Causal Association Between the Mucosal and Luminal Microbiotas from the Gastrointestinal Tract of Weaned Piglets Using Bayesian Network
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Feeding Conditions
2.2. Dissection Procedure
2.3. Analysis of the Mucosal and Digesta Microbiotas by 16S rRNA Gene Amplicon Sequencing
2.4. Statistical Analysis
3. Results
3.1. Diversity in Gut Microbiota Across Within-Segment and Between-Segment Comparisons
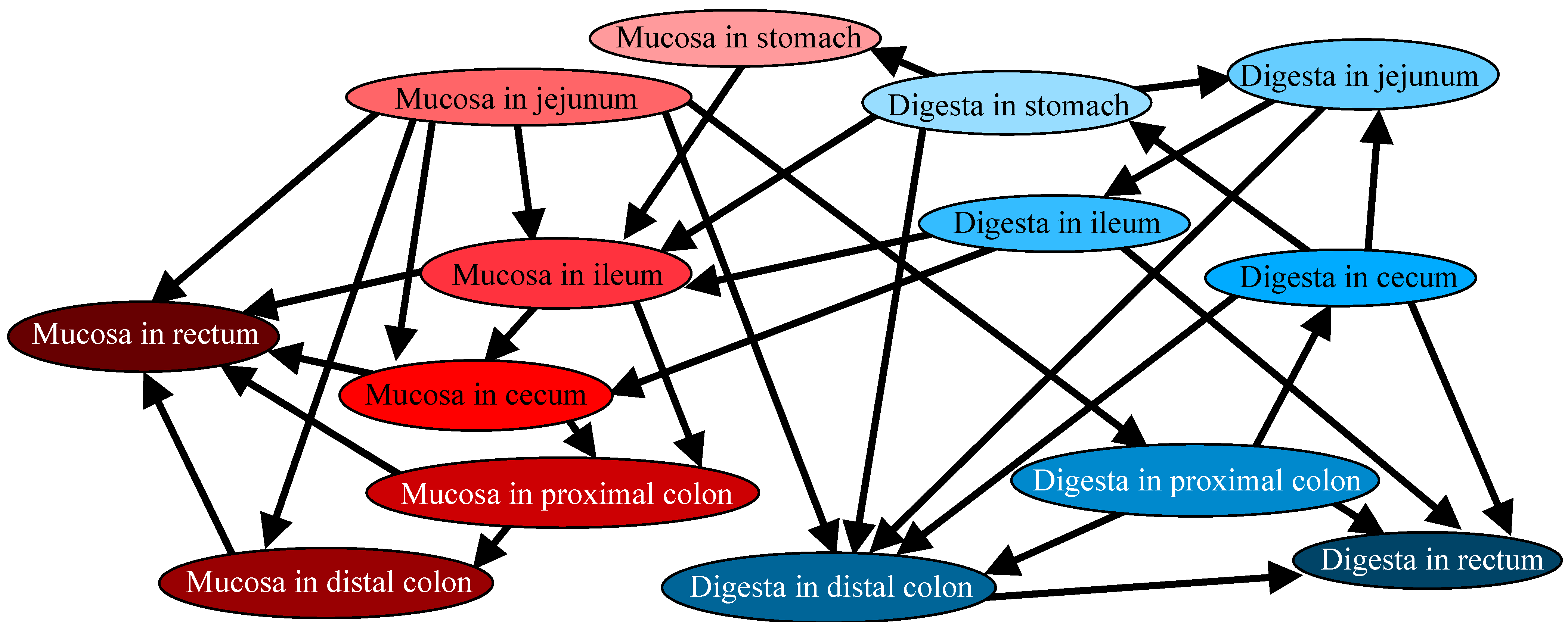
3.2. Directional Relations Among Microbiotas
3.3. Genera Abundance in Gut Microbiota Within and Between Segments
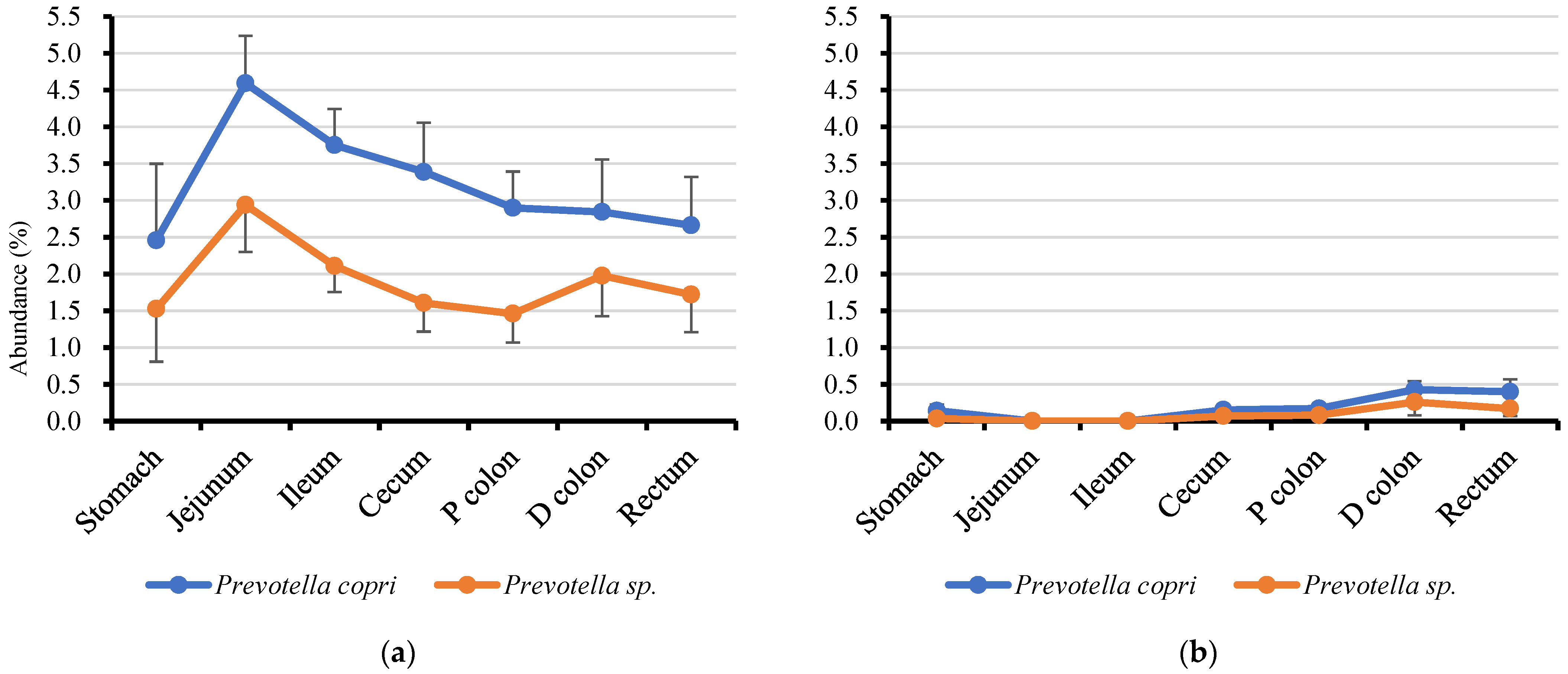
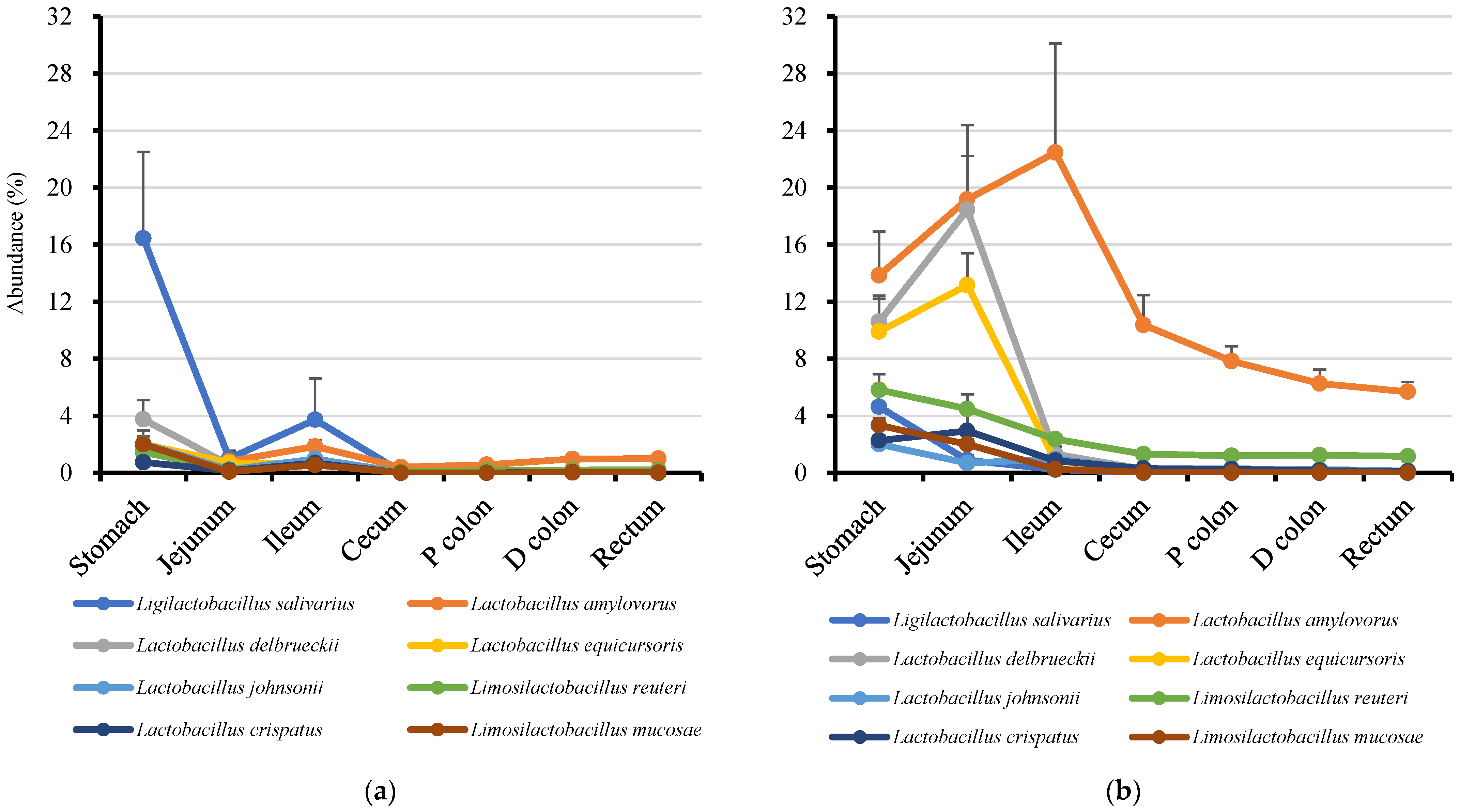
3.4. Genus Prevotella and Lactobacilli
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef]
- Fouhse, J.M.; Zijlstra, R.T.; Willing, B.P. The role of gut microbiota in the health and disease of pigs. Anim. Front. 2016, 6, 30–36. [Google Scholar] [CrossRef]
- Ramayo-Caldas, Y.; Mach, N.; Lepage, P.; Levenez, F.; Denis, C.; Lemonnier, G.; Leplat, J.-J.; Billon, Y.; Berri, M.; Doré, J.; et al. Phylogenetic network analysis applied to pig gut microbiota identifies an ecosystem structure linked with growth traits. ISME J. 2016, 10, 2973–2977. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, M.R.; Jian, C.; Uddin, M.K.; Huhtinen, M.; Salonen, A.; Peltoniemi, O.; Venhoranta, H.; Oliviero, C. Impact of intestinal microbiota on growth performance of suckling and weaned piglets. Microbiol. Spectr. 2023, 11, e0374422. [Google Scholar] [CrossRef]
- Yang, H.; Yang, M.; Fang, S.; Huang, X.; He, M.; Ke, S.; Gao, J.; Wu, J.; Zhou, Y.; Fu, H.; et al. Evaluating the profound effect of gut microbiome on host appetite in pigs. BMC Microbiol. 2018, 18, 215. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Lan, W.; Liu, G.; Ni, H.; Gu, J.-D. Exploring possible associations of the intestine bacterial microbiome with the pre-weaned weight gaining performance of piglets in intensive pig production. Sci. Rep. 2019, 9, 15534. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Huang, X.; Fang, S.; Xin, W.; Huang, L.; Chen, C. Uncovering the composition of microbial community structure and metagenomics among three gut locations in pigs with distinct fatness. Sci. Rep. 2016, 6, 27427. [Google Scholar] [CrossRef]
- Ye, Q.; Luo, T.; Han, L.; Chen, Y.; Hu, Y.; Jiang, H.; Xu, X.; Yan, X. Multi-omics analysis reveals the dominant intestinal microbial strains and metabolites related to the reproductive performance in pregnant sows. Anim. Nutr. 2024, 1, e5. [Google Scholar] [CrossRef]
- Uryu, H.; Tsukahara, T.; Ishikawa, H.; Oi, M.; Otake, S.; Yamane, I.; Inoue, R. Comparison of productivity and fecal microbiotas of sows in commercial farms. Microorganisms 2020, 8, 1469. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Bai, M.; Liu, H.; Duan, Y.; Zhou, X.; Wu, X.; Liao, P.; Li, T.; Yin, Y. Gut microbiota and blood metabolomics in weaning multiparous sows: Associations with oestrous. J. Anim. Physiol. Anim. Nutr. 2020, 104, 1155–1168. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.-L.; Lee, Y.-K. Microflora of the gastrointestinal tract. Methods Mol. Biol. 2004, 268, 12. [Google Scholar] [CrossRef]
- Johansson, M.E.V.; Phillipson, M.; Petersson, J.; Velcich, A.; Holm, L.; Hansson, G.C. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 15064–15069. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, B.; Kim, S.; Kwon, Y. Characterization of microbiota associated with digesta and mucosa in different regions of gastrointestinal tract of nursery pigs. Int. J. Mol. Sci. 2019, 20, 1630. [Google Scholar] [CrossRef]
- Zhang, Z.; Geng, J.; Tang, X.; Fan, H.; Xu, J.; Wen, X.; Ma, Z.; Shi, P. Spatial heterogeneity and co-occurrence patterns of human mucosal-associated intestinal microbiota. ISME J. 2014, 8, 881–893. [Google Scholar] [CrossRef]
- Lu, J.; Zhu, M.; Cao, H.; Zhang, X.; Wang, Z.; Zhang, X.; Li, X.; Hu, J.; Yang, G.; Shi, X. Impact of fermented corn–soybean meal on gene expression of immunity in the blood, level of secretory immunoglobulin A, and mucosa-associated bacterial community in the intestine of grower–finisher pigs. Front. Vet. Sci. 2020, 7, 246. [Google Scholar] [CrossRef]
- Gardiner, G.E.; Metzler-Zebeli, B.U.; Lawlor, P.G. Impact of intestinal microbiota on growth and feed efficiency in pigs: A review. Microorganisms 2020, 8, 1886. [Google Scholar] [CrossRef] [PubMed]
- De Rodas, B.; Youmans, B.P.; Danzeisen, J.L.; Tran, H.; Johnson, T.J. Microbiome profiling of commercial pigs from farrow to finish. J. Anim. Sci. 2018, 96, 1778–1794. [Google Scholar] [CrossRef] [PubMed]
- Maglogiannis, I.; Zafiropoulos, E.; Platis, A.; Lambrinoudakis, C. Risk analysis of a patient monitoring system using Bayesian network modeling. J. Biomed. Inform. 2006, 39, 637–647. [Google Scholar] [CrossRef]
- Tsukahara, T.; Kishino, E.; Inoue, R.; Nakanishi, N.; Nakayama, K.; Ito, T.; Ushida, K. Correlation between villous height and the disaccharidase activity in the small intestine of piglets from nursing to growing. Anim. Sci. J. 2013, 84, 54–59. [Google Scholar] [CrossRef]
- Tsukahara, T.; Inoue, R.; Nakayama, K.; Inatomi, T. Inclusion of Bacillus amyloliquefaciens strain TOA5001 in the diet of broilers suppresses the symptoms of coccidiosis by modulating intestinal microbiota. Anim. Sci. J. 2018, 89, 679–687. [Google Scholar] [CrossRef] [PubMed]
- Inoue, R.; Sawai, T.; Sawai, C.; Nakatani, M.; Romero-Pérez, G.A.; Ozeki, M.; Nonomura, K.; Tsukahara, T. A preliminary study of gut dysbiosis in children with food allergy. Biosci. Biotechnol. Biochem. 2017, 81, 2396–2399. [Google Scholar] [CrossRef] [PubMed]
- Janssen, S.; McDonald, D.; Gonzalez, A.; Navas-Molina, J.A.; Jiang, L.; Xu, Z.Z.; Winker, K.; Kado, D.M.; Orwoll, E.; Manary, M.; et al. Phylogenetic placement of exact amplicon sequences improves associations with clinical information. Msystems 2018, 3, e00021-18. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Duarte, M.E.; Kim, S.W. Significance of mucosa-associated microbiota and its impacts on intestinal health of pigs challenged with F18+ E. coli. Pathogens 2022, 11, 589. [Google Scholar] [CrossRef]
- Toutain, P.-L.; Ferran, A.A.; Bousquet-Melou, A.; Pelligand, L.; Lees, P. Veterinary medicine needs new green antimicrobial drugs. Front. Microbiol. 2016, 7, 1196. [Google Scholar] [CrossRef]
- Zeineldina, M.; Aldridge, B.; Blair, B.; Kancer, K.; Lowe, J. Impact of parenteral antimicrobial administration on the structure and diversity of the fecal microbiota of growing pigs. Microb. Pathog. 2018, 118, 220–229. [Google Scholar] [CrossRef]
- Amat, S.; Lantz, H.; Munyaka, P.M.; Willing, B.P. Prevotella in pigs: The positive and negative associations with production and health. Microorganisms 2020, 8, 1584. [Google Scholar] [CrossRef]
- Hu, J.; Chen, J.; Ma, L.; Hou, Q.; Zhang, Y.; Kong, X.; Huang, X.; Tang, Z.; Wei, H.; Wang, X.; et al. Characterizing core microbiota and regulatory functions of the pig gut microbiome. ISME J. 2024, 18, wrad037. [Google Scholar] [CrossRef]
- Wright, D.P.; Rosendale, D.I.; Roberton, A.M. Prevotella enzymes involved in mucin oligosaccharide degradation and evidence for a small operon of genes expressed during growth on mucin. FEMS Microbiol. Lett. 2000, 190, 73–79. [Google Scholar] [CrossRef]
- Sundin, O.H.; Mendoza-Ladd, A.; Zeng, M.; Diaz-Arévalo, D.; Morales, E.; Fagan, B.M.; Ordoñez, J.; Velez, P.; Antony, N.; McCallum, R.W. The human jejunum has an endogenous microbiota that differs from those in the oral cavity and colon. BMC Microbiol. 2017, 17, 160. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tsai, T.; Deng, F.; Wei, X.; Chai, J.; Knapp, J.; Apple, J.; Maxwell, C.V.; Lee, J.A.; Li, Y.; et al. Longitudinal investigation of the swine gut microbiome from birth to market reveals stage and growth performance associated bacteria. Microbiome 2019, 7, 109. [Google Scholar] [CrossRef] [PubMed]
- Valeriano, V.D.V.; Balolong, M.P.; Kang, D.-K. Probiotic roles of Lactobacillus sp. in swine: Insights from gut microbiota. J. Appl. Microbiol. 2016, 122, 554–567. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Deng, J.; Wang, Z.; Che, C.; Li, Y.; Yang, Q. Modulatory effects of Lactobacillus salivarius on intestinal mucosal immunity of piglets. Curr. Microbiol. 2011, 62, 1623–1631. [Google Scholar] [CrossRef] [PubMed]
- Hynönen, U.; Kant, R.; Lähteinen, T.; Pietilä, T.E.; Beganović, J.; Smidt, H.; Uroić, K.; Åvall-Jääskeläinen, S.; Palva, A. Functional characterization of probiotic surface layer protein-carrying Lactobacillus amylovorus strains. BMC Microbiol. 2014, 14, 199. [Google Scholar] [CrossRef] [PubMed]
- Trevisi, P.; Priori, D.; Jansman, A.J.M.; Luise, D.; Koopmans, S.-J.; Hynonen, U.; Palva, A.; van der Meulen, J.; Bosi, P. Molecular networks affected by neonatal microbial colonization in porcine jejunum, luminally perfused with enterotoxigenic Escherichia coli, F4ac fimbria or Lactobacillus amylovorus. PLoS ONE 2018, 13, e0202160. [Google Scholar] [CrossRef]
- Holman, D.B.; Brunelle, B.W.; Trachsel, J.; Allen, H.K. Meta-analysis to define a core microbiota in the swine gut. mSystems 2017, 2, e00004-17. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoshimura, S.; Tsukahara, T.; Takahashi, T.; Miura, H.; Morishima, S.; Kise, M.; Shin, J.; Yahara, Y.; Inoue, R. Causal Association Between the Mucosal and Luminal Microbiotas from the Gastrointestinal Tract of Weaned Piglets Using Bayesian Network. Microorganisms 2025, 13, 256. https://doi.org/10.3390/microorganisms13020256
Yoshimura S, Tsukahara T, Takahashi T, Miura H, Morishima S, Kise M, Shin J, Yahara Y, Inoue R. Causal Association Between the Mucosal and Luminal Microbiotas from the Gastrointestinal Tract of Weaned Piglets Using Bayesian Network. Microorganisms. 2025; 13(2):256. https://doi.org/10.3390/microorganisms13020256
Chicago/Turabian StyleYoshimura, Shu, Takamitsu Tsukahara, Toru Takahashi, Hiroto Miura, So Morishima, Masaaki Kise, Jiye Shin, Yoshihiro Yahara, and Ryo Inoue. 2025. "Causal Association Between the Mucosal and Luminal Microbiotas from the Gastrointestinal Tract of Weaned Piglets Using Bayesian Network" Microorganisms 13, no. 2: 256. https://doi.org/10.3390/microorganisms13020256
APA StyleYoshimura, S., Tsukahara, T., Takahashi, T., Miura, H., Morishima, S., Kise, M., Shin, J., Yahara, Y., & Inoue, R. (2025). Causal Association Between the Mucosal and Luminal Microbiotas from the Gastrointestinal Tract of Weaned Piglets Using Bayesian Network. Microorganisms, 13(2), 256. https://doi.org/10.3390/microorganisms13020256






