Plant Growth-Promoting Effect and Complete Genomic Sequence Analysis of the Beneficial Rhizosphere Streptomyces sp. GD-4 Isolated from Leymus secalinus
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Isolation and Subculture of Bacteria
2.3. Plant Growth-Promoting Assay of Streptomyces sp. GD-4
2.4. Root Colonization of Streptomyces sp. GD-4
2.5. Ammonia Production
2.6. Comparative Genome Analysis
2.7. Library Construction and Genome Sequencing
2.8. Genome Assembly and Annotation
3. Results
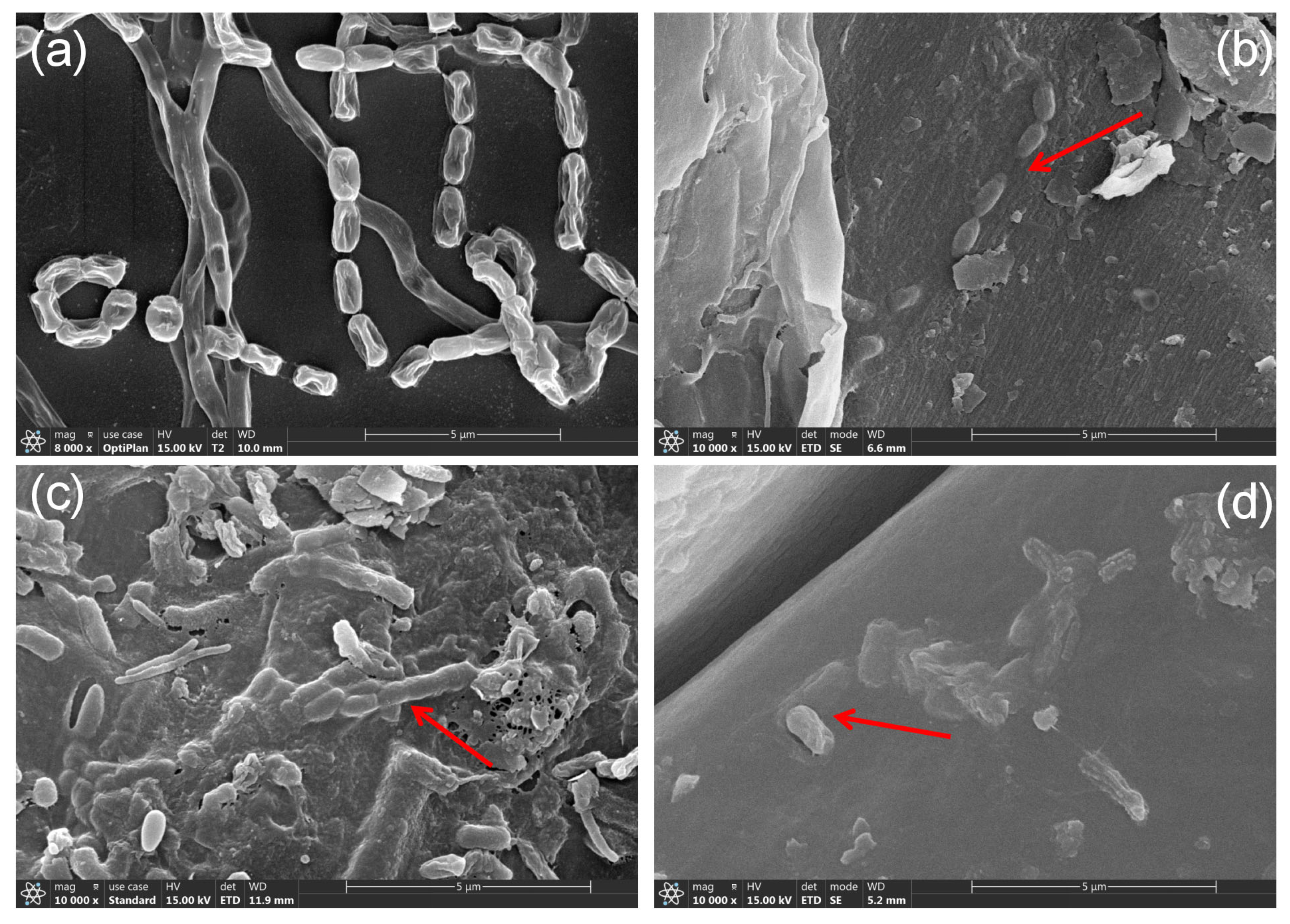
3.1. SEM Observation of the Morphology and Colonization of Strain GD-4
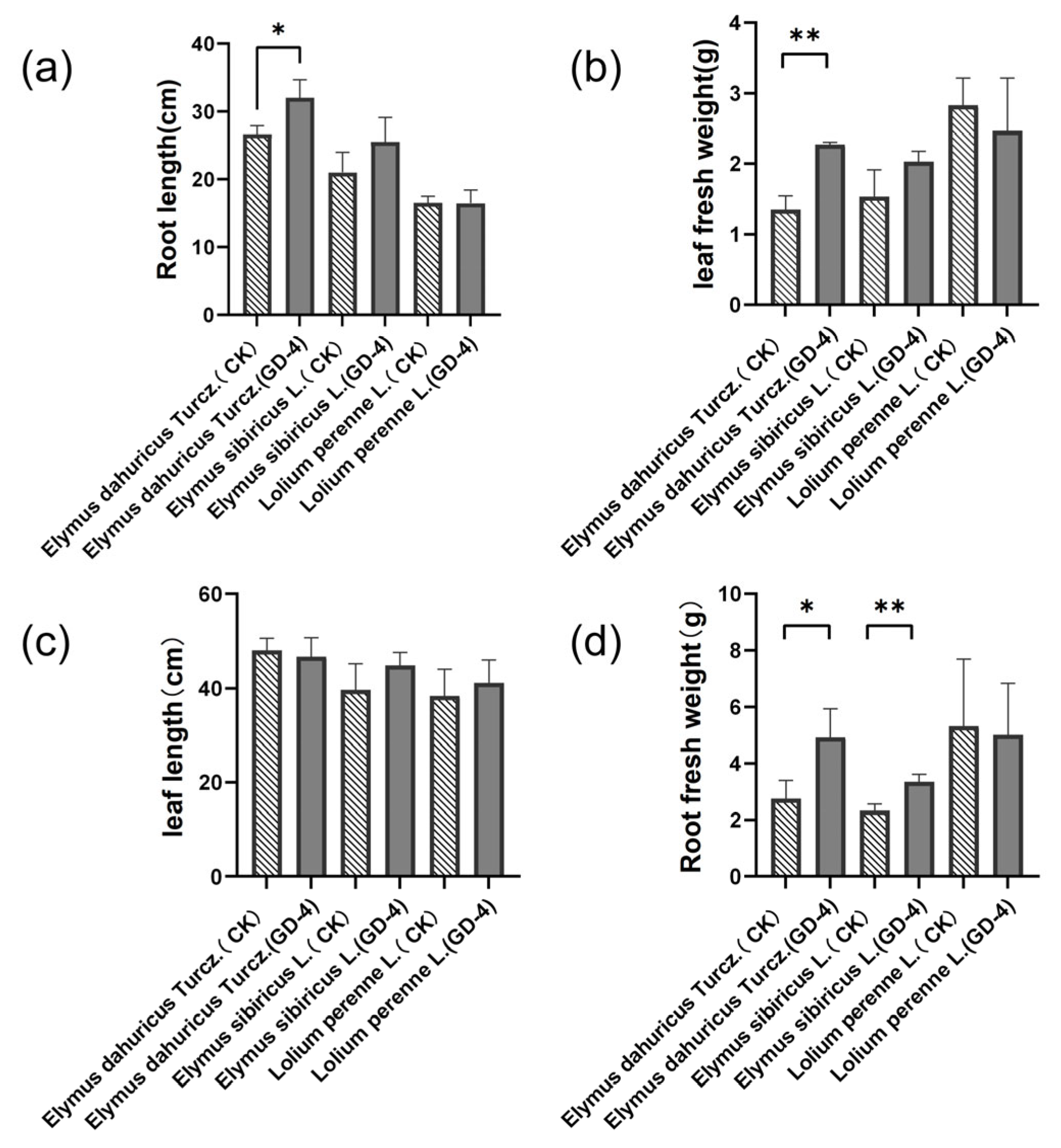
3.2. Effect of Strain GD-4 Inoculation on the Physiological Index of Plants
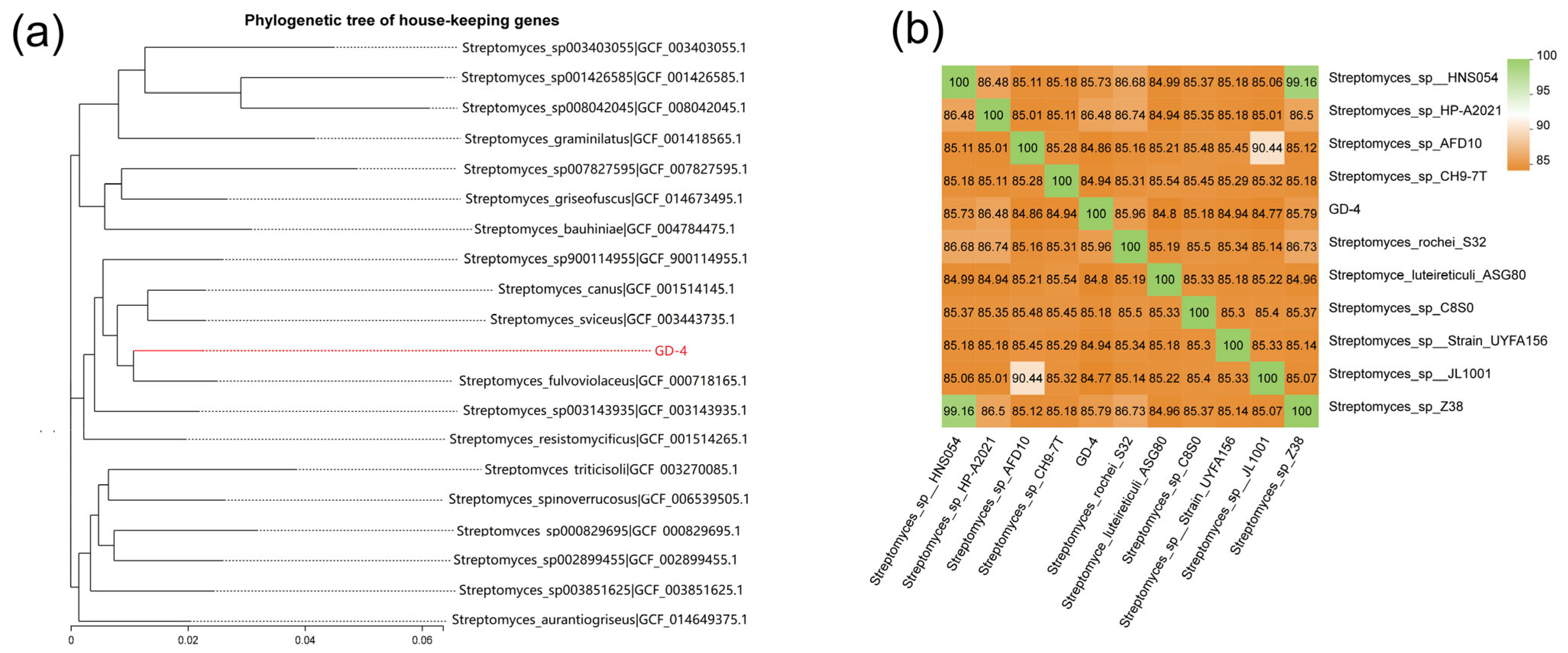
3.3. Comparative Genomics and Phylogenetic Analysis
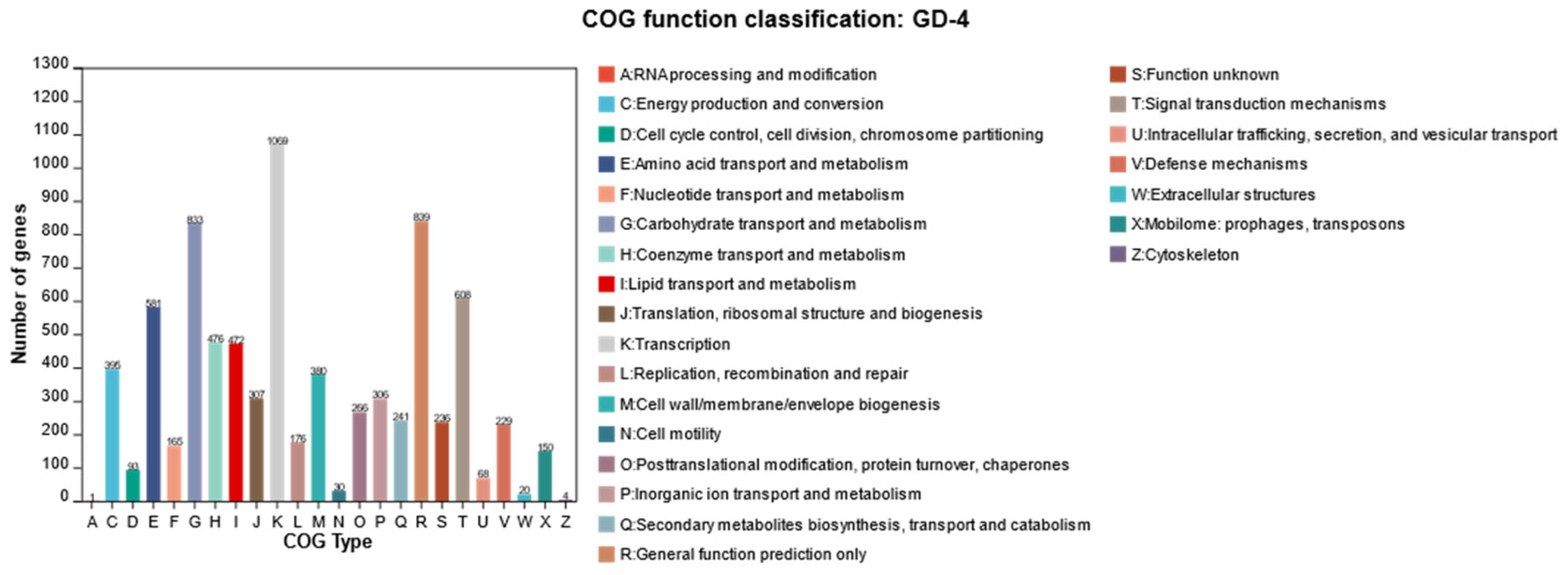
3.4. Genome Characteristics of Strain GD-4
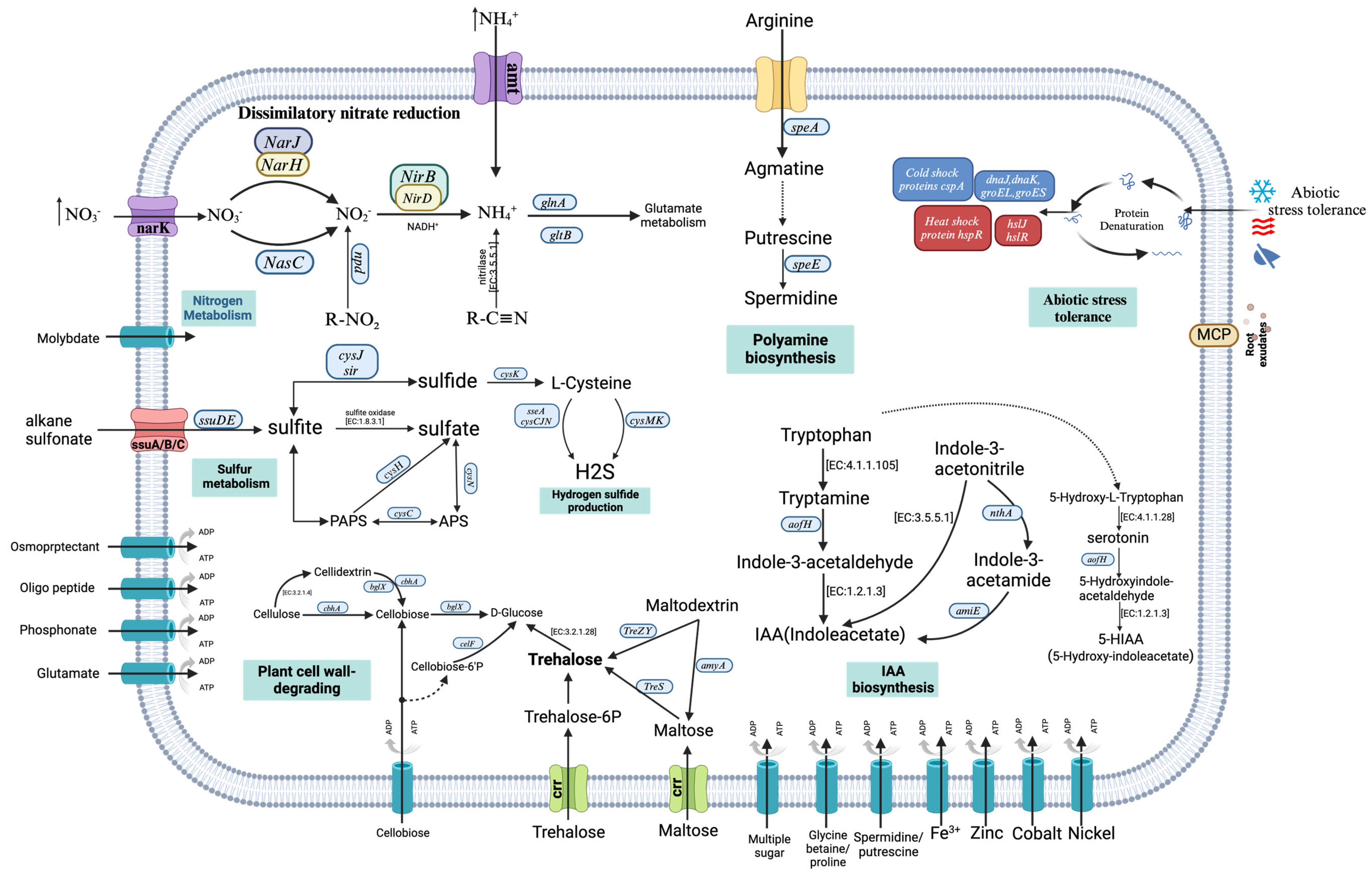
3.5. Genes Associated with Plant Growth Promotion in GD-4 Genome
3.5.1. Nitrogen Metabolism
3.5.2. Sulfur Metabolism
3.5.3. Phosphate Solubilization
3.5.4. Auxin Biosynthesis
3.5.5. Identification of Genes Responsible for Bacterial Biocontrol
3.6. Abiotic Stress Tolerance
3.7. Specific Gene Clusters in Streptomyces sp. GD-4
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Name | Source of Isolation | Genome Size | CD’s | G + C (%) | Potential Function | Accession No. | Reference |
|---|---|---|---|---|---|---|---|
| Streptomyces rochei S32 | soil | 8.0 Mb | 7486 | 72.5 | Plant growth promotion, nitrogen fixation, production of bioactive substances | CP133098 | [85] |
| Streptomyces griseoincarnatus HNS054 | Marine sponge | 7.5 Mb | 6678 | 72.3 | Heterologous expression host for secondary metabolites, salinity tolerance | CP139576 | [86] |
| Streptomyces sp. AFD10 | desert soil | 7.9 Mb | 7246 | 72.6 | Antibiotic production, bioactive secondary metabolite production | JASSVZ000000000 | [87] |
| Streptomyces sp. Z38 | root tissues | 7.3 Mb | 6446 | 72.4 | Plant growth promotion, heavy metal resistance, bio-nanoparticle synthesis | WPIR00000000 | [88] |
| Streptomyces luteireticuli ASG80 | Sisal roots | 8.7 Mb | 7867 | 71.77 | Biocontrol agent against Phytophthora diseases | CP167927 | [89] |
| Streptomyces sp. UYFA156 | seeds | 7.1 Mb | - | 73.4 | Plant growth promotion | CP040466 | [90] |
| Streptomyces sp. C8S0 | sediment | 6.9 Mb | 6,841 | 73.5 | Bioactive metabolite production | CP045031 | [91] |
| Streptomyces sp. JL1001 | Rhizosphere soil | 7.9 Mb | 7315 | 71.71 | Production of novel bioactive natural products, leinamycin-type gene cluster | CP136798 | [92] |
| Streptomyces sp.CH9-7T | Rhizosphere soil | 8.8 Mb | 7664 | 71.7 | Nocardamine production, bioactive secondary metabolite production | JAERRI000000000 | [93] |
| Streptomyces_sp_HP-A2021 | Rhizosphere soil | 9.6 Mb | 8534 | 71.07 | Bioactive secondary metabolite production, antimicrobial activity | CP094344.1 | [94] |
| Activity Description | Gene ID | KO Name | KO ID | KO Description | Enzyme |
|---|---|---|---|---|---|
| Nitrogen metabolism | gene0092 | cynT | K01673 | carbonic anhydrase | [EC:4.2.1.1] |
| gene1739 | glnA | K01915 | glutamine synthetase | [EC:6.3.1.2] | |
| gene1760 | gdhA | K00261 | glutamate dehydrogenase (NAD(P)+) | [EC:6.3.1.2] | |
| gene2048 | narH | K00371 | nitrate reductase/nitrite oxidoreductase, beta subunit | [EC:1.7.5.1 1.7.99.-] | |
| gene2544 | gltB | K00265 | glutamate synthase (NADPH) large chain | [EC:1.4.1.13] | |
| gene4307 | narK | K02575 | MFS transporter, NNP family, nitrate/nitrite transporter | - | |
| gene4308 | nasC | K00372 | assimilatory nitrate reductase catalytic subunit | [EC:1.7.99.-] | |
| gene4309 | nasB | K00360 | assimilatory nitrate reductase electron transfer subunit | [EC:1.7.99.-] | |
| gene5295 | nirK | K00368 | nitrite reductase (NO-forming) | [EC:1.7.2.1] | |
| gene5356 | - | K15371 | glutamate dehydrogenase | [EC:1.4.1.2] | |
| gene5868 | npd | K00459 | nitronate monooxygenase | [EC:1.13.12.16] | |
| gene5942 | nirD | K00363 | nitrite reductase (NADH), small subunit | [EC:1.7.1.15] | |
| gene5943 | nirB | K00362 | nitrite reductase (NADH), large subunit | [EC:1.7.1.15] | |
| gene6510 | gltD | K00266 | glutamate synthase (NADPH), small chain | [EC:1.4.1.13] | |
| gene9104 | - | K01501 | nitrilase | [EC:3.5.5.1] | |
| gene6662 | iscU | K04488 | nitrogen fixation protein NifU and related proteins | - | |
| gene0326 | narL | K07684 | two-component system, NarL family, nitrate/nitrite response regulator NarL | - | |
| gene2048 | narH | K00371 | nitrate reductase/nitrite oxidoreductase, beta subunit | [EC:1.7.5.1/1.7.99.-] | |
| gene2049 | narJ | K00373 | nitrate reductase molybdenum cofactor assembly chaperone NarJ/NarW | - | |
| gene4268 | narA | K25150 | polyether ionophore transport system ATP-binding protein | - | |
| gene4269 | narB | K25149 | polyether ionophore transport system permease protein | - | |
| Molybdenum cofactor | gene4499 | mobA | K03752 | molybdenum cofactor guanylyltransferase | [EC:2.7.7.77] |
| gene4379 | moeB | K21029 | molybdopterin-synthase adenylyltransferase | [EC:2.7.7.80] | |
| gene0517 | moaC | K03637 | cyclic pyranopterin monophosphate synthase | [EC:4.6.1.17] | |
| gene3219 | moaE | K03635 | molybdopterin synthase catalytic subunit | [EC:2.8.1.12] | |
| gene3830 | moaA | K03639 | GTP 3′,8-cyclase | [EC:4.1.99.22] | |
| gene4835 | moaD | K03636 | sulfur-carrier protein | - | |
| Nitrogen transporters | gene0085 | nasT | K07183 | two-component system, response regulator/RNA-binding antiterminator | - |
| gene2809 | amt | K03320 | ammonium transporter, Amt family | - | |
| gene1739 | glnA | K01915 | glutamine synthetase | [EC:6.3.1.2] | |
| gene2807 | glnD | K00990 | [protein-PII] uridylyltransferase | [EC:2.7.7.59] | |
| gene2808 | glnB | K04751 | nitrogen regulatory protein P-II 1 | - | |
| gene6281 | glnE | K00982 | [glutamine synthetase] adenylyltransferase/[glutamine synthetase]-adenylyl-L-tyrosine phosphorylase | [EC:2.7.7.42/2.7.7.89] | |
| gene4307 | narK | K02575 | MFS transporter, NNP family, nitrate/nitrite transporter | - |
| Activity Description | Gene ID | KO Name | KO ID | KO Description | Enzyme |
|---|---|---|---|---|---|
| Phosphate solubilization | gene1931 | zwf | K00036 | glucose-6-phosphate 1-dehydrogenase | [EC:1.1.1.49/1.1.1.363] |
| gene4131 | ppa | K01507 | inorganic pyrophosphatase | [EC:3.6.1.1] | |
| gene0144 | pqqE | K06139 | PqqA peptide cyclase | [EC:1.21.98.4] | |
| gene2585 | pqqL | K07263 | zinc protease | [EC:3.4.24.-] | |
| gene0729 | gdh | K00034 | glucose 1-dehydrogenase | [EC:1.1.1.47] | |
| gene1760 | gdhA | K00261 | glutamate dehydrogenase(NAD(P)+) | [EC:1.4.1.3] | |
| gene0424 | phoD | K01113 | alkaline phosphatase D | [EC:3.1.3.1] | |
| gene8518 | phoP | K07658 | two-component system, OmpR family, alkaline phosphatase synthesis response regulator PhoP | - | |
| gene3368 | phoH2 | K07175 | PhoH-like ATPase | - | |
| gene4887 | phoU | K02039 | phosphate transport system protein | - | |
| gene5884 | phoH | K06217 | phosphate starvation-inducible protein PhoH and related proteins | - | |
| gene6104 | phoA | K01077 | alkaline phosphatase | [EC:3.1.3.1] | |
| Phosphate transport | gene4252 | pstS | K02040 | phosphate transport systemsubstrate-binding protein | - |
| gene4803 | pstB | K02036 | phosphate transport systemATP-binding protein | [EC:7.3.2.1] | |
| gene4804 | pstA | K02038 | phosphate transport systempermease protein | - | |
| gene4805 | pstC | K02037 | phosphate transport systempermease protein | - | |
| gene6940 | pstI | K20754 | aqualysin 1 | [EC:3.4.21.111] | |
| gene1103 | phnB | K04750 | PhnB protein | - | |
| gene1789 | phnW | K03430 | 2-aminoethylphosphonate-pyruvate transaminase | [EC:2.6.1.37] | |
| gene4587 | phnO | K09994 | (aminoalkyl)phosphonate N-acetyltransferase | [EC:2.3.1.280] | |
| gene7793 | phnA | K19670 | phosphonoacetate hydrolase | [EC:3.11.1.2] |
| Activity Description | Gene ID | KO Name | KO ID | KO Description | Enzyme |
|---|---|---|---|---|---|
| IAA biosynthesis | gene0221 | paaF | K01692 | enoyl-CoA hydratase | [EC:4.2.1.17] |
| gene0256 | amiE | K01426 | amidase | [EC:3.5.1.4] | |
| gene0482 | gcdH | K00252 | glutaryl-CoA dehydrogenase | [EC:1.3.8.6] | |
| gene0485 | atoB | K00626 | acetyl-CoA C-acetyltransferase | [EC:2.3.1.9] | |
| gene0620 | - | K00128 | aldehyde dehydrogenase (NAD+) | [EC:1.2.1.3] | |
| gene1290 | - | K01593 | aromatic-L-amino-acid/L-tryptophan decarboxylase | [EC:4.1.1.28/4.1.1.105] | |
| gene1432 | cypD_E | K14338 | cytochrome P450/NADPH-cytochrome P450 reductase | [EC:1.14.14.1/1.6.2.4] | |
| gene1474 | pdhD | K00382 | dihydrolipoyl dehydrogenase | [EC:1.8.1.4] | |
| gene1502 | - | K03392 | aminocarboxymuconate-semialdehyde decarboxylase | [EC:4.1.1.45] | |
| gene1818 | fadJ | K01782 | 3-hydroxyacyl-CoA dehydrogenase/enoyl-CoA hydratase/3-hydroxybutyryl-CoA epimerase | [EC:1.1.1.35/4.2.1.17/5.1.2.3] | |
| gene2069 | katE | K03781 | catalase | [EC:1.11.1.6] | |
| gene2370 | phsA | K20219 | o-aminophenol oxidase | [EC:1.10.3.4] | |
| gene2538 | katG | K03782 | catalase-peroxidase | [EC:1.11.1.21] | |
| gene4279 | kynU | K01556 | kynureninase | [EC:3.7.1.3] | |
| gene4282 | kynA | K00453 | tryptophan 2,3-dioxygenase | [EC:1.13.11.11] | |
| gene4301 | aofH | K00274 | monoamine oxidase | [EC:1.4.3.4] | |
| gene5439 | nthB | K20807 | nitrile hydratase subunit beta | [EC:4.2.1.84] | |
| gene5441 | nthA | K01721 | nitrile hydratase subunit alpha | [EC:4.2.1.84] | |
| gene7158 | - | K22450 | aralkylamine N-acetyltransferase | [EC:2.3.1.87] | |
| gene9104 | - | K01501 | nitrilase | [EC:3.5.5.1] | |
| gene6498 | trpA | K01695 | tryptophan synthase alpha chain | [EC:4.2.1.20] | |
| gene6497 | trpB | K01696 | tryptophan synthase beta chain | [EC:4.2.1.20] | |
| gene6496 | trpC | K01609 | indole-3-glycerol phosphate synthase | [EC:4.1.1.48] | |
| gene6381 | trpD | K00766 | anthranilate phosphoribosyltransferase | [EC:2.4.2.18] | |
| gene6492 | trpE | K01657 | anthranilate synthase component I | [EC:4.1.3.27] | |
| gene4607 | hisC | K00817 | histidinol-phosphate aminotransferase | [EC:2.6.1.9] | |
| ACC Deaminase | gene2178 | map | K01265 | methionyl aminopeptidase | [EC:3.4.11.18] |
| gene1750 | kbl | K00639 | glycine C-acetyltransferase | [EC:2.3.1.29] | |
| gene1790 | pgsA | K00995 | CDP-diacylglycerol---glycerol-3-phosphate 3-phosphatidyltransferase | [EC:2.7.8.5] | |
| gene2753 | glpQ | K01126 | glycerophosphoryl diester phosphodiesterase | [EC:3.1.4.46] |
| Activity Description | Gene ID | KO Name | KO ID | KO Description | Enzyme |
|---|---|---|---|---|---|
| Enediyne antibiotics | gene1384 | ncsB3 | K20420 | 2-hydroxy-5-methyl-1-naphthoate 7-hydroxylase | [EC:1.14.15.31] |
| gene2301 | mdpB3 | K21193 | acetyltransferase/esterase | - | |
| gene2929 | ncsB4 | K21209 | acyltransferase | - | |
| gene3244 | ncsC4 | K21214 | NDP-hexose 4-ketoreductase | - | |
| gene4917 | sgcE6 | K21185 | flavin reductase | - | |
| gene4990 | sgcE11 | K21167 | enediyne biosynthesis protein E11 | - | |
| gene6898 | sgcF | K21159 | epoxide hydrolase | - | |
| gene7800 | cepH | K16431 | FAD-dependent halogenase | [EC:1.14.19.-] | |
| gene8258 | sgcD3 | K21177 | cytochrome P450 hydroxylase | [EC:1.14.-.-] | |
| gene8273 | calE5 | K21172 | enediyne biosynthesis protein CalE5 | - | |
| Ansamycins | gene1929 | tktA | K00615 | transketolase | [EC:2.2.1.1] |
| gene8433 | asm10 | K16038 | N-methyltransferase | [EC:2.1.1.-] | |
| Vancomycin-group antibiotics | gene0345 | evaC | K16437 | methylation protein EvaC | - |
| gene1329 | rfbB | K01710 | dTDP-glucose 4,6-dehydratase | [EC:4.2.1.46] | |
| gene7800 | cepH | K16431 | FAD-dependent halogenase | [EC:1.14.19.-] | |
| gene7828 | cepJ | K16434 | thioesterase CepJ | - | |
| gene8312 | nocN | K16422 | 4-hydroxymandelate oxidase | [EC:1.1.3.46] | |
| pA_gene0134 | cepA | K16428 | nonribosomal peptide synthetase CepA | - | |
| Tetracycline | gene1025 | tetX | K18221 | tetracycline 11a-monooxygenase, tetracycline resistance protein | [EC:1.14.13.231] |
| gene3288 | oxyQ | K14254 | aminotransferase | - | |
| gene5733 | oxyS | K14256 | anhydrotetracycline 6-monooxygenase/5a,11a-dehydrotetracycline 5-monooxygenase | [EC:1.14.13.38/1.14.13.234] | |
| gene8337 | oxyF | K14251 | C-methyltransferase | [EC:2.1.1.-] | |
| pA_gene0186 | oxyA | K05551 | minimal PKS ketosynthase (KS/KS alpha) | [EC:2.3.1.-/2.3.1.260/2.3.1.235] | |
| pA_gene0187 | oxyB | K05552 | minimal PKS chain-length factor (CLF/KS beta) | [EC:2.3.1.-2.3.1.260 2.3.1.235] | |
| pA_gene0188 | ctcP | K14257 | tetracycline 7-halogenase/FADH2 O2-dependent halogenase | [EC:1.14.19.49/1.14.19.-] | |
| pA_gene0199 | oxyC | K05553 | minimal PKS acyl carrier protein | - | |
| pA_gene0200 | oxyJ | K12420 | ketoreductase | [EC:1.1.1.-] | |
| Phenazine | gene2251 | phzS | K20940 | 5-methylphenazine-1-carboxylate 1-monooxygenase | [EC:1.14.13.218] |
| gene6250 | phzF | K06998 | trans-2,3-dihydro-3-hydroxyanthranilate isomerase | [EC:5.3.3.17] | |
| gene7406 | phzF | K06998 | trans-2,3-dihydro-3-hydroxyanthranilate isomerase | [EC:5.3.3.17] | |
| gene6410 | phzE | K13063 | 2-amino-4-deoxychorismate synthase | [EC:2.6.1.86] |
| Activity Description | Gene ID | KO Name | KO ID | KO Description | Enzyme |
|---|---|---|---|---|---|
| Root colonization Motility | gene8106 | flgS | K02482 | two-component system, NtrC family, sensor kinase | [EC:2.7.13.3] |
| gene2789 | fliA | K02405 | RNA polymerase sigma factor FliA | - | |
| gene2607 | rpoD | K03086 | RNA polymerase primary sigma factor | - | |
| Root colonization and interactions | gene3612 | mdh | K00024 | malate dehydrogenase | [EC:1.1.1.37] |
| gene0159 | xerD | K04763 | integrase/recombinase XerD | - | |
| Chemotaxis | gene0567 | - | K11354 | two-component system, chemotaxis family, sensor kinase Cph1 | [EC:2.7.13.3] |
| gene0888 | rbsB | K10439 | ribose transport system substrate-binding protein | - | |
| gene8627 | mcp | K03406 | methyl-accepting chemotaxis protein | - | |
| gene3596 | cheR | K00575 | chemotaxis protein methyltransferase CheR | [EC:2.1.1.80] | |
| Cellulose degradation | gene0446 | bglX | K05349 | beta-glucosidase | [EC:3.2.1.21] |
| gene0445 | - | K01179 | endoglucanase | [EC:3.2.1.4] | |
| gene0447 | cbhA | K19668 | cellulose 1,4-beta-cellobiosidase | [EC:3.2.1.91] | |
| gene1904 | celF | K01222 | 6-phospho-beta-glucosidase | [EC:3.2.1.86] | |
| Exopolysaccharide biosynthesis | gene1005 | exoZ | K16568 | exopolysaccharide production protein ExoZ | - |
| gene1536 | exoY | K16566 | exopolysaccharide production protein ExoY | - | |
| gene3072 | wecA | K02851 | UDP-GlcNAc:undecaprenyl-phosphate/decaprenyl-phosphate GlcNAc-1-phosphate transferase | [EC:2.7.8.33/2.7.8.35] | |
| gene5303 | exoA | K16557 | succinoglycan biosynthesis protein ExoA | [EC:2.4.-.-] | |
| gene5659 | gumD | K13656 | undecaprenyl-phosphate glucose phosphotransferase | [EC:2.7.8.31] | |
| gene5724 | cysE | K00640 | serine O-acetyltransferase | [EC:2.3.1.30] | |
| Biofilm formation | gene1169 | oxyR | K04761 | LysR family transcriptional regulator, hydrogen peroxide-inducible gene activator | - |
| gene1609 | glgC | K00975 | glucose-1-phosphate adenylyltransferase | [EC:2.7.7.27] | |
| gene2789 | fliA | K02405 | RNA polymerase sigma factor FliA | - | |
| gene2982 | glgP | K00688 | glycogen phosphorylase | [EC:2.4.1.1] | |
| gene3721 | bcsA | K00694 | cellulose synthase (UDP-forming) | [EC:2.4.1.12] | |
| gene3765 | crp | K10914 | CRP/FNR family transcriptional regulator, cyclic AMP receptor protein | - | |
| gene4594 | gcvA | K03566 | LysR family transcriptional regulator, glycine cleavage system transcriptional activator | - | |
| gene5656 | pgaB | K11931 | poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | [EC:3.5.1.-] | |
| gene6457 | dksA | K06204 | RNA polymerase-binding transcription factor | ||
| gene7372 | crr | K02777 | sugar PTS system EIIA component | [EC:2.7.1.-] | |
| gene8276 | rcdA | K23778 | TetR/AcrR family transcriptional regulator, regulator of biofilm formation and stress response | - | |
| gene4092 | icaR | K21453 | TetR/AcrR family transcriptional regulator, biofilm operon repressor | - |
| Activity Description | Gene ID | KO Name | KO ID | KO Description | Enzyme |
|---|---|---|---|---|---|
| Sulfur metabolism | gene0895 | sfnG | K17228 | dimethylsulfone monooxygenase | [EC:1.14.14.35] |
| gene0932 | - | K00387 | sulfite oxidase | [EC:1.8.3.1] | |
| gene1472 | cysJ | K00380 | sulfite reductase (NADPH) flavoprotein alpha-component | [EC:1.8.1.2] | |
| gene2332 | sir | K00392 | sulfite reductase (ferredoxin) | [EC:1.8.7.1] | |
| gene2334 | cysH | K00390 | phosphoadenosine phosphosulfate reductase | [EC:1.8.4.8/1.8.4.10] | |
| gene2335 | cysC | K00860 | adenylylsulfate kinase | [EC:2.7.1.25] | |
| gene2336 | cysD | K00957 | sulfate adenylyltransferase subunit 2 | [EC:2.7.7.4] | |
| gene2337 | cysN | K00956 | sulfate adenylyltransferase subunit 1 | [EC:2.7.7.4] | |
| gene2574 | sseA | K01011 | thiosulfate/3-mercaptopyruvate sulfurtransferase | [EC:2.8.1.1/2.8.1.2] | |
| gene3444 | metB | K01739 | cystathionine gamma-synthase | [EC:2.5.1.48] | |
| gene3652 | doxD | K16937 | thiosulfate dehydrogenase (quinone) large subunit | [EC:1.8.5.2] | |
| gene4896 | doxD | K16937 | thiosulfate dehydrogenase (quinone) large subunit | [EC:1.8.5.2] | |
| gene3878 | aprA | K00394 | adenylylsulfate reductase, subunit A | [EC:1.8.99.2] | |
| gene4304 | metX | K00641 | homoserine O-acetyltransferase/O-succinyltransferase | [EC:2.3.1.31/2.3.1.46] | |
| gene4469 | dmdC | K20035 | 3-(methylsulfanyl)propanoyl-CoA dehydrogenase | [EC:1.3.99.41] | |
| gene4657 | dmdB | K20034 | 3-(methylthio)propionyl---CoA ligase | [EC:6.2.1.44] | |
| gene5061 | ssuD | K04091 | alkanesulfonate monooxygenase | [EC:1.14.14.5/ 1.14.14.34] | |
| gene5724 | cysE | K00640 | serine O-acetyltransferase | [EC:2.3.1.30] | |
| gene5725 | cysK | K01738 | cysteine synthase | [EC:2.5.1.47] | |
| gene6420 | thiS | K03154 | sulfur carrier protein | - | |
| gene6037 | iscR | K13643 | Rrf2 family transcriptional regulator, iron–sulfur cluster assembly transcription factor | - | |
| gene6382 | sufS | K11717 | cysteine desulfurase/selenocysteine lyase | [EC:2.8.1.7/4.4.1.16] | |
| gene6657 | sufB | K09014 | Fe-S cluster assembly protein SufB | - | |
| gene6658 | sufD | K09015 | Fe-S cluster assembly protein SufD | - | |
| gene6660 | sufC | K09013 | Fe-S cluster assembly ATP-binding protein | - | |
| Sulfur transport | gene2339 | ssuA | K15553 | sulfonate transport system substrate-binding protein | - |
| gene2340 | ssuB | K15555 | sulfonate transport system ATP-binding protein | [EC:7.6.2.14] | |
| gene2341 | ssuC | K15554 | sulfonate transport system permease protein | - | |
| gene4388 | tauC | K15552 | taurine transport system permease protein | - | |
| gene4389 | tauA | K15551 | taurine transport system substrate-binding protein | - | |
| gene4390 | tauB | K10831 | taurine transport system ATP-binding protein | [EC:7.6.2.7] | |
| gene4391 | tauD | K03119 | taurine dioxygenase | [EC:1.14.11.17] |
| Activity Description | Gene ID | KO Name | KO ID | KO Description | Enzyme |
|---|---|---|---|---|---|
| Siderophore biosynthesis | gene7262 | aroC | K01736 | chorismate synthase | [EC:4.2.3.5] |
| gene6884 | aroH | K06208 | chorismate mutase | [EC:5.4.99.5] | |
| gene2570 | hemH | K01772 | protoporphyrin/coproporphyrin ferrochelatase | [EC:4.98.1.14.99.1.9] | |
| gene7265 | aroK | K00891 | shikimate kinase | [EC:2.7.1.71] | |
| gene6413 | bfr | K03594 | bacterioferritin | [EC:1.16.3.1] | |
| gene6412 | bfd | K02192 | bacterioferritin-associated ferredoxin | - | |
| gene3085 | lysA | K01586 | diaminopimelate decarboxylase | [EC:4.1.1.20] | |
| gene6037 | iscR | K13643 | Rrf2 family transcriptional regulator, iron–sulfur cluster assembly transcription factor | - | |
| gene2932 | iscS | K04487 | cysteine desulfurase | [EC:2.8.1.7] | |
| gene6243 | efeU | K07243 | high-affinity iron transporter | - | |
| gene7987 | pvdQ | K07116 | acyl-homoserine-lactone acylase | [EC:3.5.1.97] | |
| gene0300 | mbtH | K05375 | MbtH protein | - | |
| gene4994 | mbtN | K00257 | acyl-ACP dehydrogenase | [EC:1.3.99.-] | |
| gene7826 | mbtI | K04781 | salicylate synthetase | [EC:5.4.4.2/4.2.99.21] | |
| gene7834 | mbtB | K04788 | mycobactin phenyloxazoline synthetase | - | |
| gene2630 | iucB | K03896 | acetyl CoA:N6-hydroxylysine acetyl transferase | [EC:2.3.1.102] | |
| gene5568 | iucC | K03895 | aerobactin synthase | [EC:6.3.2.39] | |
| gene5570 | iucD | K03897 | lysine N6-hydroxylase | [EC:1.14.13.59] | |
| gene0303 | dhbF | K04780 | glyine—[glycyl-carrierprotein]ligase | [EC:6.2.1.66] | |
| gene2631 | asbA | K24108 | spermidine-citrateligase | [EC:6.3.2.-] | |
| gene5422 | entF | K02364 | L-serine—[L-seryl-carrierprotein]ligase | [EC:6.3.2.1/46.2.1.72] | |
| gene7825 | entE | K02363 | 2,3-dihydroxybenzoate—[aryl-carrierprotein]ligase | [EC:6.3.2.1/46.2.1.71] | |
| gene8325 | menF | K02552 | menaquinone-specificisochorismatesynthase | [EC:5.4.4.2] | |
| Iron uptake and transport | gene8395 | fhuB | K23228 | ferric hydroxamate transport system permease protein | - |
| gene8396 | fhuD | K23227 | ferric hydroxamate transport system substrate-binding protein | - | |
| gene8397 | fhuC | K10829 | ferric hydroxamate transport system ATP-binding protein | [EC:7.2.2.16] | |
| gene3465 | fepD | K23186 | iron-siderophore transport system permease protein | - | |
| gene3466 | fepG | K23187 | iron-siderophore transport system permease protein | - | |
| gene6856 | fepC | K23188 | iron-siderophore transport system ATP-binding protein | [EC:7.2.2.1/77.2.2.-] | |
| gene5573 | desE | K25287 | iron-desferrioxamine transport system substrate-binding protein | - | |
| gene5604 | entS | K08225 | MFS transporter, ENTS family, enterobactin (siderophore) exporter | - | |
| gene6244 | efeB | K16301 | deferrochelatase/peroxidase EfeB | [EC:1.11.1.-] | |
| gene6245 | efeO | K07224 | iron uptake system component EfeO | - | |
| gene7837 | pvdA | K10531 | L-ornithine N5-monooxygenase | [EC:1.14.13.1951.14.13.196] |
| Activity Description | Gene ID | KO Name | KO ID | KO Description | Enzyme |
|---|---|---|---|---|---|
| Heat tolerance | gene4731 | hslJ | K03668 | heat-shock protein HslJ | - |
| gene3770 | htpX | K03799 | heat-shock protein HtpX | [EC:3.4.24.-] | |
| gene6571 | hslR | K04762 | ribosome-associated heat-shock protein Hsp15 | - | |
| gene5866 | hrcA | K03705 | heat-inducible transcriptional repressor | - | |
| Cold-shock protein | gene4329 | dnaJ | K03686 | molecular chaperone DnaJ | - |
| gene0076 | dnaK | K04043 | molecular chaperone DnaK | - | |
| gene0819 | cspA | K03704 | cold-shock protein | - | |
| gene3683 | groEL | K04077 | chaperonin GroEL | [EC:5.6.1.7] | |
| gene3684 | groES | K04078 | chaperonin GroES | - | |
| Salinity tolerance | gene1089 | betB | K00130 | betaine-aldehyde dehydrogenase | [EC:1.2.1.8] |
| gene1092 | betA | K00108 | choline dehydrogenase | [EC:1.1.99.1] | |
| gene1761 | betI | K02167 | TetR/AcrR family transcriptional regulator, transcriptional repressor of bet genes | - | |
| gene1086 | proX | K02002 | glycine betaine/proline transport system substrate-binding protein | - | |
| gene1087 | proW | K02001 | glycine betaine/proline transport system permease protein | - | |
| gene1088 | proV | K02000 | glycine betaine/proline transport system ATP-binding protein | [EC:7.6.2.9] | |
| gene1866 | proP | K03762 | MFS transporter, MHS family, proline/betaine transporter | - | |
| gene2716 | proS | K01881 | prolyl-tRNA synthetase | [EC:6.1.1.15] | |
| gene4068 | proC | K00286 | pyrroline-5-carboxylate reductase | [EC:1.5.1.2] | |
| gene4311 | proA | K00147 | glutamate-5-semialdehyde dehydrogenase | [EC:1.2.1.41] | |
| gene5828 | proB | K00931 | glutamate 5-kinase | [EC:2.7.2.11] | |
| gene2892 | putR | K23253 | PucR family transcriptional regulator, proline-responsive transcriptional activator | - | |
| Oxidative stress tolerance | gene2069 | katE | K03781 | catalase | [EC:1.11.1.6] |
| gene2538 | katG | K03782 | catalase-peroxidase | [EC:1.11.1.21] | |
| gene2163 | ggt | K00681 | gamma-glutamyltranspeptidase/glutathione hydrolase | [EC:2.3.2.2] [EC:3.4.19.13] | |
| gene0778 | trxA | K03671 | thioredoxin 1 | - | |
| gene4564 | trxB | K00384 | thioredoxin reductase (NADPH) | [EC:1.8.1.9] | |
| Polyamine biosynthesis | gene4295 | speE | K00797 | spermidine synthase | [EC:2.5.1.16] |
| gene4902 | speG | K00657 | diamine N-acetyltransferase | [EC:2.3.1.57] | |
| gene8127 | speA | K01585 | arginine decarboxylase | [EC:4.1.1.19] | |
| gene0359 | argS | K01887 | arginyl-tRNA synthetase | [EC:6.1.1.19] | |
| gene7176 | argH | K01755 | argininosuccinate lyase | [EC:4.3.2.1] | |
| gene7525 | argO | K22477 | N-acetylglutamate synthase | [EC:2.3.1.1] | |
| gene2745 | potC | K11070 | spermidine/putrescine transport system permease protein | - | |
| gene2746 | potB | K11071 | spermidine/putrescine transport system permease protein | - | |
| gene2747 | potA | K11072 | spermidine/putrescine transport system ATP-binding protein | [EC:7.6.2.11] | |
| gene2748 | potD | K11069 | spermidine/putrescine transport system substrate-binding protein | - | |
| Trehalose | gene2358 | treY | K06044 | (1->4)-alpha-D-glucan 1-alpha-D-glucosylmutase | [EC:5.4.99.15] |
| gene2363 | treZ | K01236 | maltooligosyltrehalose trehalohydrolase | [EC:3.2.1.141] | |
| gene2985 | treS | K05343 | maltose alpha-D-glucosyltransferase/alpha-amylase | [EC:5.4.99.16] [EC:3.2.1.1] | |
| gene1426 | otsB | K01087 | trehalose 6-phosphate phosphatase | [EC:3.1.3.12] | |
| gene4926 | otsA | K00697 | trehalose 6-phosphate synthase | [EC:2.4.1.15/2.4.1.347] | |
| Tolerance against metal toxicity | gene1479 | chrR | K19784 | chromate reductase, NAD(P)H dehydrogenase (quinone) | - |
| gene8824 | chrA | K07240 | chromate transporter | - | |
| gene0716 | cusR | K07665 | two-component system, OmpR family, copper resistance phosphate regulon response regulator CusR | - | |
| gene5624 | copA | K17686 | P-type Cu+ transporter | [EC:7.2.2.8] | |
| gene5625 | copZ | K07213 | copper chaperone | - | |
| gene0790 | arsR | K03892 | ArsR family transcriptional regulator, arsenate/arsenite/antimonite-responsive transcriptional repressor | - | |
| gene1363 | arsB | K03893 | arsenical pump membrane protein | - | |
| gene4206 | arsA | K01551 | arsenite/tail-anchored protein-transporting ATPase | [EC:7.3.2.7/7.3.-.-] | |
| gene6310 | arsC | K00537 | arsenate reductase (glutaredoxin) | [EC:1.20.4.1] | |
| gene8604 | arsB | K03325 | arsenite transporter | - | |
| gene5916 | znuB | K09816 | zinc transport system permease protein | - | |
| gene8791 | znuB | K09816 | zinc transport system permease protein | - | |
| gene5917 | znuC | K09817 | zinc transport system ATP-binding protein | [EC:7.2.2.20] | |
| gene5918 | znuA | K09815 | zinc transport system substrate-binding protein | - | |
| gene0517 | moaC | K03637 | cyclic pyranopterin monophosphate synthase | [EC:4.6.1.17] | |
| gene5137 | moaC | K03637 | cyclic pyranopterin monophosphate synthase [EC:4.6.1.17] | - | |
| gene3219 | moaE | K03635 | molybdopterin synthase catalytic subunit | [EC:2.8.1.12] | |
| gene3830 | moaA | K03639 | GTP 3′,8-cyclase | [EC:4.1.99.22] | |
| gene6814 | moaA | K03639 | GTP 3′,8-cyclase [EC:4.1.99.22] | - | |
| gene8622 | moaA | K03639 | GTP 3′,8-cyclase [EC:4.1.99.22] | - | |
| gene4835 | moaD | K03636 | sulfur-carrier protein | - | |
| Carotenoid biosynthesis | gene0366 | crtQ | K00514 | zeta-carotene desaturase | [EC:1.3.5.6] |
| gene0713 | crtO | K02292 | beta-carotene ketolase (CrtO type) | - | |
| gene1782 | crtB | K02291 | 15-cis-phytoene synthase | [EC:2.5.1.32] | |
| gene2161 | crtI | K10027 | phytoene desaturase | [EC:1.3.99.26] [EC:1.3.99.28 ] [EC:1.3.99.29 ] [EC:1.3.99.31] | |
| gene4907 | crtU | K09879 | carotenoid phi-ring synthase/carotenoid chi-ring synthase | [EC:1.3.99.39] [EC:1.3.99.40] | |
| gene6428 | crtP | K10210 | diapolycopene oxygenase | [EC:1.14.99.44] | |
| gene7872 | cruC | K14597 | chlorobactene glucosyltransferase | - |
| Activity Description | Gene ID | KO Name | KO ID | KO Description | Enzyme |
|---|---|---|---|---|---|
| General secretory (Sec) | gene3731 | secY | K03076 | preprotein translocase subunit SecY | - |
| gene3781 | secE | K03073 | preprotein translocase subunit SecE | - | |
| gene5351 | secA | K03070 | preprotein translocase subunit SecA | [EC:7.4.2.8] | |
| gene6638 | secG | K03075 | preprotein translocase subunit SecG | - | |
| gene7245 | secD | K03072 | preprotein translocase subunit SecD | - | |
| gene7246 | secF | K03074 | preprotein translocase subunit SecF | - | |
| gene8418 | secDF | K12257 | SecD/SecF fusion protein | - | |
| Twin-arginine translocation system | gene2382 | tatA | K03116 | sec-independent protein translocase protein TatA | - |
| gene7067 | tatA | K03116 | sec-independent protein translocase protein TatA | - | |
| gene8221 | tatA | K03116 | sec-independent protein translocase protein TatA | - | |
| gene3274 | tatB | K03117 | sec-independent protein translocase protein TatB | - | |
| gene5174 | tatD | K03424 | TatD DNase family protein | [EC:3.1.21.-] | |
| gene7068 | tatC | K03118 | sec-independent protein translocase protein TatC | - |

References
- Hu, G.; Yu, L.; Dong, Z.; Lu, J.; Li, J.; Wang, Y.; Lai, Z. Holocene Aeolian Activity in the Zoige Basin, Northeastern Tibetan Plateau, China. CATENA 2018, 160, 321–328. [Google Scholar] [CrossRef]
- Zhu, Q.; Chen, H.; Peng, C.; Liu, J.; Piao, S.; He, J.-S.; Wang, S.; Zhao, X.; Zhang, J.; Fang, X.; et al. An Early Warning Signal for Grassland Degradation on the Qinghai-Tibetan Plateau. Nat. Commun. 2023, 14, 6406. [Google Scholar] [CrossRef]
- Xia, Y.; He, R.; Xu, W.; Zhang, J. The Zoige Pioneer Plant Leymus secalinus Has Different Endophytic Bacterial Community Structures to Adapt to Environmental Conditions. PeerJ 2023, 11, e15363. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Zhao, W.; Zhao, M. Remediation of Blowouts by Clonal Plants in Maqu Degraded Alpine Grasslands of Northwest China. J. Plant Res. 2017, 130, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Ghoreshizadeh, S.; Calvo-Peña, C.; Ruiz-Muñoz, M.; Otero-Suárez, R.; Coque, J.J.R.; Cobos, R. Pseudomonas Taetrolens ULE-PH5 and Pseudomonas sp. ULE-PH6 Isolated from the Hop Rhizosphere Increase Phosphate Assimilation by the Plant. Plants 2024, 13, 402. [Google Scholar] [CrossRef] [PubMed]
- Myo, E.M.; Ge, B.; Ma, J.; Cui, H.; Liu, B.; Shi, L.; Jiang, M.; Zhang, K. Indole-3-Acetic Acid Production by Streptomyces Fradiae NKZ-259 and Its Formulation to Enhance Plant Growth. BMC Microbiol. 2019, 19, 155. [Google Scholar] [CrossRef]
- Chieb, M.; Gachomo, E.W. The Role of Plant Growth Promoting Rhizobacteria in Plant Drought Stress Responses. BMC Plant Biol. 2023, 23, 407. [Google Scholar] [CrossRef]
- Lu, P.; Hao, T.; Li, X.; Wang, H.; Zhai, X.; Tian, Q.; Bai, W.; Stevens, C.; Zhang, W. Ambient Nitrogen Deposition Drives Plant-diversity Decline by Nitrogen Accumulation in a Closed Grassland Ecosystem. J. Appl. Ecol. 2021, 58, 1888–1898. [Google Scholar] [CrossRef]
- Moreau, D.; Bardgett, R.D.; Finlay, R.D.; Jones, D.L.; Philippot, L. A Plant Perspective on Nitrogen Cycling in the Rhizosphere. Funct. Ecol. 2019, 33, 540–552. [Google Scholar] [CrossRef]
- Kane, J.L.; Schartiger, R.G.; Daniels, N.K.; Freedman, Z.B.; McDonald, L.M.; Skousen, J.G.; Morrissey, E.M. Bioenergy Crop Miscanthus x Giganteus Acts as an Ecosystem Engineer to Increase Bacterial Diversity and Soil Organic Matter on Marginal Land. Soil Biol. Biochem. 2023, 186, 109178. [Google Scholar] [CrossRef]
- Zhu, X.; Liu, D.; Yin, H. Roots Regulate Microbial N Processes to Achieve an Efficient NH4+ Supply in the Rhizosphere of Alpine Coniferous Forests. Biogeochemistry 2021, 155, 39–57. [Google Scholar] [CrossRef]
- Arsyadi, A.; Guo, Y.; Ebihara, A.; Sakagami, N.; Sakoda, M.; Tago, K.; Kamijo, T.; Ohta, H.; Nishizawa, T. A Nitrate-Transforming Bacterial Community Dominates in the Miscanthus Rhizosphere on Nitrogen-Deficient Volcanic Deposits of Miyake-Jima. Microorganisms 2023, 11, 260. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Du, Q.; Wu, Y.; Li, R.; Yan, X.; Li, N.; Wang, X. Rapid Dissimilatory Nitrate Reduction to Ammonium Conserves Bioavailable Nitrogen in Organic Deficient Soils. Soil Biol. Biochem. 2023, 177, 108923. [Google Scholar] [CrossRef]
- Ren, H.; Xu, Z.; Isbell, F.; Huang, J.; Han, X.; Wan, S.; Chen, S.; Wang, R.; Zeng, D.-H.; Jiang, Y.; et al. Exacerbated Nitrogen Limitation Ends Transient Stimulation of Grassland Productivity by Increased Precipitation. Ecol. Monogr. 2017, 87, 457–469. [Google Scholar] [CrossRef]
- Kraft, B.; Tegetmeyer, H.E.; Sharma, R.; Klotz, M.G.; Ferdelman, T.G.; Hettich, R.L.; Geelhoed, J.S.; Strous, M. The Environmental Controls That Govern the End Product of Bacterial Nitrate Respiration. Science 2014, 345, 676–679. [Google Scholar] [CrossRef]
- Cameron, K.C.; Di, H.J.; Moir, J.L. Nitrogen Losses from the Soil/Plant System: A Review. Ann. Appl. Biol. 2013, 162, 145–173. [Google Scholar] [CrossRef]
- Kuypers, M.M.M.; Marchant, H.K.; Kartal, B. The Microbial Nitrogen-Cycling Network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef]
- Pandey, C.B. DNRA: A Short-Circuit in Biological N-Cycling to Conserve Nitrogen in Terrestrial Ecosystems. Sci. Total Environ. 2020, 738, 139710. [Google Scholar] [CrossRef]
- Hamada, M.A.; Soliman, E.R.S. Characterization and Genomics Identification of Key Genes Involved in Denitrification-DNRA-Nitrification Pathway of Plant Growth-Promoting Rhizobacteria (Serratia Marcescens OK482790). BMC Microbiol. 2023, 23, 210. [Google Scholar] [CrossRef]
- Li, Y. Enhancing Wheat Yield Through Microbial Organic Fertilizer Substitution for Partial Chemical Fertilization: Regulation of Nitrogen Conversion and Utilization. J. Soil Sci. Plant Nutr. 2024, 24, 935–943. [Google Scholar] [CrossRef]
- Moon, N.J.; Henk, W.G. Progression of Epiphytic Microflora in Wheat and Alfalfa Silages as Observed by Scanning Electron Microscopy. Appl. Environ. Microbiol. 1980, 40, 1122–1129. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Shi, C.; Liu, L.; Han, J.; Yang, Q.; Wang, Y.; Li, X.; Fu, W.; Gao, H.; Huang, H.; et al. Majorbio Cloud 2024: Update Single-cell and Multiomics Workflows. iMeta 2024, 3, e217. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Besemer, J.; Borodovsky, M. GeneMark: Web Software for Gene Finding in Prokaryotes, Eukaryotes and Viruses. Nucleic Acids Res. 2005, 33, W451–W454. [Google Scholar] [CrossRef]
- Kollmar, M. (Ed.) Gene Prediction: Methods and Protocols; Springer: New York, NY, USA, 2019; Volume 1962, ISBN 978-1-4939-9172-3. [Google Scholar]
- Van Dommelen, A.; Keijers, V.; Somers, E.; Vanderleyden, J. Cloning and Characterisation of the Azospirillum Brasilense glnD Gene and Analysis of a glnD Mutant. Mol. Gen. Genom. 2002, 266, 813–820. [Google Scholar] [CrossRef]
- Tremblay, P.; Hallenbeck, P.C. Of Blood, Brains and Bacteria, the Amt/Rh Transporter Family: Emerging Role of Amt as a Unique Microbial Sensor. Mol. Microbiol. 2009, 71, 12–22. [Google Scholar] [CrossRef]
- Forchhammer, K.; Selim, K.A.; Huergo, L.F. New Views on PII Signaling: From Nitrogen Sensing to Global Metabolic Control. Trends Microbiol. 2022, 30, 722–735. [Google Scholar] [CrossRef]
- Kompaniiets, D.; He, L.; Wang, D.; Zhou, W.; Yang, Y.; Hu, Y.; Liu, B. Structural Basis for Transcription Activation by the Nitrate-Responsive Regulator NarL. Nucleic. Acids. Res. 2024, 52, 1471–1482. [Google Scholar] [CrossRef]
- De Boer, A.P.N.; Van Der Oost, J.; Reijnders, W.N.M.; Westerhoff, H.V.; Stouthamer, A.H.; Van Spanning, R.J.M. Mutational Analysis of the Nor Gene Cluster Which Encodes Nitric-Oxide Reductase from Paracoccus Denitrificans. Eur. J. Biochem. 1996, 242, 592–600. [Google Scholar] [CrossRef]
- Thakur, M.; Anand, A. Hydrogen sulfide: An Emerging Signaling Molecule Regulating Drought Stress Response in Plants. Physiol. Plant. 2021, 172, 1227–1243. [Google Scholar] [CrossRef]
- Jaiswal, S.; Singh, S.P.; Singh, S.; Gupta, R.; Tripathi, D.K.; Corpas, F.J.; Singh, V.P. Hydrogen Sulphide: A Key Player in Plant Development and Stress Resilience. Plant Cell Environ. 2024. early view. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.S.; Islam, F.; Ye, Y.; Ashline, M.; Wang, D.; Zhao, B.; Fu, Z.Q.; Chen, J. The Interplay between Hydrogen Sulfide and Phytohormone Signaling Pathways under Challenging Environments. Int. J. Mol. Sci. 2022, 23, 4272. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Sun, L.; Yin, Z.; Cui, S.; Huang, W.; Xie, Z. Insights into Genomic Evolution and the Potential Genetic Basis of Klebsiella variicola subsp. Variicola ZH07 Reveal Its Potential for Plant Growth Promotion and Autotoxin Degradation. Microbiol. Spectr. 2022, 10, e00846-22. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Chater, K.F.; Chandra, G.; Niu, G.; Tan, H. Molecular Regulation of Antibiotic Biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 2013, 77, 112–143. [Google Scholar] [CrossRef]
- Mavrodi, D.V.; Bonsall, R.F.; Delaney, S.M.; Soule, M.J.; Phillips, G.; Thomashow, L.S. Functional Analysis of Genes for Biosynthesis of Pyocyanin and Phenazine-1-Carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 2001, 183, 6454–6465. [Google Scholar] [CrossRef] [PubMed]
- Subhadra, B.; Surendran, S.; Lim, B.R.; Yim, J.S.; Kim, D.H.; Woo, K.; Kim, H.-J.; Oh, M.H.; Choi, C.H. The Osmotic Stress Response Operon betIBA Is under the Functional Regulation of BetI and the Quorum-Sensing Regulator AnoR in Acinetobacter nosocomialis. J. Microbiol. 2020, 58, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Fourie, K.R.; Wilson, H.L. Understanding GroEL and DnaK Stress Response Proteins as Antigens for Bacterial Diseases. Vaccines 2020, 8, 773. [Google Scholar] [CrossRef]
- Istamycin Aminoglycosides Profiling and Their Characterization in Streptomyces Tenjimariensis ATCC 31603 Culture Using High-performance Liquid Chromatography with Tandem Mass Spectrometry. J. Sep. Sci. 2016, 39, 24.
- Kaewkla, O.; Perkins, M.; Thamchaipenet, A.; Saijuntha, W.; Sukpanoa, S.; Suriyachadkun, C.; Chamroensaksri, N.; Chumroenphat, T.; Franco, C.M.M. Description of Streptomyces Naphthomycinicus Sp. Nov., an Endophytic Actinobacterium Producing Naphthomycin A and Its Genome Insight for Discovering Bioactive Compounds. Front. Microbiol. 2024, 15, 1353511. [Google Scholar] [CrossRef]
- Nofiani, R.; Rudiyansyah; Ardiningsih, P.; Rizky; Zahra, S.T.A.; Sukito, A.; Weisberg, A.J.; Chang, J.H.; Mahmud, T. Genome Features and Secondary Metabolite Potential of the Marine Symbiont Streptomyces Sp. RS2. Arch. Microbiol. 2023, 205, 244. [Google Scholar] [CrossRef]
- Iftime, D.; Kulik, A.; Härtner, T.; Rohrer, S.; Niedermeyer, T.H.J.; Stegmann, E.; Weber, T.; Wohlleben, W. Identification and Activation of Novel Biosynthetic Gene Clusters by Genome Mining in the Kirromycin Producer Streptomyces collinus Tü 365. J. Ind. Microbiol. Biotechnol. 2016, 43, 277–291. [Google Scholar] [CrossRef]
- Bursy, J.; Kuhlmann, A.U.; Pittelkow, M.; Hartmann, H.; Jebbar, M.; Pierik, A.J.; Bremer, E. Synthesis and Uptake of the Compatible Solutes Ectoine and 5-Hydroxyectoine by Streptomyces coelicolor A3(2) in Response to Salt and Heat Stresses. Appl. Environ. Microbiol. 2008, 74, 7286–7296. [Google Scholar] [CrossRef] [PubMed]
- Seyedsayamdost, M.R.; Traxler, M.F.; Zheng, S.-L.; Kolter, R.; Clardy, J. Structure and Biosynthesis of Amychelin, an Unusual Mixed-Ligand Siderophore from Amycolatopsis sp. AA4. J. Am. Chem. Soc. 2011, 133, 11434–11437. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, P.N.; Jha, D.K. Plant Growth-Promoting Rhizobacteria (PGPR): Emergence in Agriculture. World J. Microbiol. Biotechnol. 2012, 28, 1327–1350. [Google Scholar] [CrossRef]
- De Andrade, L.A.; Santos, C.H.B.; Frezarin, E.T.; Sales, L.R.; Rigobelo, E.C. Plant Growth-Promoting Rhizobacteria for Sustainable Agricultural Production. Microorganisms 2023, 11, 1088. [Google Scholar] [CrossRef]
- Gupta, R.; Khan, F.; Alqahtani, F.M.; Hashem, M.; Ahmad, F. Plant Growth–Promoting Rhizobacteria (PGPR) Assisted Bioremediation of Heavy Metal Toxicity. Appl. Biochem. Biotechnol. 2024, 196, 2928–2956. [Google Scholar] [CrossRef]
- Narsing Rao, M.P.; Lohmaneeratana, K.; Bunyoo, C.; Thamchaipenet, A. Actinobacteria–Plant Interactions in Alleviating Abiotic Stress. Plants 2022, 11, 2976. [Google Scholar] [CrossRef]
- Zhu, X.; Meng, L.; Jiang, C.; Nie, W.; Cao, Y.; Lian, B. The Mechanism of a Multifunctional Strain of Streptomyces sp. on the Growth of Pinus massoniana Seedlings. J. Soil Sci. Plant Nutr. 2024, 24, 8121–8136. [Google Scholar] [CrossRef]
- Youseif, S.H.; El-Megeed, F.H.A.; Salous, M.S.; Mohamed, A.H. Streptomyces Biostimulants: An Effective Sustainable Approach to Reduce Inorganic N Input and Maintain High Yield of Wheat Crop in Different Soil Types. J. Appl. Microbiol. 2023, 134, lxad156. [Google Scholar] [CrossRef]
- Qin, S.; Feng, W.-W.; Wang, T.-T.; Ding, P.; Xing, K.; Jiang, J.-H. Plant Growth-Promoting Effect and Genomic Analysis of the Beneficial Endophyte Streptomyces sp. KLBMP 5084 Isolated from Halophyte Limonium Sinense. Plant Soil 2017, 416, 117–132. [Google Scholar] [CrossRef]
- Boukelloul, I.; Aouar, L.; Cherb, N.; Carvalho, M.F.; Oliveira, R.S.; Akkal, S.; Nieto, G.; Zellagui, A.; Necib, Y. Actinobacteria Isolated from Soils of Arid Saharan Regions Display Simultaneous Antifungal and Plant Growth Promoting Activities. Curr. Microbiol. 2024, 81, 327. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Hu, L.; Hu, H.; Zhou, F.; Wu, L.; Wang, S.; Rozhkova, T.; Li, C. Identification of a Novel Streptomyces Sp. Strain HU2014 Showing Growth Promotion and Biocontrol Effect Against Rhizoctonia spp. in Wheat. Plant Dis. 2023, 107, 1139–1150. [Google Scholar] [CrossRef]
- Nonthakaew, N.; Panbangred, W.; Songnuan, W.; Intra, B. Plant Growth-Promoting Properties of Streptomyces spp. Isolates and Their Impact on Mung Bean Plantlets’ Rhizosphere Microbiome. Front. Microbiol. 2022, 13, 967415. [Google Scholar] [CrossRef] [PubMed]
- Stephens, B.B.; Loar, S.N.; Alexandre, G. Role of CheB and CheR in the Complex Chemotactic and Aerotactic Pathway of Azospirillum brasilense. J. Bacteriol. 2006, 188, 4759–4768. [Google Scholar] [CrossRef] [PubMed]
- Kanungpean, D.; Kakuda, T.; Takai, S. Participation of CheR and CheB in the Chemosensory Response of Campylobacter Jejuni. Microbiology 2011, 157, 1279–1289. [Google Scholar] [CrossRef]
- García-Fontana, C.; Reyes-Darias, J.A.; Muñoz-Martínez, F.; Alfonso, C.; Morel, B.; Ramos, J.L.; Krell, T. High Specificity in CheR Methyltransferase Function. J. Biol. Chem. 2013, 288, 18987–18999. [Google Scholar] [CrossRef]
- Augimeri, R.V.; Varley, A.J.; Strap, J.L. Establishing a Role for Bacterial Cellulose in Environmental Interactions: Lessons Learned from Diverse Biofilm-Producing Proteobacteria. Front. Microbiol. 2015, 6, 1282. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, J.; Kong, Z. Cellular Basis of Legume–Rhizobium Symbiosis. Plant Commun. 2024, 5, 101045. [Google Scholar] [CrossRef]
- Zhang, Y.; Ku, Y.-S.; Cheung, T.-Y.; Cheng, S.-S.; Xin, D.; Gombeau, K.; Cai, Y.; Lam, H.-M.; Chan, T.-F. Challenges to Rhizobial Adaptability in a Changing Climate: Genetic Engineering Solutions for Stress Tolerance. Microbiol. Res. 2024, 288, 127886. [Google Scholar] [CrossRef]
- Frungillo, L. Getting to the Root of Nodulation: How Legumes and Rhizobia Use Nitrate Uptake to Control Symbiosis. Plant Cell 2022, 34, 1443–1444. [Google Scholar] [CrossRef]
- Koch, H. The Microbial-Driven Nitrogen Cycle and Its Relevance for Plant Nutrition. J. Exp. Bot. 2024, 75, 5547–5556. [Google Scholar] [CrossRef] [PubMed]
- Putz, M.; Schleusner, P.; Rütting, T.; Hallin, S. Relative Abundance of Denitrifying and DNRA Bacteria and Their Activity Determine Nitrogen Retention or Loss in Agricultural Soil. Soil Biol. Biochem. 2018, 123, 97–104. [Google Scholar] [CrossRef]
- Wei, Z. Biochar Amendment Alters the Partitioning of Nitrate Reduction by Significantly Enhancing DNRA in a Paddy Field. Biochar 2022, 4, 44. [Google Scholar] [CrossRef]
- Friedl, J.; De Rosa, D.; Rowlings, D.W.; Grace, P.R.; Müller, C.; Scheer, C. Dissimilatory Nitrate Reduction to Ammonium (DNRA), Not Denitrification Dominates Nitrate Reduction in Subtropical Pasture Soils upon Rewetting. Soil Biol. Biochem. 2018, 125, 340–349. [Google Scholar] [CrossRef]
- Huang, X.; Luoluo; Xie, D.; Li, Z. Dissimilatory Nitrate Reduction to Ammonium in Four Pseudomonas spp. under Aerobic Conditions. Heliyon 2023, 9, e14983. [Google Scholar] [CrossRef]
- Reay, M.K.; Marsden, K.A.; Powell, S.; Rivera, L.M.; Chadwick, D.R.; Jones, D.L.; Evershed, R.P. The Soil Microbial Community and Plant Biomass Differentially Contribute to the Retention and Recycling of Urinary-N in Grasslands. Soil Biol. Biochem. 2023, 180, 109011. [Google Scholar] [CrossRef]
- Makino, K.; Shinagawa, H.; Amemura, M.; Kawamoto, T.; Yamada, M.; Nakata, A. Signal Transduction in the Phosphate Regulon of Escherichia coli Involves Phosphotransfer between PhoR and PhoB Proteins. J. Mol. Biol. 1989, 210, 551–559. [Google Scholar] [CrossRef]
- Eichhorn, E.; Van Der Ploeg, J.R.; Leisinger, T. Deletion Analysis of the Escherichia coli Taurine and Alkanesulfonate Transport Systems. J. Bacteriol. 2000, 182, 2687–2695. [Google Scholar] [CrossRef]
- Tang, J.; Li, Y.; Zhang, L.; Mu, J.; Jiang, Y.; Fu, H.; Zhang, Y.; Cui, H.; Yu, X.; Ye, Z. Biosynthetic Pathways and Functions of Indole-3-Acetic Acid in Microorganisms. Microorganisms 2023, 11, 2077. [Google Scholar] [CrossRef]
- Salwan, R.; Sharma, V.; Sharma, A.; Singh, A. Molecular Imprints of Plant Beneficial Streptomyces sp. AC30 and AC40 Reveal Differential Capabilities and Strategies to Counter Environmental Stresses. Microbiol. Res. 2020, 235, 126449. [Google Scholar] [CrossRef]
- Etesami, H.; Glick, B.R. Bacterial Indole-3-Acetic Acid: A Key Regulator for Plant Growth, Plant-Microbe Interactions, and Agricultural Adaptive Resilience. Microbiol. Res. 2024, 281, 127602. [Google Scholar] [CrossRef] [PubMed]
- Cen, X.; Li, H.; Zhang, Y.; Huang, L.; Luo, Y. Isolation and Plant Growth Promotion Effect of Endophytic Siderophore-Producing Bacteria: A Study on Halophyte Sesuvium Portulacastrum. Plants 2024, 13, 2703. [Google Scholar] [CrossRef] [PubMed]
- Timofeeva, A.M.; Galyamova, M.R.; Sedykh, S.E. Plant Growth-Promoting Soil Bacteria: Nitrogen Fixation, Phosphate Solubilization, Siderophore Production, and Other Biological Activities. Plants 2023, 12, 4074. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y. Screening of Siderophore-Producing Bacteria and Their Effects on Promoting the Growth of Plants. Curr. Microbiol. 2022, 79, 150. [Google Scholar] [CrossRef]
- Codd, R.; Richardson-Sanchez, T.; Telfer, T.J.; Gotsbacher, M.P. Advances in the Chemical Biology of Desferrioxamine B. ACS Chem. Biol. 2018, 13, 11–25. [Google Scholar] [CrossRef]
- Wang, W.; Qiu, Z.; Tan, H.; Cao, L. Siderophore Production by Actinobacteria. Biometals 2014, 27, 623–631. [Google Scholar] [CrossRef]
- Cruz-Morales, P.; Ramos-Aboites, H.E.; Licona-Cassani, C.; Selem-Mójica, N.; Mejía-Ponce, P.M.; Souza-Saldívar, V.; Barona-Gómez, F. Actinobacteria Phylogenomics, Selective Isolation from an Iron Oligotrophic Environment and Siderophore Functional Characterization, Unveil New Desferrioxamine Traits. FEMS Microbiol. Ecol. 2017, 93, fix086. [Google Scholar] [CrossRef]
- Araujo, R.; Dunlap, C.; Barnett, S.; Franco, C.M.M. Decoding Wheat Endosphere–Rhizosphere Microbiomes in Rhizoctonia Solani–Infested Soils Challenged by Streptomyces Biocontrol Agents. Front. Plant Sci. 2019, 10, 1038. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, T.; Xue, Z.; Liu, Y.; Li, Y.; Li, Y.; Chen, Q. Streptomyces Application Triggers Reassembly and Optimization of the Rhizosphere Microbiome of Cucumber. Diversity 2021, 13, 413. [Google Scholar] [CrossRef]
- Hu, D.; Li, S.; Li, Y.; Peng, J.; Wei, X.; Ma, J.; Zhang, C.; Jia, N.; Wang, E.; Wang, Z. Streptomyces sp. Strain TOR3209: A Rhizosphere Bacterium Promoting Growth of Tomato by Affecting the Rhizosphere Microbial Community. Sci. Rep. 2020, 10, 20132. [Google Scholar] [CrossRef]
- Richter, A.A.; Mais, C.-N.; Czech, L.; Geyer, K.; Hoeppner, A.; Smits, S.H.J.; Erb, T.J.; Bange, G.; Bremer, E. Biosynthesis of the Stress-Protectant and Chemical Chaperon Ectoine: Biochemistry of the Transaminase EctB. Front. Microbiol. 2019, 10, 2811. [Google Scholar] [CrossRef] [PubMed]
- Bagnoli, F.; Rappuoli, R. Protein and Sugar Export and Assembly in Gram-Positive Bacteria; Springer International Publishing: Basel, Switzerland, 2017; pp. 69–94. [Google Scholar]
- Tseng, T.-T.; Tyler, B.M.; Setubal, J.C. Protein Secretion Systems in Bacterial-Host Associations, and Their Description in the Gene Ontology. BMC Microbiol. 2009, 9, S2. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Jiao, M.; Nie, X.; Wang, C.; Yu, X.; Liu, Y.; Wei, X. Genomic and Metabolomic Profiling Reveal Streptomyces rochei S32 Contributes to Plant Growth by Nitrogen Fixation and Production of Bioactive Substances. Plant Soil 2024, 501, 343–360. [Google Scholar] [CrossRef]
- Wang, Q.; Zhao, J.; Liu, Z.; Ding, S.; Huang, Z.; Chen, J. Genomic Insights and Synthetic Biology Applications of Marine Actinomycete Streptomyces Griseoincarnatus HNS054. Int. J. Mol. Sci. 2024, 25, 3127. [Google Scholar] [CrossRef]
- Fatima, A.; Abbas, M.; Nawaz, S.; Rehman, Y.; Rehman, S.U.; Sajid, I. Whole Genome Sequencing (WGS) and Genome Mining of Streptomyces sp. AFD10 for Antibiotics and Bioactive Secondary Metabolites Biosynthetic Gene Clusters (BGCs). Gene Rep. 2024, 37, 102050. [Google Scholar] [CrossRef]
- Dávila Costa, J.S.; Hoskisson, P.A.; Paterlini, P.; Romero, C.M.; Alvarez, A. Whole Genome Sequence of the Multi-Resistant Plant Growth-Promoting Bacteria Streptomyces sp. Z38 with Potential Application in Agroindustry and Bio-Nanotechnology. Genomics 2020, 112, 4684–4689. [Google Scholar] [CrossRef]
- Xu, G.; Wu, W.; Zhu, L.; Liang, Y.; Liang, M.; Tan, S.; Chen, H.; Huang, X.; He, C.; Lu, Y.; et al. Whole Genome Sequencing and Biocontrol Potential of Streptomyces Luteireticuli ASG80 Against Phytophthora Diseases. Microorganisms 2024, 12, 2255. [Google Scholar] [CrossRef]
- Vaz Jauri, P.; Beracochea, M.; Fernández, B.; Battistoni, F. Whole-Genome Sequencing of Streptomyces sp. Strain UYFA156, a Cultivar-Specific Plant Growth-Promoting Endophyte of Festuca Arundinacea. Microbiol. Resour. Announc. 2019, 8, e00722-19. [Google Scholar] [CrossRef]
- Gallegos-Lopez, S.; Mejia-Ponce, P.M.; Gonzalez-Salazar, L.A.; Rodriguez-Orduña, L.; Souza-Saldivar, V.; Licona-Cassani, C. Draft Genome Sequence of Streptomyces sp. Strain C8S0, Isolated from a Highly Oligotrophic Sediment. Microbiol. Resour. Announc. 2020, 9, e01441-19. [Google Scholar] [CrossRef]
- Jiang, L.; Zeng, Z.; Wang, Z.; Tang, M.; Jiang, S.; Ma, Q.; Wang, Z.; Peng, D.; Li, S.; Pu, H. Genomic Investigation of a Rhizosphere Isolate, Streptomyces sp. JL1001, Associated with Polygonatum Cyrtonema Hua. Curr. Microbiol. 2024, 81, 368. [Google Scholar] [CrossRef]
- Duangupama, T.; Pittayakhajonwut, P.; Intaraudom, C.; Suriyachadkun, C.; Tadtong, S.; Kuncharoen, N.; He, Y.-W.; Tanasupawat, S.; Thawai, C. Description of Streptomyces Siderophoricus sp. nov., a Promising Nocardamine-Producing Species Isolated from the Rhizosphere Soil of Mangifera Indica. J. Antibiot. 2024, 77, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Li, L.-F.; Wu, Q.-X.; Wu, H.; Li, Y.; Peng, Q.; Han, R.-H.; Zhang, D.-H.; Yu, W.-D.; Xu, R.; Wang, J.; et al. Complete Genome Sequence of Streptomyces Sp. HP-A2021, a Promising Bacterium for Natural Product Discovery. Biochem. Genet. 2023, 61, 2042–2055. [Google Scholar] [CrossRef] [PubMed]
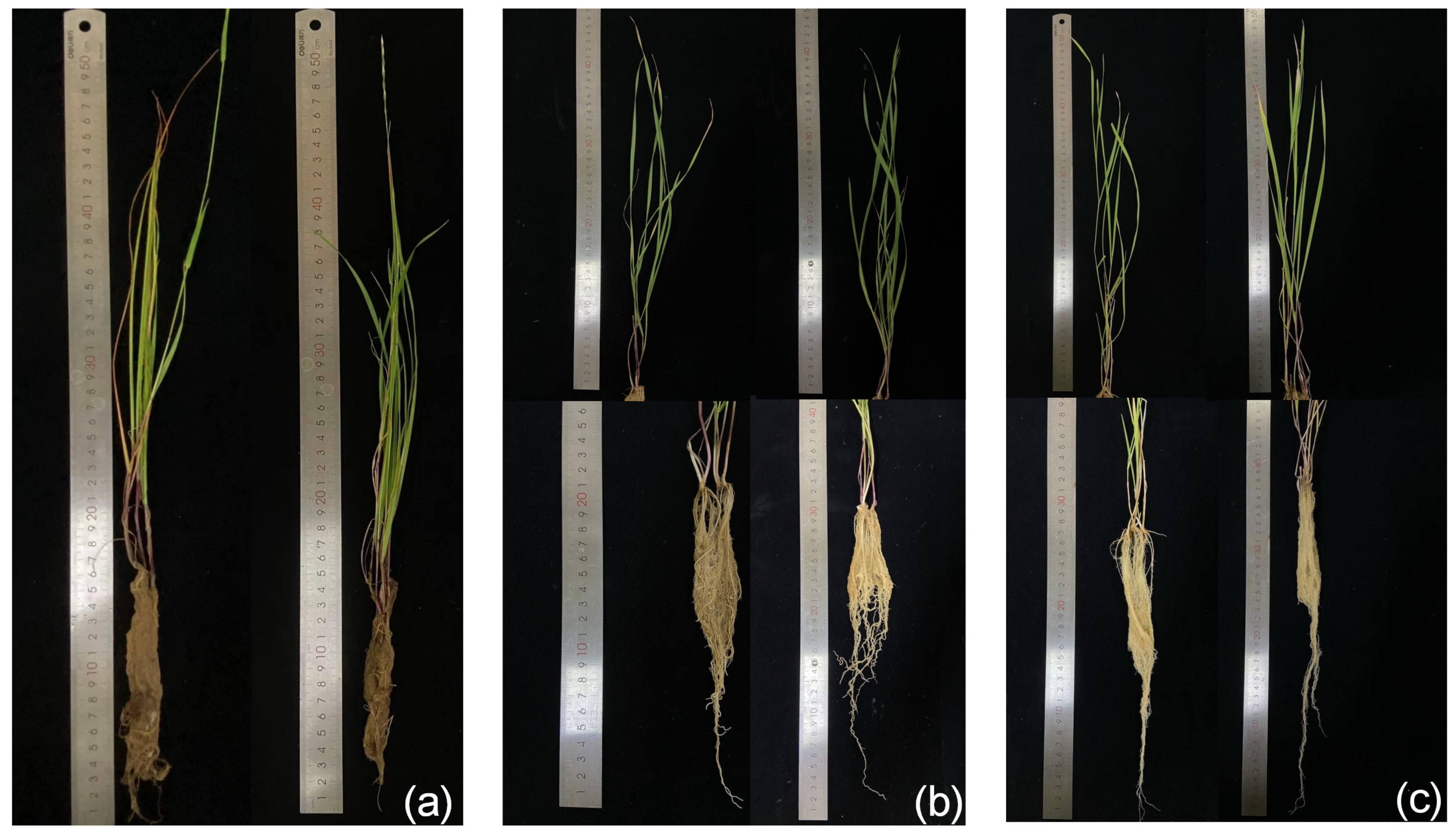


| Features | Chromosome |
|---|---|
| Total size of the contigs (In Megabases) | 9,994,786 bp |
| Number of protein-coding genes | 9352 |
| Number of rRNA genes | 18 |
| Number of tRNA genes | 71 |
| Number of sRNA genes | 62 |
| G + C% | 70.46% |
| Signal transduction | 138 |
| Prophage regions | 1 |
| Functional Annotations | Number of Protein-Coding Genes (CDs) | Percentage (%) |
|---|---|---|
| Total | 9352 | 100 |
| COG | 7009 | 74.9 |
| KEGG | 6107 | 65.3 |
| GO | 5435 | 63.66 |
| NR | 9258 | 58.11 |
| Swiss-port | 5902 | 63.1 |
| Pfam | 7426 | 79.4 |
| Function | Gene ID | KO Name | KO ID | KO Description | Enzyme |
|---|---|---|---|---|---|
| Nitrogen metabolism | gene2544 | gltB | K00265 | glutamate synthase (NADPH), large chain | [EC:1.4.1.13] |
| gene5942 | nirD | K00363 | nitrite reductase (NADH), small subunit | [EC:1.7.1.15] | |
| gene5943 | nirB | K00362 | nitrite reductase (NADH), large subunit | [EC:1.7.1.15] | |
| gene4307 | narK | K02575 | MFS transporter, NNP family, nitrate/nitrite transporter | - | |
| gene0326 | narL | K07684 | nitrate/nitrite response regulator NarL, two-component system, NarL family | - | |
| gene2048 | narH | K00371 | nitrate reductase/nitrite oxidoreductase, beta subunit | [EC:1.7.5.11.7.99.-] | |
| gene2049 | narJ | K00373 | nitrate reductase molybdenum cofactor assembly chaperone NarJ/NarW | - | |
| gene2544 | gltB | K00265 | glutamate synthase (NADPH), large chain | [EC:1.4.1.13] | |
| gene6510 | gltD | K00266 | glutamate synthase (NADPH), small chain | [EC:1.4.1.13] | |
| gene1739 | glnA | K01915 | glutamine synthetase | [EC:6.3.1.2] | |
| gene2808 | glnB | K04751 | nitrogen regulatory protein P-II 1 | - | |
| gene2807 | glnD | K00990 | [protein-PII] uridylyltransferase | [EC:2.7.7.59] | |
| gene6281 | glnE | K00982 | [glutamine synthetase] adenylyltransferase/[glutamine synthetase]-adenylyl-L-tyrosine phosphorylase | [EC:2.7.7.42 2.7.7.89] | |
| gene6662 | iscU | K04488 | nitrogen fixation protein NifU and related proteins | - | |
| gene2809 | amt | K03320 | ammonium transporter, Amt family | - | |
| gene0085 | nasT | K07183 | two-component system, response regulator/RNA-binding antiterminator | - | |
| gene0517 | moaC | K03637 | cyclic pyranopterin monophosphate synthase | [EC:4.6.1.17] | |
| gene3219 | moaE | K03635 | molybdopterin synthase catalytic subunit | [EC:2.8.1.12] | |
| gene3830 | moaA | K03639 | GTP 3′,8-cyclase | [EC:4.1.99.22] | |
| gene4835 | moaD | K03636 | sulfur-carrier protein | - | |
| gene8041 | urtE | K11963 | urea transport system ATP-binding protein | - | |
| gene8042 | urtD | K11962 | urea transport system ATP-binding protein | - | |
| gene8043 | urtC | K11961 | urea transport system permease protein | - | |
| gene8044 | urtB | K11960 | urea transport system permease protein | - | |
| gene8045 | urtA | K11959 | urea transport system substrate-binding protein | - | |
| sulfur metabolism | gene1472 | cysJ | K00380 | sulfite reductase (NADPH) flavoprotein alpha-component | [EC:1.8.1.2] |
| gene2334 | cysH | K00390 | phosphoadenosine phosphosulfate reductase | [EC:1.8.4.8 1.8.4.10] | |
| gene2335 | cysC | K00860 | adenylylsulfate kinase | [EC:2.7.1.25] | |
| gene2336 | cysD | K00957 | sulfate adenylyltransferase subunit 2 | [EC:2.7.7.4] | |
| gene2337 | cysN | K00956 | sulfate adenylyltransferase subunit 1 | [EC:2.7.7.4] | |
| gene2339 | ssuA | K15553 | sulfonate transport system substrate-binding protein | - | |
| gene2340 | ssuB | K15555 | sulfonate transport system ATP-binding protein | [EC:7.6.2.14] | |
| gene2341 | ssuC | K15554 | sulfonate transport system permease protein | - | |
| gene5061 | ssuD | K04091 | alkanesulfonate monooxygenase | [EC:1.14.14.5 1.14.14.34] | |
| gene2574 | sseA | K01011 | thiosulfate/3-mercaptopyruvate sulfurtransferase | [EC:2.8.1.1 2.8.1.2] | |
| gene4389 | tauA | K15551 | taurine transport system substrate-binding protein | - | |
| gene4390 | tauB | K10831 | taurine transport system ATP-binding protein | [EC:7.6.2.7] | |
| gene4388 | tauC | K15552 | taurine transport system permease protein | - | |
| gene4391 | tauD | K03119 | taurine dioxygenase | [EC:1.14.11.17] | |
| gene6420 | thiS | K03154 | sulfur carrier protein | - | |
| Phosphorus metabolism | gene4333 | pstC | K02037 | phosphate transport system permease protein | - |
| gene4334 | pstA | K02038 | phosphate transport system permease protein | - | |
| gene4335 | pstB | K02036 | phosphate transport system ATP-binding protein | [EC:7.3.2.1] | |
| gene1629 | phoD | K01113 | alkaline phosphatase D | [EC:3.1.3.1] | |
| gene6104 | phoA | K01077 | alkaline phosphatase | [EC:3.1.3.1] | |
| gene4080 | ppx-gppA | K01524 | exopolyphosphatase/guanosine-5′-triphosphate,3′-diphosphate pyrophosphatase [EC:3.6.1.11 3.6.1.40] | - | |
| gene4131 | ppa | K01507 | inorganic pyrophosphatase | [EC:3.6.1.1] | |
| gene3124 | phoP | K07658 | two-component system, OmpR family, alkaline phosphatase synthesis response regulator PhoP | - | |
| gene3254 | phoH | K06217 | phosphate starvation-inducible protein PhoH and related proteins | - | |
| gene4251 | phoU | K02039 | phosphate transport system protein | - | |
| gene5771 | phoH2 | K07175 | PhoH-like ATPase | - | |
| gene0729 | gdh | K00034 | glucose 1-dehydrogenase | [EC:1.1.1.47] |
| Function | Gene ID | KO Name | KO ID | KO Description | Enzyme |
|---|---|---|---|---|---|
| Auxin biosynthesis | gene6498 | trpA | K01695 | tryptophan synthase alpha chain | [EC:4.2.1.20] |
| gene6497 | trpB | K01696 | tryptophan synthase beta chain | [EC:4.2.1.20] | |
| gene6496 | trpC | K01609 | indole-3-glycerol phosphate synthase | [EC:4.1.1.48] | |
| gene6381 | trpD | K00766 | anthranilate phosphoribosyltransferase | [EC:2.4.2.18] | |
| gene6492 | trpE | K01657 | anthranilate synthase component I | [EC:4.1.3.27] | |
| gene5439 | nthB | K20807 | nitrile hydratase subunit beta | [EC:4.2.1.84] | |
| gene5441 | nthA | K01721 | nitrile hydratase subunit alpha | [EC:4.2.1.84] | |
| gene0620 | - | K00128 | aldehyde dehydrogenase (NAD+) | [EC:1.2.1.3] | |
| gene1290 | - | K01593 | aromatic-L-amino-acid/L-tryptophan decarboxylase | [EC:4.1.1.28 4.1.1.105] | |
| gene9104 | - | K01501 | nitrilase | [EC:3.5.5.1] | |
| gene4607 | hisC | K00817 | histidinol-phosphate aminotransferase | [EC:2.6.1.9] | |
| Abiotic stress tolerance | gene2632 | ectB | K00836 | diaminobutyrate-2-oxoglutarate transaminase | [EC:2.6.1.76] |
| gene6739 | ectD | K10674 | ectoine hydroxylase | [EC:1.14.11.55] | |
| gene6740 | ectC | K06720 | L-ectoine synthase | [EC:4.2.1.108] | |
| gene6742 | ectA | K06718 | L-2,4-diaminobutyric acid acetyltransferase | [EC:2.3.1.178] | |
| gene3683 | groEL | K04077 | chaperonin GroEL | [EC:5.6.1.7] | |
| gene3684 | groES | K04078 | chaperonin GroES | - | |
| gene3608 | betB | K00130 | betaine-aldehyde dehydrogenase [EC:1.2.1.8] | - | |
| gene1092 | betA | K00108 | choline dehydrogenase [EC:1.1.99.1] | - | |
| gene1761 | betI | K02167 | TetR/AcrR family transcriptional regulator, transcriptional repressor of bet genes | ||
| gene5096 | cspA | K03704 | cold-shock protein | - | |
| gene4328 | hspR | K13640 | MerR family transcriptional regulator, heat-shock protein HspR | - | |
| gene4731 | hslJ | K03668 | heat-shock protein HslJ | - | |
| gene3770 | htpX | K03799 | heat-shock protein HtpX | [EC:3.4.24.-] | |
| gene6571 | hslR | K04762 | ribosome-associated heat-shock protein Hsp15 | - | |
| gene5866 | hrcA | K03705 | heat-inducible transcriptional repressor | - | |
| gene1087 | proW | K02001 | glycine betaine/proline transport system permease protein | - | |
| gene1088 | proV | K02000 | glycine betaine/proline transport system ATP-binding protein | [EC:7.6.2.9] | |
| gene1866 | proP | K03762 | MFS transporter, MHS family, proline/betaine transporter | - | |
| gene2716 | proS | K01881 | prolyl-tRNA synthetase | [EC:6.1.1.15] | |
| gene4311 | proA | K00147 | glutamate-5-semialdehyde dehydrogenase | [EC:1.2.1.41] | |
| gene5828 | proB | K00931 | glutamate 5-kinase | [EC:2.7.2.11] | |
| gene2163 | ggt | K00681 | gamma-glutamyltranspeptidase/glutathione hydrolase | [EC:2.3.2.2 3.4.19.13] | |
| gene0962 | gst | K00799 | glutathione S-transferase | [EC:2.5.1.18] | |
| gene7747 | trxA | K03671 | thioredoxin 1 | - | |
| gene4564 | trxB | K00384 | thioredoxin reductase (NADPH) | [EC:1.8.1.9] | |
| gene2356 | treX | K01214 | isoamylase | [EC:3.2.1.68] | |
| gene2358 | treY | K06044 | (1->4)-alpha-D-glucan 1-alpha-D-glucosylmutase | [EC:5.4.99.15] | |
| gene2985 | treS | K05343 | maltose alpha-D-glucosyltransferase/alpha-amylase | [EC:5.4.99.16 3.2.1.1] | |
| gene4925 | otsB | K01087 | trehalose 6-phosphate phosphatase | [EC:3.1.3.12] | |
| gene4926 | otsA | K00697 | trehalose 6-phosphate synthase | [EC:2.4.1.15 2.4.1.347] | |
| Iron uptake and transport | gene8395 | fhuB | K23228 | ferric hydroxamate transport system permease protein | - |
| gene8396 | fhuD | K23227 | ferric hydroxamate transport system substrate-binding protein | - | |
| gene8397 | fhuC | K10829 | ferric hydroxamate transport system ATP-binding protein | [EC:7.2.2.16] | |
| gene3465 | fepD | K23186 | iron-siderophore transport system permease protein | - | |
| gene3466 | fepG | K23187 | iron-siderophore transport system permease protein | - | |
| gene6856 | fepC | K23188 | iron-siderophore transport system ATP-binding protein | [EC:7.2.2.177.2.2.-] | |
| gene5573 | desE | K25287 | iron-desferrioxamine transport system substrate-binding protein | - | |
| gene5604 | entS | K08225 | MFS transporter, ENTS family, enterobactin (siderophore) exporter | - |
| Cluster ID | Type | Similar Cluster | Similarity (%) | Gene No. |
|---|---|---|---|---|
| cluster1 | butyrolactone | salinomycin | 4 | 11 |
| cluster2 | butyrolactone | merochlorin A/merochlorin B | 78 | 118 |
| cluster1 | NRPS | griseochelin | 100 | 85 |
| cluster2 | RiPP-like | informatipeptin | 37 | 6 |
| cluster3 | terpene | hopene | 92 | 22 |
| cluster4 | siderophore | grincamycin | 8 | 13 |
| cluster5 | NRPS-like | s56-p1 | 11 | 35 |
| cluster6 | terpene | geosmin | 100 | 18 |
| cluster7 | RiPP-like | - | - | 12 |
| cluster8 | siderophore | - | - | 7 |
| cluster9 | terpene | albaflavenone | 100 | 21 |
| cluster10 | terpene | naphthomycin A | 9 | 22 |
| cluster11 | NRPS-like | A40926 | 7 | 41 |
| cluster12 | siderophore | desferrioxamin B/desferrioxamine E | 83 | 9 |
| cluster13 | melanin | istamycin | 5 | 11 |
| cluster14 | NRPS-like | cyphomycin | 2 | 35 |
| cluster15 | ectoine | ectoine | 100 | 10 |
| cluster16 | NAPAA | - | - | 32 |
| cluster17 | NRPS | herboxidiene | 4 | 36 |
| cluster18 | T1PKS | amychelin | 81 | 74 |
| cluster19 | melanin | melanin | 71 | 9 |
| cluster20 | T1PKS | spore pigment | 83 | 130 |
| cluster21 | terpene | 2-methylisoborneol | 100 | 17 |
| cluster22 | NRPS | lasalocid | 9 | 75 |
| cluster23 | NRPS | tylactone | 6 | 21 |
| cluster24 | RRE-containing | mycotrienin I | 7 | 30 |
| cluster25 | terpene | - | - | 21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, W.; Liu, Y.; Cheng, Y.; Zhang, J. Plant Growth-Promoting Effect and Complete Genomic Sequence Analysis of the Beneficial Rhizosphere Streptomyces sp. GD-4 Isolated from Leymus secalinus. Microorganisms 2025, 13, 286. https://doi.org/10.3390/microorganisms13020286
Xu W, Liu Y, Cheng Y, Zhang J. Plant Growth-Promoting Effect and Complete Genomic Sequence Analysis of the Beneficial Rhizosphere Streptomyces sp. GD-4 Isolated from Leymus secalinus. Microorganisms. 2025; 13(2):286. https://doi.org/10.3390/microorganisms13020286
Chicago/Turabian StyleXu, Wanru, Yimeng Liu, Yiping Cheng, and Jie Zhang. 2025. "Plant Growth-Promoting Effect and Complete Genomic Sequence Analysis of the Beneficial Rhizosphere Streptomyces sp. GD-4 Isolated from Leymus secalinus" Microorganisms 13, no. 2: 286. https://doi.org/10.3390/microorganisms13020286
APA StyleXu, W., Liu, Y., Cheng, Y., & Zhang, J. (2025). Plant Growth-Promoting Effect and Complete Genomic Sequence Analysis of the Beneficial Rhizosphere Streptomyces sp. GD-4 Isolated from Leymus secalinus. Microorganisms, 13(2), 286. https://doi.org/10.3390/microorganisms13020286





