| −0.5 ± 0.20 | −0.9–[−0.12] | ||
| −0.3 ± 0.16 | −0.6–[−0.01] | ||
| —Feed conversion ratio | |||
| 11.5 ± 1.39 | 8.7–14.3 | 0.723 | |
| 0.1 ± 0.38 | −0.6–0.88 | ||
| 0.0 ± 0.00 | 0.0–0.00 | ||
| —Residual feed intake | |||
| 10.0 ± 0.50 | 8.4–11.7 | 0.365 | |
| −0.3 ± 0.36 | −1.0–0.36 | ||
| −0.2 ± 0.22 | −0.6–0.21 | ||
| CH4/DMI ratio | |||
| 10.4 ± 0.86 | 8.6–12.2 | 0.706 | |
| 0.0 ± 0.03 | −0.1–0.05 | ||
| 0.0 ± 0.04 | −0.1–0.07 | ||
| ADG—Average daily gain | |||
| 10.8 ± 1.00 | 8.7–12.8 | 0.398 | |
| −0.5 ± 0.55 | −1.6–0.54 | ||
| −0.3 ± 0.47 | −1.2–0.61 |
| Predictors | Estimates | 95% Confidence Intervals (Lower–Upper Limits) | p-Value |
|---|---|---|---|
| —Dry matter intake | |||
| 8.9 ± 1.25 | 6.4–11.46 | 0.365 | |
| 0.0 ± 0.11 | −0.1–0.29 | ||
| 0.0 ± 0.09 | −0.1–0.21 | ||
| —Forage NDF intake | |||
| 18.6 ± 0.94 | 16.7–20.62 | 0.422 | |
| 0.0 ± 0.31 | −0.5–0.67 | ||
| −0.3 ± 0.24 | −0.8–0.17 | ||
| intake | |||
| 9.1 ± 0.69 | 7.7–10.60 | 0.401 | |
| 0.1 ± 0.22 | −0.2–0.62 | ||
| 0.0 ± 0.18 | −0.3–0.41 | ||
| —Feed conversion ratio | |||
| 10.7 ± 1.06 | 8.6–12.88 | 0.215 | |
| −0.1 ± 0.22 | −0.5–0.16 | ||
| −0.2 ± 0.16 | −0.5–0.10 | ||
| —Residual feed intake | |||
| 9.5 ± 0.27 | 8.6–10.39 | 0.417 | |
| −0.4 ± 0.35 | −1.1–0.23 | ||
| −0.0 ± 0.21 | −0.4–0.42 | ||
| CH4/DMI ratio | |||
| 9.8 ± 0.75 | 8.3–11.39 | 0.235 | |
| 0.0 ± 0.03 | 0.0–0.06 | ||
| 0.0 ± 0.04 | −0.1–0.05 | ||
| ADG—Average daily gain | |||
| 8.5 ± 0.89 | 6.7–10.28 | 0.203 | |
| 0.7 ± 0.54 | −0.3–1.83 | ||
| 0.4 ± 0.46 | −0.4–1.37 |
| Predictors | Estimates | 95% Confidence Intervals (Lower–Upper Limits) | p-Value |
|---|---|---|---|
| —Dry matter intake | |||
| 20.1 ± 0.54 | 19.0–21.26 | 0.530 | |
| 0.0 ± 0.04 | −0.1–0.05 | ||
| 0.0 ± 0.04 | −0.1–0.05 | ||
| —Forage NDF intake | |||
| 19.8 ± 0.43 | 18.9–20.73 | 0.170 | |
| 0.2 ± 0.12 | 0.0–0.48 | ||
| 0.0 ± 0.09 | −0.1–0.18 | ||
| intake (?) | |||
| 20.0 ± 0.33 | 19.3–20.77 | 0.324 | |
| −0.1 ± 0.09 | −0.3–0.06 | ||
| 0.0 ± 0.07 | −0.2–0.08 | ||
| —Feed conversion ratio | |||
| 19.1 ± 0.48 | 18.2–20.16 | 0.313 | |
| 0.0 ± 0.07 | 0.0–0.24 | ||
| 0.1 ± 0.07 | 0.0–0.24 | ||
| —Residual feed intake | |||
| 19.7 ± 0.17 | 19.2–20.35 | 0.672 | |
| 0.1 ± 0.15 | −0.1–0.44 | ||
| 0.0 ± 0.09 | −0.2–0.16 | ||
| CH4/DMI ratio | |||
| 19.5 ± 0.34 | 18.8–20.29 | 0.550 | |
| 0.0 ± 0.01 | 0.0–0.04 | ||
| 0.0 ± 0.02 | 0.0–0.05 | ||
| ADG—Average daily gain | |||
| 20.3 ± 0.41 | 19.5–21.21 | 0.211 | |
| −0.4 ± 0.24 | −0.8–0.06 | ||
| −0.3 ± 0.20 | −0.7–0.10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neves, A.L.A.; Vieira, R.A.M.; Vargas-Bello-Pérez, E.; Chen, Y.; McAllister, T.; Ominski, K.H.; Lin, L.; Guan, L.L. Impact of Feed Composition on Rumen Microbial Dynamics and Phenotypic Traits in Beef Cattle. Microorganisms 2025, 13, 310. https://doi.org/10.3390/microorganisms13020310
Neves ALA, Vieira RAM, Vargas-Bello-Pérez E, Chen Y, McAllister T, Ominski KH, Lin L, Guan LL. Impact of Feed Composition on Rumen Microbial Dynamics and Phenotypic Traits in Beef Cattle. Microorganisms. 2025; 13(2):310. https://doi.org/10.3390/microorganisms13020310
Chicago/Turabian StyleNeves, André L. A., Ricardo Augusto Mendonça Vieira, Einar Vargas-Bello-Pérez, Yanhong Chen, Tim McAllister, Kim H. Ominski, Limei Lin, and Le Luo Guan. 2025. "Impact of Feed Composition on Rumen Microbial Dynamics and Phenotypic Traits in Beef Cattle" Microorganisms 13, no. 2: 310. https://doi.org/10.3390/microorganisms13020310
APA StyleNeves, A. L. A., Vieira, R. A. M., Vargas-Bello-Pérez, E., Chen, Y., McAllister, T., Ominski, K. H., Lin, L., & Guan, L. L. (2025). Impact of Feed Composition on Rumen Microbial Dynamics and Phenotypic Traits in Beef Cattle. Microorganisms, 13(2), 310. https://doi.org/10.3390/microorganisms13020310







